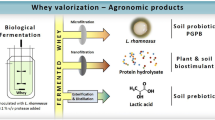
Abstract
The l-lactic acid obtained from a whey waste valorization process decreased pH and increased phosphate solubilization of a Plagic Antrosol soil. This behavior was reversible due to the complete and rapid degradation of lactic acid by soil microorganisms, leading to a strong soil biological stimulation. Both dehydrogenase (DHA) and acid phosphomonoesterase (APA) activities were also significantly stimulated, due to the stimulation of microbial activity, whereas the β-glucosidase activity was unaffected. There was also a shift in bacterial biodiversity with growth of bacterial strains using lactic acid as a C source. The bacteria enriched belonged to the plant growth-promoting bacteria (PGPb), such as Rhizobium and Azotobacter genus, involved in N2 fixation, and Pseudomonas and Bacillus genera, involved in phosphorous bioavailability. We conclude that lactic acid application improves fertility parameters on soils. Further research at field scale is needed to confirm these results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plants produce large amounts of exudates such as carbohydrates, carboxylic acids, and amino acids (Cawthray 2003; Strobel 2001). Organic C released from these exudates stimulates microbial growth and activity in the rhizosphere (Baudoin et al. 2003).
The root exudates can also affect the composition of microbial communities of the rhizosphere (Baudoin et al. 2003; Henry et al. 2008; Renella et al. 2006; Shi et al. 2011). In addition, low-molecular-weight organic acids (LMWOAs) present in the root exudates can carry out important processes, such as releasing cations from minerals and thus increase their availability in rhizosphere, and increase phosphate solubility by dissolving phosphate precipitates (Strobel 2001). On the other hand, they serve as C source for rhizosphere microorganisms, and thus, they may stimulate the activity of microorganisms of agronomic interest (Haoliang et al. 2007).
Phosphorus (P) is an essential macronutrient for plant growth. It is generally present in soil as an insoluble form (Kim et al. 1998). Phosphorus fertilization has often caused an accumulation of inorganic P in soil that is not bioavailable. P fertilizers have been decreasing both in quantity and in quality, which has led to the development of technologies aimed at making better use of P in soil. This would lead to lower energy inputs and a subsequent decrease in economic investment by the farmer (Delgado and Torrent 2000; Jasinski 2010; Van Vuuren et al. 2010).
Using LMWOAs such as oxalic, citric, butyric, malonic, lactic, succinic, malic, gluconic, acetic, fumaric, adipic, indoleacetic, and 2-ketogluconic acids produced by rhizobacteria is an approach to solubilize phosphate precipitated as calcium phosphates, thus making P available for plant nutrition (Drouillon and Merckx 2003; Paredes-Mendoza and Espinosa-Victoria 2010; Strobel 2001). Also, organic acids increase the availability of P in soils mainly through both decreased adsorption of P and increased solubilization of P compounds (Bolan et al. 1994). LMWOAs not only increase P availability due to the acidification of rhizosphere soil but also form stable complexes with some metals, such as Al and Fe (Paredes-Mendoza and Espinosa-Victoria 2010; Strobel 2001).
Soil enzyme activities respond immediately to changes in soil fertility status, and therefore, they can be used as soil quality indicators (Bandick and Dick 1999; Masciandaro et al. 2004; Nannipieri et al. 1990).
Several microbial species can excrete lactic acid which solubilizes soil tricalcium phosphate, the most insoluble calcium phosphate, thus increasing P availability in soil (Chen et al. 2006; Kaur and Reddy 2015; Pereira and Castro 2014). However, the effects of the direct application of lactic acid on P solubilization, activity, and composition of microbial communities of soil are unknown. Therefore, the aim of this research was to get more insights on these effects by measuring enzyme activities and bacterial diversity. The l-lactic acid was obtained from cheesemaking whey by a fermentation process that gave high yields. The use of whey waste is crucial due to the large volumes produced and the environmental legislation that bans its dumping because of its high COD and the recalcitrance of some of its components that have an environmental impact.
