Abstract
Anthropogenic effects on soil fungi have been poorly investigated in peaty soils, where they have a crucial role in the maintenance of soil fertility and in the regulation of nutrient cycles. In this study, we assessed the effects of land-use intensification on the composition of fungal communities in Mediterranean peaty soils drained for agricultural purposes. To this end, a continuous maize cropping system was compared with an extensive grassland and an agricultural soil left abandoned for 15 years. Molecular diversity and community composition of total soil fungi and arbuscular mycorrhizal fungi (AMF) were assessed, as well as soil chemical properties potentially responsible for fungal shifts. The relative roles of intensification and soil chemical properties were also quantified by applying variation partitioning analysis. Multivariate analyses show that: (i) land-use intensification shapes the composition of the community of total soil fungi and AMF in soil and roots; (ii) base saturation (Bas Sat) and exchangeable calcium (ExchCa) in soil are the significant soil chemical drivers of the composition of the total soil fungal community; (iii) Bas Sat is the only significant chemical parameter shaping the soil AMF community; and (iv) no soil chemical properties affect root AMF. Based on variation partitioning, which highlights a large overlap between land-use intensification and Bas Sat, we can assert that land-use intensification is well-correlated with Bas Sat in shaping the total soil fungal community composition, as well as the AMF. By contrast, intensification acts as a major driver with respect to ExchCa in shaping the composition of the total soil fungal communities.
Similar content being viewed by others

Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil microorganisms play a crucial role in maintaining soil fertility due to their influence on regulation of nutrient cycles (Nannipieri et al. 2003). Among these, soil fungi represent the majority of soil microflora in many habitats (Joergensen and Wichern 2008). They have key direct and indirect functions in lignin and soil organic matter (SOM) decomposition, carbon (C) sequestration, soil aggregation, and in nutrient mineralization and immobilization (Gadd 2006). It has been widely suggested that fungi are more important than bacteria in decomposing organic matter, especially in acidic ecosystems such as peatlands (Thormann 2006). Fungi metabolize C with higher assimilation efficiency than bacteria, which means that C is mainly retained in fungal biomass instead of respired as CO2. Fungal decomposers, such as white-rot fungi belonging to Basidiomycota, have developed unique pathways to degrade lignin, cellulose, and hemicellulose (Bahri et al. 2006). Among fungi, arbuscular mycorrhizal fungi (AMF; phylum Glomeromycota; Schuessler et al. 2001) form a root symbiosis with up to 80 % of land plant species and agricultural crops (Smith and Read 2008). AMF enhance plant mineral nutrient uptake, mainly phosphorus (P), in exchange for photosynthetically fixed C (Bago et al. 2000; Hodge et al. 2001) and protection against root fungal pathogens and drought stress (Newsham et al. 1995; Augé 2001). AMF may stimulate SOM decomposition in AMF-active zones (Cheng et al. 2012) and play important roles in limiting the uptake of heavy metals (Leyval et al. 1997). In addition, AMF drive the diversity of plant communities (van der Heijden et al. 1998) and ameliorate soil aggregate stability and macroaggregate formation both through the extraradical hyphal development and the production of coagulating substances, such as glomalin, thereby limiting soil erosion (Rillig 2004; Bedini et al. 2009).
Several studies have shown that soil fungal community composition and functionality are affected by soil environment changes due to land-use intensifications and by edaphic factors (Bardgett et al. 2005). Soil fungal abundances and communities are sensitive to the intensification of the management practices, such as tillage, herbicide application (García-Orenes et al. 2013), mineral fertilization (Bardgett et al. 1996), and manure addition (Bittman et al. 2005), with negative effects on fungal agroecosystem functions and services (Bardgett et al. 2005). Regarding edaphic factors, fungal community compositions are more strongly related to changes in soil nutrient availability, namely, nitrogen (N) and P, than in soil pH and texture (Lauber et al. 2008; Rousk et al. 2010).
As regards AMF, land-use intensification through tillage, fertilizers, biocides, and crop rotations (i.e., monocropping or rotations with nonmycorrhizal crops) negatively modify AMF richness and community composition promoting the unique occurrence of members of Glomeraceae (i.e., Helgason et al. 1998; Jansa et al. 2002; Oehl et al. 2003; Fitter et al. 2005; Gosling et al. 2006). Specifically, AMF richness is adversely affected by an increasing of land-use intensity both in soil (Lumini et al. 2010; Verbruggen et al. 2012) and roots (Helgason et al. 1998; Hijri et al. 2006; Schnoor et al. 2011), although no changes have been observed by other studies (Jansa et al. 2002; Mathimaran et al. 2007; Galvan et al. 2009; Dai et al. 2013; Moora et al. 2014).
As regards AMF community composition, with the application of high-throughput sequencing techniques, it has recently been observed that in tilled habitats also members of Archaeosporaceae, Claroideoglomeraceae and Diversisporaceae largely occurred (Lumini et al. 2010; Moora et al. 2014; Xiang et al. 2014). Concerning edaphic factors, soil pH, P, texture, and C:N ratio were shown, together with host identity, to be key drivers of the AMF colonizing roots in grassland, woodland and agricultural soils (Dumbrell et al. 2010; Torrecillas et al. 2012; Holland et al. 2014; Jansa et al. 2014).
Among management practices, grazing can generally increase soil fungal abundance (Bardgett et al. 1996; Ford et al. 2013), while an increased mowing frequency did not alter soil fungal abundance or functionality (Denef et al. 2009). As regards AMF, a decrease in root colonization due to grazing was observed (Barto and Rillig 2010), while as regards mowing, contrasting effects were reported, from any effect on AMF extraradical mycelium (Eom et al. 1999) to a stimulation of soil AMF biomass and species richness (Antonsen and Olsson 2005).
Peaty soils are widely known for high fertility due to their high C content and for their fundamental role as C sink (Verhoeven and Setter 2010). In the past, most peatlands were reclaimed for agricultural purposes with consequences on their ecological functions. To date, the effect of anthropogenic influences, such as the impact of land-use intensification on the composition of the fungal communities, has been poorly investigated in peaty soils (Andersen et al. 2013). In particular, a unique study focusing on AMF and CO2 emissions was conducted on peaty soils located in Mediterranean areas (Pellegrino et al. 2014).
In this study, we assessed for the first time the effects of land-use intensification on the composition of fungal communities in Mediterranean peaty soils drained for agricultural purposes. To this end, an intensively cultivated peaty soil represented by a continuous maize (Zea mays L.) cropping system was compared with an extensive grassland and an agricultural soil left abandoned for 15 years. We monitored soil chemical properties and total soil fungal and AMF molecular composition. We hypothesized that in peaty soils, the intensification of land-use would negatively affect the composition of fungal communities in soil and of AMF in soil and roots. We also hypothesized that soil properties are important drivers influencing the community of both fungi and AMF although less powerful respect to land-use intensification in determining their shifts. Our results may have important implications for the management of soil fertility in peaty soils aiming to achieve cropping systems improving or preserving soil services.