Material and methods
l-Lactic acid production and analysis
Whey fermentation was performed using Lactobacillus rhamnosus under optimized fermentation conditions, and the produced l-lactic acid was then purified by physical/chemical processes. The first step consisted in microfiltering the fermented whey to separate insoluble matter containing the microbial biomass. Then, peptides were separated by nanofiltration (200 Da), achieving a highly concentrated lactic acid and a salt product. Lactic acid was finally detached from salts using esterification-distillation technology (Kwak et al. 2012).
The purity of l-lactic acid obtained from whey, as well as the amount of l-lactic acid in soil, was determined by reverse phase chromatography using an AKTA purifier (GE Healthcare) and a Nova-Pak C18 4 μm, 4.6 × 150-mm column (Waters). Samples were loaded into a 50-μl loop connected to the AKTA purifier system. The column was equilibrated and eluted with a buffer composed of 90% formic acid 0.01% and 10% methanol in isocratic mode at a flow rate of 0.8 ml min−1. The organic acids were detected at 215 nm with a UV900 module attached to the GE Healthcare elution column. The optical properties of l-lactic acid were evaluated with a commercial kit supplied by Megazyme.
Treatment and incubation procedure
The main properties of soil, a Plagic Antrosol (IUSS Working Group WRB 2015), are shown in Table 1 and were determined as described by Tejada et al. (2010).
Two hundred grams of soil was preincubated at 25 °C for 7 days at 30–40% of their water-holding capacity, according to Tejada (2009). After this preincubation period, soil samples were mixed with l-lactic acid at three different concentrations: L1 (0.1% w/w), L2 (0.5% w/w), and L3 (1% w/w). The soil without l-lactic acid was used as control (L0). Each treatment was replicated three times. Distilled water was added to each soil to reach 60% water-holding capacity. All treated soil samples were placed in semi-closed microcosms and incubated in the incubation chamber under darkness at 25 °C for 0, 1, 5, 7, 12, 21, and 28 days. The samples were withdrawn for the incubation microcosm at each incubation time.
Ten grams of soil was taken from every sample, pooled, and stored in sealed polyethylene bags at 4 °C for chemical and biochemical analyses, whereas 2 g of soil subsamples was stored at −20 °C prior to determining the bacterial diversity.
Soil chemical and biochemical analyses
Soil pH was determined in distilled water with a glass electrode (soil/H2O ratio 1:2.5) (Métodos Oficiales de Análisis 1986).
l-Lactic acid was extracted by incubating 1 g of soil with 10 ml of extraction buffer (H2SO4 0.1 N) for 1 h (Bolan et al. 1994). The samples were centrifuged at 12,000×g for 15 min at 4 °C to remove insoluble particles; then, the supernatant was passed through a 0.2-μm filter of regenerated cellulose and analyzed for its l-lactic acid content as described before.
The Olsen P in soil was determined in soils taken at 5, 12, and 28 days according to Olsen et al. (1954).
Soil dehydrogenase activity was determined using 0.5 g of soil and the reduction of 2-[4-iodophenyl]-3-[4-nitrophenyl]-5-phenyltetrazolium chloride (INT) to p-iodonitrotetrazolium formazan (INTF) as described by Tabatabai (1994). Samples were incubated for 30 min at room temperature. The product concentration was measured at 485 nm using a GeneQuant 1300 spectrophotometer (GE Healthcare Bio-Sciences AB, USA).
Soil acid phosphomonoesterase activity was measured using p-nitrophenyl phosphate as the substrate (Tabatabai 1994). Of soil, 0.5 g was incubated for 30 min with modified universal buffer (MUB) substrate buffer (pH 6.5). Then, 0.5 M CaCl2 and 0.5 M NaOH were added to stop the reaction and to extract the product, p-nitrophenol, whose concentration was determined photometrically at 410 nm.