Materials and methods
Field site and experimental setup
The experimental site is located in the southern portion of the Massaciuccoli Lake basin (43° 49′ N–10° 19′ E; Pisa, Italy; Pistocchi et al. 2012). The soil is classified as Histosol, according to the USDA system (Soil Survey Staff 1975), and defined as peaty soil (IPCC 2006). The climate is Mediterranean (Csa), according to Köppen classification.
The long-term experiment (15 years long) was a completely randomized design with land-use intensification as the treatment and three replicates (n = 3; field replicates of 0.7 ha). The land-use types were: (i) a maize monoculture (high intensity, HI), (ii) an extensive grassland (low intensity, LI), and (iii) an agricultural soil left abandoned (zero intensity, ZI). In HI, field replicates were ploughed (30-35 cm), harrowed, and sown with maize at the beginning of June. Crops were harvested in late September. Chemical and mechanical postemergence weed control was also applied. No pest management was applied. Details of the sowing and fertilization are found in Pellegrino et al. (2014). In LI natural vegetation was mainly composed of Portulaca oleraceae L., Capsella bursa-pastoris (L.) Med., Juncus bufonius L., Polygonum persicaria L., Sorghum halepense (L.) Pers., Cynodon dactylon (L.) Pers., Datura stramonium L., Rumex crispus L., Echinochloa crus-galli (L.) Beauv., Calystegia sp., and Amaranthus retroflexus L. Vegetation was mowed and removed twice a year with no further fertilization or pest management application. In ZI, natural succession vegetation was left to develop and no fertilizers or other agricultural practices were applied. The most abundant plant species was Phragmites australis (Cav.) Trin. ex Steud (common reed). A detailed list of the plant species composition is given by Pellegrino et al. (2014). Further details on soil parameters and climate conditions are provided in Online Resource 1.
Sampling and analyses
In late July 2011, one soil sample resulting from pooling seven soil bulk cores was collected (0–30 cm in depth) from each of the three field replicates per treatment (a total of nine soil bulk samples) to cover chemical and fungal spatial variability. Sampling was carried out only once in July since mid-summer is the best choice, because soil sampling should not be close to soil treatments and because the variability in chemical parameters changes slightly during the year (Pellegrino et al. 2011, 2014). These facts along with the fact that fungi and AMF consistently maintain the same patterns of variability in differently managed systems, although with seasonal changes due to abiotic conditions (soil moisture and temperature), were taken into account when choosing July as a single sampling time (Vandenkoornhuyse et al. 2002; Oehl et al. 2010; Hannula et al. 2012; Di Bene et al. 2013). In the laboratory, from each of the nine soil samples, roots were carefully plucked with forceps, washed, and stored at 4 °C for root genomic DNA extraction. Then, from each of the nine soil samples, a subsample was oven dried at 60 °C and sieved at 2 mm for chemical parameter determinations, and a subsample was sieved at 2 mm and stored at 4 °C for soil genomic DNA extraction and clones library construction. The chemical properties of soil under the three land-use treatments are shown in Table S1.
Soil DNA was extracted from 0.5 g of soil using the PowerSoil® MO BIO kit (MO BIO Laboratories Inc., Carlsbad, CA, USA), while root DNA was extracted from 100-mg fresh root samples using the DNeasy® Plant Mini Kit (QIAGEN, Germantown, MD, USA). The fungal small subunit ribosomal RNA (SSU rRNA) region was amplified by the primer pair NS1 and EF3 for the first PCR (≈1,750 bp) and by the primer pair NS1 and FR1 for the second PCR (≈1,650 bp), following Hoshino and Morimoto (2008).
Although the internal transcribed spacer (ITS) region is commonly used as barcode for determining fungal diversity due to its resolution power till species separation (Schoch et al. 2012; Herr et al. 2015), its variability is too high to address the phylogeny of higher ranks (Lindahl et al. 2013). For this reason, the SSU rRNA region is considered more suitable for comparing and for discerning the highest fungal phylogenetic ranks (i.e., phyla and orders), and therefore, it is useful for an initial assessment of the total soil fungal community (Hunt et al. 2004) also when using the most recent high-throughput sequencing techniques (Kuramae et al. 2013; Lienhard et al. 2014). Moreover, the SSU rRNA region is known to be a successful marker for Chytridiomycota identification (Freeman et al. 2009), and it generally has a higher discriminatory power for basal fungi (including Zygomycota) compared to both the ITS region and the nuclear ribosomal large subunit (LSU; Schoch et al. 2012). Finally, another reason to target the SSU rRNA region is the availability of the SILVA database, which is a well-curated, and annotated database for such a gene (Kuramae et al. 2013).
Regarding AMF in both soil and root samples, PCR amplification was performed using the primer pair NS31 and AM1 (≈550 bp; Simon et al. 1992; Helgason et al. 1998). Although longer and more highly discriminating regions are available (Krüger et al. 2012; Pellegrino et al. 2012), the NS31/AM1 SSU rRNA region was targeted because most Glomeromycota diversity is obtained using this region (Öpik et al. 2010).
After quality checking, the PCR amplicons were ligated into the pGem®-T Easy vector (Promega Corporation, Madison, WI, USA) and used to transform XL10-Gold® Ultracompetent Escherichia coli cells (Stratagene®, La Jolla, CA, USA). At least 25 recombinant clones per library were screened (9 and 18 libraries for total soil fungi and AMF, respectively). For each library, all colonies containing inserts of the correct size were sequenced using the NS1/FR1 primers and AM1 primer for total soil fungi and AMF, respectively. Details on the quality check of extracted DNA and PCR amplicons, PCR protocols, and the Sanger sequencer are available in Online Resource 1.
Phylogenetic analysis
The sequences’ affiliation with fungi was verified using the BLAST tool in GenBank. BLAST searches were performed separately, with the sequences corresponding to the forward fragment (NS1; 645 bp) and with the sequences corresponding to the reverse fragment (FR1; 640 bp). The GenBank sequences most similar (>97 %) to our fungal sequences were included in the alignment to construct the phylogenetic tree. First, the two alignments were performed separately and then concatenated and trimmed to the length of ca. 1,265 bp in SeaView version 4.2.5. The concatenated alignment consisted of 178 fungal sequences (143 newly generated sequences, 34 from GenBank and Meristolohmannia meristacaroides as the outgroup). Regarding AMF, after a BLAST check for the sequences’ affiliation with Glomeromycota, they were aligned with an updated AMF reference dataset (Online Resource 1). An alignment of 339 AMF sequences (139 newly generated sequences, 160 sequences retrieved from Pellegrino et al. 2014, 27 from the reference dataset, 12 from GenBank and Corallochytrium limacisporum as the outgroup) was trimmed to the same length (ca. 490 bp). All alignments were performed using the online version of MAFFT 7.
Phylogenetic analyses were conducted using MEGA version 5.1 and the Kimura two-parameter model. Neighbor-joining (NJ) trees were constructed with 1,000 replicates to produce bootstrap values. We used Mothur to assign sequences to molecular operational taxonomic units (MOTUs) after calculating the Jukes–Kantor pairwise distances by PHYLIP. Similar sequences of total soil fungi and AMF were clustered into MOTUs using 99 and 97 % identity thresholds, respectively.
MOTU richness, Shannon index (H′), and Pielou evenness (J′) were calculated using Primer v6. Community sampling effort was verified by Coleman rarefaction curves in EstimateS version 9.1 (Online Resource 1). All newly generated nucleotide sequences were submitted to the EMBL nucleotide sequence database, and the accession numbers assigned are LN555148–LN555522 and LN555530–LN555579. References of the software used are reported in Online Resource 1.
Statistical analyses
Dependent variables were analyzed by analysis of covariance (ANCOVA) with land-use intensification as a fixed factor and the spatial coordinates of the plots (latitude/longitude) as covariables. All data were transformed when necessary to fulfil assumptions of the ANCOVA. LSD significant difference tests were used for comparisons. All analyses were performed in SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). When assumptions for the ANCOVA were not fulfilled, data were analyzed using the Kruskal–Wallis nonparametric test, followed by Mann–Whitney U post hoc tests.
Multivariate analyses based on constrained ordination [partial redundancy analysis (pRDA) and partial canonical correspondence analysis (pCCA)] were used to investigate the influence of land uses or soil chemical properties (used as explanatory variables) on the community composition (relative abundance of MOTUs) of total soil fungi and AMF communities (used as response variables). Further details are provided in Online Resource 1. The variation partitioning test (VarPart) was performed to assess the unique and shared contribution of land-use intensification and of each soil chemical property in explaining the community composition of total soil fungi and AMF. For the VarPart, we used the approach described in Legendre (2008). Monte Carlo permutation tests were used to determine statistical significance. Cocorrespondence analyses (CoCa) were applied to explore relations between AMF community composition in the roots and the soil (ter Braak and Schaffers 2004). All multivariate analyses were performed in CANOCO 5 (ter Braak and Smilauer 2012).
Results
Effect of land-use intensification on phylogenetic diversity and abundance of total soil fungi
We screened and sequenced approximately 200 clones obtained from the clone libraries (69, 88, and 43 clones from HI, LI, and ZI, respectively). Total soil fungal sequences were grouped into 16 MOTUs (Fig. 1; Fig. S1; Table S2). A total of 10, 13, and 9 MOTUs were retrieved in the HI, LI, and ZI, respectively (Fig. 1). The orders retrieved exclusively in each land use were Gomphales (11) in HI, Pleosporales (7) and Dothideomycetes (8) in LI, and Xylariales (4) and Corticiales (12) in ZI (Fig. 1). Rarefaction curves (Fig. S2) revealed that the sampling effort was sufficient.
Neighbor-joining (NJ) tree of total soil fungal sequences derived from soil under three levels of intensification of land use: high intensity (HI; maize monoculture), low intensity (LI; extensive grassland), and zero intensity (ZI; agricultural soil left abandoned). NJ is based on the fungal 18S rRNA gene (≈1,650 bp; NS1/FR1 fragment) and involved 143 newly detected nucleotide sequences plus 25 reference sequences retrieved from the NCBI database. The tree is rooted with Meristolohmannia meristacaroides. In the collapsed NJ tree, black triangles represent the 16 fungal molecular operational taxonomic units (MOTUs) retrieved in the study. The sequences obtained were affiliated with the orders Hypocreales (1 and 2), Sordariales (3), Xylariales (4), Chaetothyriales (5), Eurotiales (6), Pleosporales (7), Dothideomycetes (8), Helotiales (9), Agaricales (10), Gomphales (11), Corticiales (12), Filobasidiales (13), Tremellales (14), and to two classes Chytridiomycota (15), and Zygomycota (16). For each MOTU, the proportions of sequences from each land-use type (HI, red; LI, yellow; ZI, green) are shown in pie charts. Accession numbers of sequences obtained in the present study are shown in Fig. S1. Bootstrap values (based on 1,000 replicates) are shown at the nodes. The scale bar indicates substitutions per site
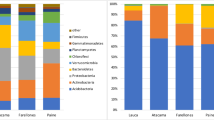
Although no differences were found among land uses, at the phylum level, Ascomycota, with a mean relative abundance of 49.7 ± 5.7 %, was the most abundant phylum (P < 0.001), followed by Basidiomycota (29.9 ± 5.4 %) and then Zygomycota and Chytridiomycota (mean of 10.2 ± 2.8 %; Fig. 2a). At the class level (Fig. 2b, c), only Eurotiomycetes (Ascomycota) was significantly different among land uses, with higher abundance in ZI (57.4 %) than in HI/LI (mean of 26.0 %; P = 0.04.
Soil fungal community diversity shown as relative abundances of 16 molecular operational taxonomic units (MOTUs) retrieved in the soil under three levels of intensification of land use: high intensity (HI; maize monoculture), low intensity (LI; extensive grassland), and zero intensity (ZI; agricultural soil left abandoned). Relative abundances were calculated for a fungi, b Ascomycota, and c Basidiomycota
Relative abundance of the fungal MOTUs is reported in Table S3. MOTU richness ranged from 6.3 to 9.0 and H′ from 1.57 to 2.02 in HI and LI, respectively (Table S4). J′ was significantly affected by land use, with lower values observed in HI (0.85) compared to other land uses (mean of 0.95). Mean richness based on fungal classes was 5.3 ± 0.3 for each land use.
Effect of land-use intensification on phylogenetic diversity and abundance of AMF
From the clone libraries obtained from soil and root DNA, approximately 166 (49, 94, and 46 clones from HI, LI, and ZI) and 100 clones (14, 60, and 26 from HI, LI, and ZI) were sequenced, respectively. AMF sequences were grouped into 14 and 10 different MOTUs in soil and roots, respectively (Fig. 3; Fig.S3). MOTUs were affiliated with Funneliformis sp. (12, 13), Rhizophagus sp. (5, 6, 7), Sclerocystis sp. (4), Scutellospora sp. (15), Glomus spp. (1, 2, 3, 8, 9, 10, 11, 14), and uncultured Glomeromycota (16, 17) (Table S6). Rarefaction curves showed that the sampling effort was sufficient (Fig. S2). Mean MOTU richness was 5.1 ± 0.6 and 3.0 ± 0.8 in soil and roots, respectively (Table S5).