Soil β-glucosidase activity was measured using p-nitrophenyl-β-d-glucopyranoside as substrate (Tabatabai 1994). Soil (0.25 g) was incubated for 30 min with MUB substrate buffer (pH 6). To stop the reaction and extract the product, p-nitrophenol, whose concentration was determined photometrically at 410 nm, 0.5 M CaCl2 and Tris buffer (pH 12) were added.
Extraction of DNA and denaturing gradient gel electrophoresis analysis
At 0, 5, and 12 days of the incubation period, soil DNA was extracted according to a bead beating protocol of the UltraClean™ Soil DNA Isolation Kit (MoBio Laboratories). The V3–V5 regions of 16S ribosomal DNA (rDNA) were amplified by PCR using F357-GC and R907 primers. Primer F357-GC included a GC clamp at the 5′ end (F357: 5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CCC TAC GGG AGG CAG CAG-3′ and R907: 5′-CCG TCA ATT CCT TTG AGT TT-3′). All PCRs were performed with a Mastercycler (Eppendorf, Germany). Fifty microliters of the PCR mixture contained 1.5 U of EcoTaq (Ecogen, BIOLINE), 10 mM Tris–HCl (pH 9), 1.5 mM MgCl2, each deoxynucleotide triphosphate at a concentration of 200 μM, each primer at a concentration of 0.5 μM, and 2 μl of DNA template. After 9 min of initial denaturation at 95 °C, a touchdown thermal profile protocol was used and the annealing temperature was decreased by 1 °C per cycle from 65 to 55 °C. Twenty additional cycles at 55 °C were then performed. Amplification was performed using 1 min for DNA denaturation at 94 °C, 1 min for primer annealing. and 1.5 min for primer extension at 72 °C, followed by 10 min for final primer extension. PCR products were dried prior to electrophoresis analysis. Denaturing gradient gel electrophoresis (DGGE) was performed in 6% (w/v) polyacrylamide gel that was 0.75 mm thick, and the denaturing gradient ranged from 40 to 60% (100% denaturant contained 7 M urea and 40% formamide), using a DCode Universal Mutation Detection System (Bio-Rad) at 100 V for 16 h and at 60 °C (Muyzer et al. 1993). Gels were stained for 15 min in 1× TAE buffer containing ethidium bromide and were viewed under UV light and analyzed using a UV transilluminator system. To evaluate the variability, we previously checked that diversity of 16S rDNA analyzed by DGGE was homogeneous among samples, for it has been amplified and extracted several replicates of the same soil, observing that extraction system using the bead beating protocol was highly reproducible.
For each treatment, the dominant bands of the DGGE obtained were excised from DGGE gels with a sharp scalpel and the DNA was extracted and purified using the QIAquick Gel Extraction Kit (Qiagen). Then, these bands were used as templates for a new amplification using the same primers. The resulting PCR products with a minimal size of 500 bp were sequenced using the ABI PRISM-377 DNA Sequencer software (PerkinElmer). The obtained sequences were compared with 16S rRNA sequences available from the National Center for Biotechnology Information (NCBI) database, using the BLAST program (Basic Local Alignment Search Tool).
Results and discussion
l-Lactic characterization
We got pure l-lactic from a whey biorefinery process using L. rhamnosus with yields of 35–40 g of lactic acid per liter of whey. The product only contained the l-isomer (see Fig. 1).
Evolution of soil chemical properties
After applying l-lactic acid to the soil, there was a significant decrease (p < 0.01) in pH values (Fig. 2). This decrease was greater with the higher dose of applied l-lactic acid. During the incubation period, the pH values of soils treated with l-lactic acid increased and reached the pH of the control soil (about 7.9 to 8.0). The L1 soil reached this value after 12 days, the L2 soil after 21 days, and the L3 soil at the end of the experiment.