Neighbor-joining (NJ) tree of arbuscular mycorrhizal fungal (AMF) sequences derived from roots and soil under three levels of intensification of land use: high intensity (HI; maize monoculture), low intensity (LI; extensive grassland), and zero intensity (ZI; agricultural soil left abandoned). NJ is based on SSU rRNA gene sequences (SSU ≈ 550 bp; NS31/AM1 fragment) and involved 339 sequences (139 newly detected nucleotide sequences, 160 sequences retrieved from Pellegrino et al. 2014, 27 from the reference dataset, and 12 from GenBank). In detail, HI and ZI sequences are those from Pellegrino et al. (2014). The tree is rooted with Corallochytrium limacisporum. In the collapsed NJ tree, black triangles represent the 17 AMF molecular operational taxonomic units (MOTUs) retrieved in the study. The sequences obtained were affiliated with Funneliformis sp. (12 and 13), Rhizophagus sp. (5, 6, and 7), Sclerocystis sp. (4), Scutellospora sp. (15), Glomus spp. (1, 2, 3, 8, 9, 10, 11, and 14) and uncultured Glomeromycota (16 and 17). For each MOTU, the proportions of sequences from each land-use type (HI, red; LI, yellow; ZI, green) and matrix (soil: left column; roots: right column) are shown in pie charts. Accession numbers of sequences obtained in the present study are shown in Fig. S3. Bootstrap values (based on 1,000 replicates) are shown at the nodes. The scale bar indicates substitutions per site
The AMF MOTUs retrieved exclusively in the soil of each land use were one AMF MOTU affiliated with Glomus sp. (14) in HI; four MOTUs affiliated with Glomus sp. (1), Rhizophagus sp. (7) and Funneliformis sp. (12, 13) in ZI; and three MOTUs affiliated with Glomus spp. (8, 10, 11) in LI (Fig. 3). Regarding roots, four MOTUs affiliated with Glomus sp. (9), Scutellospora sp. (15), and uncultured Glomeromycota (16, 17) were exclusively retrieved in ZI, while three unique MOTUs affiliated with Glomus sp. (1, 14) and Sclerocystis sp. (4) were found in LI. As shown by relative abundance (Fig. S4), Glo2_AMASS (3) was statistically more abundant in the soil of HI (17.0 %) than in that of LI (3.2 %), as was Sclero1_AMASS (HI = 41.1 % and LI = 29.2 %). In the root matrix, Glo1.b_AMASS (2) was more abundant in LI (52.0 %) than in ZI (6.0 %). With regard to the matrix, we observed only in HI significantly higher values (P < 0.05) of the AMF richness and diversity in the soil compared to the roots (Table S5).
Effect of land-use intensification and soil chemical properties on the community composition of total soil fungi
Partial RDA showed that land-use intensity affected the composition of total soil fungal communities (P = 0.02; Fig. 4a). The first two axes explained 37.6 % of total variance. The Monte Carlo permutation test highlighted that the soil fungal community under ZI was significantly different from those under other land uses. Cortica_AMASS (12) (Basidiomycota), Xyla_AMASS (4), and Eurotia_AMASS (6) (Ascomycota) showed preferential presence in ZI, while Unc_Zygo_AMASS (Zygomycota) and Gompha_AMASS (11) and Tremella_AMASS (14) showed preferential presence in LI and in HI, respectively. Partial RDA based on soil chemical properties showed that they explained 44.4 % (axes I and II) of total variance (Fig. 4c). Bas Sat and ExchCa were significant soil chemical drivers in the total fungal community (bold arrows, Fig. 4c). Variation partitioning determined that with Bas Sat, most of the contribution was shared with land-use intensification, while with ExchCa, the contribution of land-use intensification alone was greatest (Table 1).
Partial redundancy analysis (pRDA) biplots based on relative abundances of molecular operational taxonomic units (MOTUs) of a total soil fungi and b arbuscular mycorrhizal fungi (AMF), used as response variables, and three levels of intensification of land use, used as environmental variables. Land-use types were high intensity (HI; maize monoculture), low intensity (LI; extensive grassland), and zero intensity (ZI; agricultural soil left abandoned). The sum of the variance explained by 1st and 2nd axes accounted for 37.6 and 49.5 % of the total variance for (a) and (b), respectively. The Monte Carlo permutational tests showed that AMF and total fungal assemblages were significantly different between the soil under ZI compared to soils under HI and LI (P = 0.02). pRDA biplots at the bottom show the influence of soil chemical properties (used as explanatory variables) on the relative abundances of MOTUs of c total soil fungi and d AMF (used as response variables). The sum of the variance explained by 1st and 2nd axes accounted for 44.4 and 58.6 % of the total variance for (c) and (d), respectively. Red arrows in bold show the significant soil chemical parameters selected after the Monte Carlo permutational test and stepwise selection (P ≤ 0.01). Significant soil chemical drivers were base saturation (Bas Sat) and exchangeable calcium (ExchCa) in the total fungal community and Bas Sat in the AMF community. Spatial coordinates of the plots (latitude/longitude) were used as covariables.The affiliation of the MOTUs shown in the biplots are listed in Table S2 and S6. Relative abundances of AMF from HI and ZI land-use types are from Pellegrino et al. (2014)
Effect of land-use intensification and soil chemical properties on the community composition of AMF
Partial RDA showed that land-use intensity affected composition of soil AMF communities (P = 0.02; Fig. 4b). The first two axes explained 49.5 % of total variance. The Monte Carlo permutation test highlighted that the soil AMF community under ZI was significantly different from those under other land uses. Regarding roots, pCCA highlighted that intensification significantly affected the composition of AMF community (P = 0.002). The first two axes explained 46.1 % of total variance (Fig. 5a). Each land-use type had a different AMF community composition (Monte Carlo permutation test, P = 0.002). The arrows in the biplot (Fig. 4b; Fig. 5a) confirmed that the AMF community composition was driven by the MOTUs retrieved exclusively in each land use. Regarding the effect of the matrix, pCCA showed that the AMF community composition in roots was significantly different from that observed in soil (P = 0.002). The first two axes accounted for 42.1 % of the total variance (Fig. 5b). Partial RDA showed that soil chemical properties explained 58.6 % (axes I and II) of total variance of the AMF composition observed in the soil (Fig. 4d), whereas no effect was observed in the roots. Bas Sat was the only soil chemical parameter that significantly shaped soil AMF composition (Monte Carlo permutational test and stepwise selection, P = 0.006). Variation partitioning determined that most of the contribution was shared between land-use intensification and Bas Sat (Table 1). In addition, CoCA did not detect any relation between the AMF compositions in soil and roots (P = 0.758).
Partial canonical correspondence analysis (pCCA) biplot based on relative abundances of molecular operational taxonomic units (MOTUs) of arbuscular mycorrhizal fungi (AMF) retrieved in roots used as response variables and three levels of intensification of land use used as environmental variables (a). Land-use types were high intensity (HI; maize monoculture), low intensity (LI; extensive grassland), and zero intensity (ZI; agricultural soil left abandoned). Spatial coordinates of the plots (latitude/longitude) were used as covariables. The 1st and 2nd axes accounted for 46.1 % of the total variance. The Monte Carlo permutational tests showed that AMF assemblages were significantly different among all three land-use types (P ≤ 0.05). pCCA biplot based on the relative abundances of the MOTUs of AMF retrieved in soil and roots of all treatments used as response variables, and the matrixes, soil, and roots used as environmental variables (b). Land-use intensities (HI, LI, and ZI) were used as covariables. The 1st and 2nd axes accounted for 42.1 % of the total variance. The Monte Carlo permutational tests showed that AMF assemblages were significantly different between the two matrixes (P ≤ 0.002). Relative abundances from HI and ZI land-use types are from Pellegrino et al. (2014)
Discussion
Despite the primary role of peatlands as a major sink of C and the role of fungi in such habitats as principal microbial decomposers, we still know little about the community composition of fungi in peatlands, above all, in Mediterranean peaty soils drained for agricultural purposes. In this study, we provided for the first time insights into the contribution of land-use intensification and soil properties in shaping the composition of soil fungal communities in Mediterranean agricultural peaty soils. A continuous maize cropping system was compared to an extensive grassland and an agricultural soil left abandoned for 15 years. Multivariate analyses show that (i) land-use intensification shapes the composition of the total soil fungal community and AMF in soil and roots, (ii) base saturation and exchangeable calcium in the soil are the significant chemical drivers of the composition of the total fungal community, (iii) base saturation is the only chemical parameter that significantly shapes the soil AMF community, and (iv) no soil properties affect the AMF that inhabit roots.
Effect of land-use intensification and soil properties on phylogenetic diversity and abundance of total soil fungi
Although many studies have thoroughly investigated soil fungal diversity in different natural habitats (e.g., Anderson and Cairney 2004; Buee et al. 2009), our study was enhanced by the awareness that their diversity had been studied less in agricultural soils (Stromberger 2005; Lynch and Thorn 2006), especially peaty soils (Artz et al. 2007; Tavi et al. 2010). In agreement with previous peatland studies (Thormann 2006; Artz et al. 2007), we found an abundance of Ascomycota regardless of agricultural intensification. Similar dominance was also reported in arable soils, grasslands, and woodlands with high soil N mineral concentration and a C:N ratio (8–12) characteristic of well-humified organic matter (Lauber et al. 2008; Orgiazzi et al. 2012). Accordingly, Allison et al. (2007) and Nemergut et al. (2008) reported that N mineral fertilization significantly increased the abundance of Ascomycota compared to Basidiomycota.
Chytridiomycota were absent in the abandoned soils (ZI), whereas they were highly prevalent in the maize monoculture (HI). Our results are consistent with the description of lower abundance of Chytridiomycota by Lienhard et al. (2014) under no-till system as compared to conventional tillage, according to the ability of this phylum to have resistant structures that permit survival through periodic drying and high summer temperatures typically occurring under tilled soils (Gleason et al. 2004). Similarly, Artz et al. (2007) detected no Chytridiomycota sequences in peatlands abandoned for more than 5 years, whereas Kuramae et al. (2013) observed Chytridiomycota in buried maize leaves. Therefore, our data support the hypothesis that this phylum contributes to the decomposition of crop residues with a high C:N ratio. Zygomycota were detected in all land uses, which confirms their common occurrence in peaty soils (Thormann 2006).
Within Ascomycota, the dominance of Sordariomycetes and Leotiomycetes in HI (more than 70 % of sequences) is consistent with previous data on arable soils (Lauber et al. 2008; Klaubauf et al. 2010). In contrast, Dothideomycetes, largely observed in cultivated soils (Lauber et al. 2008), were found in grasslands (LI) but not in HI. Interestingly, Eurotiomycetes, a taxon associated with cellulose decomposition (Schneider et al. 2012), were more abundant in ZI than in the more intensively managed land uses.
Regarding Basidiomycota, in agreement with Lauber et al. (2008), Agaricomycetes abundance was higher in ZI than in HI and LI due to the abundance of Cortica_AMASS. Corticiales are usually observed in forest soils as decomposers of dead woody substrates (i.e., saprotrophs; Allison et al. 2007). The mycoparasites Tremellales were mostly found in HI, according to Kuramae et al. (2013) who found this taxon as one of the groups involved in litter decomposition of maize leaves.
Total soil fungal diversity was not significantly depleted by land-use intensification, as indicated by MOTU richness and H′ index values, which fell into ranges similar to those in previous work (Lauber et al. 2008; Klaubauf et al. 2010). In the less intensively managed areas, the higher values of the Pielou evenness index (J′) showed that the MOTUs of the total soil fungi indicate that fungi are more uniformly distributed respect to those in HI.
Effect of land-use intensification and soil properties on phylogenetic diversity and abundance of AMF
Root and soil MOTU richness, H′, and J′ were similar to values observed previously (Helgason et al. 1998; Franke-Snyder et al. 2001; Pellegrino et al. 2011) but lower than those reported by others (Jansa et al. 2002; Oehl et al. 2010). These inconsistencies might be due to differences in pedo-climatic conditions or in detection methodology. Our results showed that AMF diversity in roots was lower than that in soil and that land-use intensification did not modify the diversity and richness of AMF, but greatly altered their compositions. This is consistent with findings of other authors (Jansa et al. 2002; Pellegrino et al. 2011, 2014; Moora et al. 2014).
The study of a wider range of land-use intensifications, with respect to Pellegrino et al. (2014), strengthened the fact that AMF richness in roots and in soil is not depleted by agricultural intensification (Daniell et al. 2001; Johnson et al. 2004; Hijri et al. 2006). Focusing on the differences between soil and root, the significantly higher values of AMF MOTU richness and diversity already observed by Pellegrino et al. (2014) in soil under HI compared to roots was not more detected in the lower intensive land uses.
Glomeraceae was the family found in soil and roots regardless of land-use intensification, whereas Gigasporaceae was only found in roots of ZI. These data are in agreement with previous findings showing that the propagation of Gigasporaceae is negatively affected by the soil tillage disruption of extra radical mycelium (Helgason et al. 1998; Jansa et al. 2002).