The l-lactic acid was degraded in soil immediately after its application (Fig. 3). After 7 days, l-lactic acid concentration in L1 samples decreased by about 85%, while those in L2 and L3 soils decreased more than 50%. The l-lactic acid after 28 days was completely degraded in any sample.
The degradation of l-lactic acid was probably due to the microbial activity and the observed trend agrees with that described by Siotto et al. (2012). In both studies, lactic acid disappeared completely from soil after 25–30 days. This rapid mineralization in soil is similar to that of glucose (Van Veen et al. 1985).
This hypothesis is supported by our DGGE results (Fig. 5) that show a change in soil bacterial composition with stimulation of bacteria using lactic acid as C source for growth.
The highest Olsen P value was obtained after 5 days and with the highest dose of lactic acid applied to soil with a 20% increase in the free phosphate level. At the same day, the L2 soil showed a 17% increase and the L1 soil an increase of 13.6% (Table 2). Then, the Olsen P levels decreased, and at the end of the incubation period, the Olsen P values of the treated soils were similar to that of the L0 soil. The short-term decrease in soil pH due to the acidity of lactic acid agrees with those obtained by Chen et al. (2006), Hwangbo et al. (2003), and Strobel (2001), and it is responsible for the P solubilization. According to Kpomblekou-A and Tabatabai (1994), carboxyl groups of organic acids are responsible for chelating Ca cations, thus favoring soil P solubilization from Ca phosphates.
The application of lactic acid increases the P availability for some weeks. Therefore, it can be applied when plants need P as also shown with P applications to soils (Li et al. 2015; Zhai et al. 2015).
Soil enzyme activities
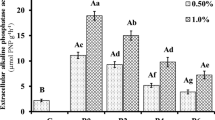
Dehydrogenase activity is related to oxidative phosphorylation processes (Trevors 1984), and its activity has been proposed as a measure of soil microbial activity (Nannipieri et al. 1990). Dehydrogenase activity was stimulated by adding lactic acid to the soil reaching the maximum after 12 days (Fig. 4a). This stimulation depended on the l-lactic acid application rate. After 12 days, dehydrogenase activity decreased progressively, and by the end of the incubation period, values of treated soils were comparable with that of the control.
The addiction of l-lactic acid to soil also stimulated acid phosphomonoesterase activity (Fig. 4b), and the peaks occurred between 5 and 12 days. After 12 days, this enzyme decreased. At the end of the incubation, the highest enzyme activity occurred in the L3 soil, followed by those of the L2 and L1 soils. Probably, the greater stimulation of acid phosphomonoesterase activity than dehydrogenase activity depended on the great demand of P, to support the increased microbial activity of the soil.
In contrast, Fig. 4c shows that l-lactic acid did not affect β-glucosidase activity in all treated soils, probably because the C source for microorganisms was provided by l-lactic acid, and thus, the activity carbohydrate hydrolases were not induced.
Composition of soil bacterial community
Bacterial species of the dominant DGGE bands (7, 8, 10, 11, 13, and 14) (Fig. 5 and Table 3) belong to genus that can use lactate as a C source for their growth (Pseudomonas, Bacillus, Azotobacter, and Rhizobium) (Bhattacharyya and Barnes 1978; Chai et al. 2009; Gao et al. 2012; Iswaran et al. 1973). These bacterial genera are known as plant growth-promoting rhizobacteria (PGPR), and therefore, they can stimulate plant growth and reduce pathogen infection and biotic or abiotic plant stress (Lugtenberg and Kamilova 2009; Pii et al. 2015; Van Loon and Bakker 2005). Since Rhizobium and Azotobacter are bacteria involved in nitrogen fixation, adding lactic acid to soil can have an important role in both agronomic and ecological terms, minimizing the need for P fertilizer application and stimulating the biological activity and growth of PGPRs.
Conclusions
l-lactic acid obtained from whey waste stimulated soil microbial activity and the release of soluble phosphates. Moreover, the application of lactic acid is involved in shaping the composition of bacterial communities in soil, leading to an enrichment of PGPRs. These results could have an important role in both agronomic and ecological terms. However, these findings need to be further confirmed by field experiments, with evaluation of grow productivity and quality parameters.