Effect of land-use intensification and soil properties on the community composition of total soil fungi and AMF
Our study showed that land-use intensification shaped the composition of community of both total fungi and AMF in the soil. Specifically, both fungal community compositions in ZI were different from those in the more intensively managed systems. This agrees well with previous findings, which indicate that undisturbed habitats are highly distinct from anthropogenic areas regardless of the type of disturbance (Lauber et al. 2008; Lumini et al. 2010; Orgiazzi et al. 2012). In contrast, AMF root communities also differed between HI and LI, which were characterized by mowing and vegetation removal. Thus, AMF-inhabiting roots are greatly influenced by both the aboveground composition of the plant species and the type of disturbance. Tillage and fertilization, as well as the removal of aboveground biomass, whether by grazing or mowing, have already been shown to greatly affect these symbionts in the roots (Titus and Lepš 2000; Barto and Rillig 2010).
Bas Sat was found to be a significant driver in shaping fungal community composition, which is in agreement with previous studies (Da Silva et al. 2014; Jansa et al. 2014). Similar to observations by Grayston et al. (2004), we found that ExchCa was an additional driver of the composition of the total soil fungal community that may affect microbes through changes in pH and influences on soil aggregation (Muneer and Oades 1989). It is notable that soil pH had no effect on structuring either of the two fungal communities. These findings support that total soil fungal abundance and diversity are unaffected by soil pH (Rousk et al. 2010), whereas recent observations (Da Silva et al. 2014) do not confirm pH as a major driver of soil AMF composition (Jansa et al. 2014).
On the basis of the variation partitioning results, which highlight a large overlap between land-use intensification and Bas Sat, we assert that these drivers are well-correlated and commonly shape soil fungal community composition. Since land-use intensification had a large individual effect and did not overlap with ExchCa, we argue that intensification drives the composition of total soil fungi without correlating with ExchCa.
Overall, our results demonstrated that land-use intensification significantly shaped the community composition of soil fungi in Mediterranean peaty soils, although it did not modify their diversity and richness. We quantified the role of soil properties by highlighting the significant and synergistic effect of base saturation with land-use intensification.
References
Allison SD, Hanson CA, Treseder KK (2007) Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol Biochem 39:1878–1887
Andersen R, Chapman SJ, Artz RRE (2013) Microbial communities in natural and disturbed peatlands: a review. Soil Biol Biochem 57:979–994
Anderson IC, Cairney JW (2004) Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6:769–779
Antonsen H, Olsson PA (2005) Relative importance of burning, mowing and species translocation in the restoration of a former boreal hayfield: responses of plant diversity and the microbial community. J Appl Ecol 42:337–347
Artz RR, Anderson IC, Chapman SJ, Hagn A, Schloter M, Potts JM, Campbell CD (2007) Changes in fungal community composition in response to vegetational succession during the natural regeneration of cutover peatlands. Microb Ecol 54:508–522
Augé RM (2001) Water relations, drought and VA mycorrhizal symbiosis. Mycorrhiza 1:3–42
Bago B, Pfeffer PE, Shachar-Hill Y (2000) Carbon metabolism and transport in in arbuscular mycorrhizas. Plant Physiol 124:949–958
Bahri H, Dignac MF, Rumpel C, Rasse DP, Chenu C, Mariotti A (2006) Lignin turnover kinetics in an agricultural soil is monomer specific. Soil Biol Biochem 38:1977–1988
Bardgett RD, Hobbs PJ, Frostegård Å (1996) Changes in soil fungal: bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol Fertil Soils 22:261–264
Bardgett RD, Usher MB, Hopkins DW (2005) Biological diversity and function in soils. Cambridge University Press, Cambridge
Barto EK, Rillig MC (2010) Does herbivory really suppress mycorrhiza? A meta‐analysis. J Ecol 98:745–753
Bedini S, Pellegrino E, Avio L, Pellegrini S, Bazzoffi P, Argese E, Giovannetti M (2009) Changes in soil aggregation and glomalin-related soil protein content as affected by the arbuscular mycorrhizal fungal species Glomus mosseae and Glomus intraradices. Soil Biol Biochem 41:1491–1496
Bittman S, Forge TA, Kowalenko CG (2005) Responses of the bacterial and fungal biomass in a grassland soil to multi-year applications of dairy manure slurry and fertilizer. Soil Biol Biochem 37:613–623
Buee M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456
Cheng L, Booker FL, Tu C, Burkey KO, Zhou L, Shew HD, Rufty TW, Hu S (2012) Arbuscular mycorrhizal fungi increase organic carbon decomposition under elevated CO2. Science 337:1084–1087
da Silva IR, de Mello CMA, Neto RAF, da Silva DKA, de Melo AL, Oehl F, Maia LC (2014) Diversity of arbuscular mycorrhizal fungi along an environmental gradient in the Brazilian semiarid. Appl Soil Ecol 84:166–175
Dai M, Bainard LD, Hamel C, Gan Y, Lynch D (2013) Impact of land use on arbuscular mycorrhizal fungal communities in rural Canada. Appl Environ Microbiol 79:6719–6729
Daniell TJ, Husband R, Fitter AH, Young JPW (2001) Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiol Ecol 36:203–209
Denef K, Roobroeck D, Manimel Wadu MC, Lootens P, Boeckx P (2009) Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol Biochem 41:144–153
Di Bene C, Pellegrino E, Debolini M, Silvestri N, Bonari E (2013) Short- and long-term effects of olive mill wastewater land spreading on soil chemical and biological properties. Soil Biol Biochem 56:21–30
Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345
Eom AH, Hartnett DC, Wilson GWT, Figge DAH (1999) The effect of fire, mowing and fertilizer amendment on arbuscular mycorrhizas in tall-grass prairie. Am Midl Nat 142:55–70
Fitter AH, Gilligan CA, Hollingworth K, Kleczkowski A, Twyman RM, Pitchford JW (2005) Biodiversity and ecosystem function in soil. Funct Ecol 19:369–377
Ford H, Rousk J, Garbutt A, Jones L, Jones DL (2013) Grazing effects on microbial community composition, growth and nutrient cycling in salt marsh and sand dune grasslands. Biol Fertil Soils 49:89–98
Franke-Snyder M, Douds DD, Galvez L, Phillips JG, Wagoner P, Drinkwater L, Morton JB (2001) Diversity of communities of arbuscular mycorrhizal (AM) fungi present in conventional versus low-input agricultural sites in eastern Pennsylvania, USA. Appl Soil Ecol 16:35–48
Freeman KR, Martin AP, Karki D, Lynch RC, Mitter MS, Meyer AF, Longcore JE, Simmons DR, Schmidt SK (2009) Evidence that chytrids dominate fungal communities in high-elevation soils. Proc Natl Acad Sci 106:18315–18320
Gadd GM (2006) Fungi in biogeochemical cycles (vol 24). Cambridge University Press, Cambridge
Galvan GA, Paradi I, Burger K, Baar J, Kuyper TW, Scholten OE, Kik C (2009) Molecular diversity of arbuscular mycorrhizal fungi in onion roots from organic and conventional farming systems in the Netherlands. Mycorrhiza 19:317–328
García-Orenes F, Morugán-Coronado A, Zornoza R, Scow K (2013) Changes in soil microbial community structure influenced by agricultural management practices in a Mediterranean agro-ecosystem. PLoS One 8:e80522
Gleason FH, Letcher PM, McGee PA (2004) Some Chytridiomycota in soil recover from drying and high temperatures. Mycol Res 108:583–589
Gosling P, Hodge A, Goodlass G, Bending GD (2006) Arbuscular mycorrhizal fungi and organic farming. Agric Ecosyst Environ 113:17–35
Grayston SJ, Campbell CD, Bardgett RD, Mawdsley JL, Clegg CD, Ritz K, Griffiths BS, Rodwell JS, Edwards SJ, Davies WJ, Elston DJ, Millard P (2004) Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl Soil Ecol 25:63–84
Hannula SE, De Boer W, Van Veen J (2012) A 3-year study reveals that plant growth stage, season and field site affect soil fungal communities while cultivar and GM-trait have minor effects. PLoS One 7:e33819
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JP (1998) Ploughing up the wood-wide web? Nature 394:431
Herr JR, Öpik M, Hibbett DS (2015) Towards the unification of sequence‐based classification and sequence‐based identification of host‐associated microorganisms. New Phytol 205:27–31
Hijri I, Sýkorová Z, Oehl F, Ineichen K, Mäder P, Wiemken A, Redecker D (2006) Communities of arbuscular mycorrhizal fungi in arable soils are not necessarily low in diversity. Mol Ecol 15:2277–2289
Hodge A, Campbell CD, Fitter AH (2001) An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413:297–299
Holland TC, Bowen P, Bogdanoff C, Hart MM (2014) How distinct are arbuscular mycorrhizal fungal communities associating with grapevines? Biol Fertil Soils 50:667–674
Hoshino YT, Morimoto S (2008) Comparison of 18S rDNA primers for estimating fungal diversity in agricultural soils using polymerase chain reaction-denaturing gradient gel electrophoresis. Soil Sci Plant Nutr 54:701–710
Hunt J, Boddy L, Randerson PF, Rogers HJ (2004) An evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microb Ecol 47:385–395
IPCC (2006) 2006 IPCC Agriculture, Forestry and other land uses. In: Eggleston HS, Buendia L, Miwa K, Ngara T, Tanabe K (eds) Guidelines for National Greenhouse Gas Inventories (vol 4) prepared by the National Greenhouse Gas Inventories Programme. IGES, Japan
Jansa J, Erb A, Oberholzer HR, Šmilauer P, Egli S (2014) Soil and geography are more important determinants of indigenous arbuscular mycorrhizal communities than management practices in Swiss agricultural soils. Mol Ecol 23:2118–2135
Jansa J, Mozafar A, Anken T, Ruh R, Sanders IR, Frossard E (2002) Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 12:225–234
Joergensen RG, Wichern F (2008) Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem 40:2977–2991
Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ (2004) Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol 161:503–515
Klaubauf S, Inselsbacher E, Zechmeister-Boltenstern S, Wanek W, Gottsberger R, Strauss J, Gorfer M (2010) Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers 44:65–75
Krüger M, Krüger C, Walker C, Stockinger H, Schüssler A (2012) Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193:970–984
Kuramae EE, Hillekens RH, Hollander M, Heijden MG, den Berg M, Straalen NM, Kowalchuk GA (2013) Structural and functional variation in soil fungal communities associated with litter bags containing maize leaf. FEMS Microbiol Ecol 84:519–531
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415
Legendre P (2008) Studying beta diversity: ecological variation partitioning by multiple regression and canonical analysis. J Plant Physiol 1:3–8
Leyval C, Turnau K, Haselwandter K (1997) Effect of heavy metal pollution on mycorrhizal colonization and function: physiological, ecological and applied aspects. Mycorrhiza 7:139–153
Lienhard P, Terrat S, Prévost-Bouré NC, Nowak V, Régnier T, Sayphoummie S, Panyasiri K, Tivet F, Mathieu O, Levêque J, Maron PA, Ranjard L (2014) Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron Sustain Dev 34:525–533
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Koljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H (2013) Fungal community analysis by high‐throughput sequencing of amplified markers—a user’s guide. New Phytol 199:288–299
Lumini E, Orgiazzi A, Borriello R, Bonfante P, Bianciotto V (2010) Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land use gradient using a pyrosequencing approach. Environ Microbiol 12:2165–2179
Lynch MD, Thorn RG (2006) Diversity of basidiomycetes in Michigan agricultural soils. Appl Environ Microbiol 72:7050–7056
Mathimaran N, Ruh R, Jama B, Verchot L, Frossard E, Jansa J (2007) Impact of agricultural management on arbuscular mycorrhizal fungal communities in Kenyan ferralsol. Agric Ecosyst Environ 119:22–32
Moora M, Davison J, Öpik M, Metsis M, Saks Ü, Jairus T, Vasar M, Zobel M (2014) Anthropogenic land use shapes the composition and phylogenetic structure of soil arbuscular mycorrhizal fungal communities. FEMS Microbiol Ecol. doi:10.1111/1574-6941.12420
Muneer M, Oades JM (1989) The role of Ca-organic interactions in soil aggregate stability: III. Mechanisms and models. Aust J Soil Res 27:411–423
Nannipieri P, Ascher J, Ceccherini M, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Nemergut DR, Townsend AR, Sattin SR, Freeman KR, Fierer N, Neff J, Bowman WD, Schadt CW, Weintraub MN, Schmidt SK (2008) The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: implications for carbon and nitrogen cycling. Environ Microbiol 10:3093–3105
Newsham KK, Fitter AH, Watkinson AR (1995) Multi-functionality and biodiversity in arbuscular mycorrhizas. Trends Ecol Evol 10:407–411
Oehl F, Laczko E, Bogenrieder A, Stahr K, Bösch R, van der Heijden M, Sieverding E (2010) Soil type and land use intensity determine the composition of arbuscular mycorrhizal fungal communities. Soil Biol Biochem 42:724–738
Oehl F, Sieverding E, Ineichen K, Mäder P, Boller T, Wiemken A (2003) Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Appl Environ Microbiol 69:2816–2824
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier U, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Orgiazzi A, Lumini E, Nilsson RH, Girlanda M, Vizzini A, Bonfante P, Bianciotto V (2012) Unravelling soil fungal communities from different Mediterranean land-use backgrounds. PLoS One 7(4):e34847