References
Bandick AK, Dick RP (1999) Field management effects on soil enzyme activities. Soil Biol Biochem 31:1471–1479
Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol Biochem 35:1183–1192
Bhattacharyya P, Barnes EM Jr (1978) Proton-coupled sodium uptake by membrane vesicles from Azotobacter vinelandii. J Biol Chem 253:3848–3851
Bolan NS, Naidu R, Mahimairaja S, Baskaran S (1994) Influence of low-molecular-weight organic acids on the solubilization of phosphates. Biol Fert Soils 18:311–319
Cawthray GR (2003) Short communication: an improved reversed-phase liquid chromatographic method for the analysis of low-molecular mass organic acids in plant root exudates. J. Chromatogr A 1011:233–240
Chai Y, Kolter R, Losick R (2009) A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J Bacteriol 191:2423–2430
Chen YP, Rekha PD, Arun AB, Shen FT, Lai W, Young CC (2006) Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol 34:33–41
Delgado A, Torrent J (2000) Phosphorus forms and desorption patterns in heavily fertilized calcareous and limed acid soils. Soil Sci Soc Am J 64:2031–2037
Drouillon M, Merckx R (2003) The role of citric acid as a phosphorus mobilization mechanism in highly P-fixing soils. Gayana Bot 60:55–62
Gao C, Hu C, Ma C, Su F, Yu H, Jiang T, Dou P, Wang Y, Qin T, Lv M, Xu P (2012) Genome sequence of the lactate-utilizing Pseudomonas aeruginosa strain XMG. J Bacteriol 194:4751–4752
Haoliang L, Chongling Y, Jingchun L (2007) Low-molecular-weight organic acids exuded by mangrove (Kandelia candel (L.) Druce) roots and their effect on cadmium species change in the rhizosphere. Environ Exp Bot 61:159–166
Henry S, Texier S, Hallet S, Bru D, Dambreville C, Chèneby D, Bizouard F, Germon J, Philippot L (2008) Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ Microbiol 10:3082–3092
Hwangbo H, Park RD, Kim YW, Rim YS, Park KH, Kim TH, Kim KY (2003) 2-ketogluconic acid production and phosphate solubilization by Enterobacter intermedium. Curr Microbiol 47:87–92
Iswaran V, Apte R, Sen A (1973) Use of sodium lactate for culture of Rhizobium spp. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 128:240–242
IUSS Working Group WRB (2015) World reference base for soil resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports No. 106. FAO, Rome
Jasinski SM (2010). Phosphate rock. In: United States Geological Survey minerals information. Mineral Commodity Summaries p 118–119
Kaur G, Reddy MS (2015) Effects of phosphate-solubilizing bacteria, rock phosphate and chemical fertilizers on maize-wheat cropping. Pedosphere 25:428–437
Kim KILY, Jordan D, Mcdonald GA (1998) Enterobacter agglomerans, phosphate solubilizing bacteria, and microbial activity in soil: effect of carbon sources. Soil Biol Biochem 30:995–1003
Kpomblekou-A K, Tabatabai MA (1994) Effect of organic acids on release of phosphorus from phosphate rocks1. Soil Sci 158:442–453
Kwak H, Hwang DW, Hwang YK, Chang JS (2012) Recovery of alkyl lactate from ammonium lactate by an advanced precipitation process. Sep Purif Technol 93:25–32
Li L, Liang X, Ye Y, Zhao Y, Zhang Y, Jin Y, Yuan J, Chen Y (2015) Effects of repeated swine manure applications on legacy phosphorus and phosphomonoesterase activities in a paddy soil. Biol Fertil Soils 51:167–181
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Masciandaro G, Ceccanti B, Benedicto S, Lee HC, Cook HF (2004) Enzyme activity and C and N pools in soil following application of mulches. Can J Soil Sci 84:19–30
Métodos Oficiales de Análisis (1986). Secretaria General Técnica del Ministerio de Agricultura, Pesca y Alimentación. Spain Government
Muyzer G, de Waal EC, Uitterlinden UG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nannipieri P, Grego S, Ceccanti C (1990) Ecological significance of the biological activity in soil. In: Bollag J-M, Stotzky G (eds) Soil biochemistry vol 6. Marcell Dekker, New York, pp 293–355
Olsen SR, Cole CV, Watanabe FS, Dean LA (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA Circular 939:1–19
Paredes-Mendoza M, Espinosa-Victoria D (2010) Organic acids produced by phosphate solubilizing rhizobacteria: a critical review. Terra Latinoamericana 28:61–70
Pereira SIA, Castro PML (2014) Phosphate-solubilizing rhizobacteria enhance Zea mays growth in agricultural P-deficient soils. Ecol Eng 73:526–535
Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C (2015) Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. Biol Fertil Soils 51(403):415
Renella G, Egamberdiyeva D, Landi L, Mench M, Nannipieri P (2006) Microbial activity and hydrolase activities during decomposition of root exudates released by an artificial root surface in Cd-contaminated soils. Soil Biol Biochem 38:702–708
Shi S, Richardson AE, O'Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011) Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77:600–610
Siotto M, Sezenna E, Saponaro S, Degli Innocenti F, Tosin M, Bonomo L, Mezzanotte V (2012) Kinetics of monomer biodegradation in soil. J Environ Manag 93:31–37
Strobel BW (2001) Influence of vegetation on low-molecular-weight carboxylic acids in soil solution—a review. Geoderma 99:169–198
Tabatabai MA (1994) Soil enzymes. In: Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A (eds) Methods of soil analysis. Part 2. Microbiological and biochemical properties. Soil Science Society of America, Madison, pp 775–833
Tejada M (2009) Evolution of soil biological properties after addition of glyphosate, diflufenican and glyphosate+diflufenican herbicides. Chemosphere 76:365–373
Tejada M, Gomez I, Hernández T, García C (2010) Response of Eisenia fetida to the application of different organic wastes in an aluminium-contaminated soil. Ecotox Environ Safe 73:1944–1949
Trevors JT (1984) Dehydrogenase activity in soil: a comparison between the INT and TTC assay. Soil Biol Biochem 16:673–674
Van Loon L, Bakker P (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: PGPR: biocontrol and biofertilization, Springer, Berlin, p 39–66
Van Veen JN, Ladd JN, Amato M (1985) Turnover of carbon and nitrogen through the microbial biomass in a sandy loam and a clay soil incubated with [14C(U)]glucose and [15N](NH4)2SO4 under different moisture regimes. Soil Biol Biochem 17:747–756
Van Vuuren DP, Bouwman AF, Beusen AHW (2010) Phosphorus demand for the 1970 – 2100 period: a scenario analysis of resource depletion. Global Environ Chang 20:428–439
Zhai L, CaiJi Z, Liu J, Wang H, Gai X, Xi B, Liu H (2015) Short-term effects of maize residue biochar on phosphorus availability in two soils with different phosphorus sorption capacities. Biol Fertil Soils 51:113–122
Acknowledgements
This work was supported by the Ministry of Science and Technology (Spain), Plan Nacional I+D CTM 2015-64354, and Council of Economy, Innovation, Science and Employment of the Junta de Andalucía (RNM-2011-7887). Thanks also to the Biology Service of Centro de Investigación, Tecnología e Innovación de la Universidad de Sevilla (CITIUS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rodríguez-Morgado, B., Jiménez, P.C., Moral, M.T. et al. Effect of l-lactic acid from whey wastes on enzyme activities and bacterial diversity of soil. Biol Fertil Soils 53, 389–396 (2017). https://doi.org/10.1007/s00374-017-1187-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-017-1187-z