Pellegrino E, Bosco S, Ciccolini V, Pistocchi C, Sabbatini T, Silvestri N, Bonari E (2014) Agricultural abandonment in Mediterranean reclaimed peaty soils: long-term effects on soil chemical properties, arbuscular mycorrhizas and CO2 flux. Agric Ecosyst Environ 199:164–175
Pellegrino E, Di Bene C, Tozzini C, Bonari E (2011) Impact on soil quality of a 10-year-old short-rotation coppice poplar stand compared with intensive agricultural and uncultivated systems in a Mediterranean area. Agric Ecosyst Environ 140:245–254
Pellegrino E, Turrini A, Gamper HA, Cafà G, Bonari E, Young JPW, Giovannetti M (2012) Establishment, persistence and effectiveness of arbuscular mycorrhizal fungal inoculants in the field revealed using molecular genetic tracing and measurement of yield components. New Phytol 194:810–822
Pistocchi C, Silvestri N, Rossetto R, Sabbatini T, Guidi M, Baneschi I, Bonari E, Trevisan D (2012) A simple model to assess nitrogen and phosphorus contamination in ungauged surface drainage networks: application to the Massaciuccoli Lake Catchment, Italy. J Environ Qual 41:544–553
Rillig MC (2004) Arbuscular mycorrhizae, glomalin, and soil aggregation. Can J Soil Sci 84:355–363
Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Schneider T, Keiblinger KM, Schmid E, Sterflinger-Gleixner K, Ellersdorfer G, Roschitzki B, Richter A, Eber L, Zechmeister-Boltenstern S, Riedel K (2012) Who is who in litter decomposition? metaproteomics reveals major microbial players and their biogeochemical functions. ISME J 6:1749–1762
Schnoor TK, Lekberg Y, Rosendahl S, Olsson PA (2011) Mechanical soil disturbance as a determinant of arbuscular mycorrhizal fungal communities in semi-natural grassland. Mycorrhiza 21:211–220
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246
Schuessler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Simon L, Lalonde M, Bruns TD (1992) Specific amplification of 18S fungal ribosomal genes from vesicular arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58:291–295
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Academic Press, Amsterdam
Soil Survey Staff (1975) Soil taxonomy: a basic system of soil classification for making and interpreting soil surveys. USDA-SCS Agric. Handb. 436. U.S. Gov Print Office, Washington
Stromberger ME (2005) Fungal communities of agroecosystems. In: Dighton J, White JF, Oudemans P (eds) The fungal community: its organization and role in the ecosystem, 3rd edn. CRC Press, Boca Raton, pp 813–832
Tavi NM, Keinänen-Toivola MM, Koponen HT, Huttunen JT, Kekki TK, Biasi C, Martikainen PJ (2010) Impact of Phalaris arundinacea cultivation on microbial community of a cutover peatland. Boreal Environ Res 15:437–445
ter Braak CJF, Schaffers AP (2004) Co-correspondence analysis: a new ordination method to relate two community compositions. Ecology 85:834–846
ter Braak CJF, Smilauer P (2012) Canoco reference manual and user’s guide: software for ordination (version 5.0). Microcomputer Power, Ithaca
Thormann MN (2006) Diversity and function of fungi in peatlands: a carbon cycling perspective. Can J Soil Sci 86:281–293
Titus JH, Lepš J (2000) The response of arbuscular mycorrhizae to fertilization, mowing, and removal of dominant species in a diverse oligotrophic wet meadow. Am J Bot 87:392–401
Torrecillas E, Alguacil MM, Roldan A (2012) Host preferences of arbuscular mycorrhizal fungi colonizing annual herbaceous plant species in semiarid Mediterranean prairies. Appl Environ Microbiol 78:6180–6186
Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JPW (2002) Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol 11:1555–1564
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:72–75
Verbruggen E, van der Heijden MGA, Weedon JT, Kowalchuk GA, Roling WFM (2012) Community assembly, species richness and nestedness of arbuscular mycorrhizal fungi in agricultural soils. Mol Ecol 21:2341–2353
Verhoeven JT, Setter TL (2010) Agricultural use of wetlands: opportunities and limitations. Ann Bot London 105:155–163
Xiang D, Verbruggen E, Hu Y, Veresoglou SD, Rillig MC, Zhou W, Xu T, Li H, Hao Z, Chen Y, Chen B (2014) Land use influences arbuscular mycorrhizal fungal communities in the farming–pastoral ecotone of northern China. New Phytol 204:968–978
Acknowledgments
This work represents part of the PhD thesis project of C.V. which was funded by Scuola Superiore Sant’Anna. The work was also supported by the “Consorzio di Bonifica Versilia-Massaciuccoli” and the “Regione Toscana” (“Restoration of a Mediterranean Drained Peatland”: https://sites.google.com/site/restomedpeat-land/home). We are grateful to Martti Vasar for his bioinformatic advices. The authors wish to thank the two anonymous reviewers and the editor for their help in improving the previous version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 24.1 kb)
ESM 2
(DOCX 70.7 kb)
Supplementary Fig. S6
Neighbor-Joining (NJ) tree of total soil fungal sequences derived from soil from three levels of intensification of land use: high intensity (diamond; HI: maize monoculture), low intensity (square; LI: extensive grassland) and zero intensity (triangle; ZI: agricultural soil left abandoned). NJ is based on the fungal 18S rRNA gene (≈ 1650 bp; NS1/FR1 fragment) and involved 143 newly detected nucleotide sequences plus 25 reference sequences retrieved from the NCBI database. The tree is rooted with Meristolohmannia meristacaroides. The sequences retrieved were classified in 16 AMF molecular operational taxonomic units (MOTUs) affiliated with the orders Hypocreales (1, 2), Sordariales (3), Xylariales (4), Chaetothyriales (5), Eurotiales (6), Pleosporales (7), Dothideomycetes (8), Helotiales (9), Agaricales (10), Gomphales (11), Corticiales (12), Filobasidiales (13), Tremellales (14) and to two classes Chytridiomycota (15) and Zygomycota (16). Bootstrap values (based on 1000 replicates) are shown at the nodes. The scale bar indicates substitutions per site. Sequences obtained in the present study are shown in bold, and their accession numbers are prefixed by the internal clone identifier (H) and the soil sample number from which they were obtained. All new sequences were submitted to the EMBL nucleotide sequence database (accession numbers: LN555148 - LN555383 and LN555530 - LN555579) (JPEG 1.37 MB)
ESM 3
(DOCX 67.4 kb)
Supplementary Fig. S7
Rarefaction curves showing the relation between sampled sequence number and the number of observed molecular operational taxonomic units (MOTUs) of (a) soil arbuscular mycorrhizal fungi (AMF), (b) root AMF and (c) total soil fungi retrieved in three levels of intensification of land use: high intensity (HI; maize monoculture; red line), low intensity (LI; extensive grassland; yellow line) and zero intensity (ZI; agricultural soil left abandoned; green line). AMF sequences from HI and ZI land-use types are from Pellegrino et al. (2014) (JPEG 162 kb)
ESM 4
(DOCX 64.0 kb)
ESM 5
(DOCX 15.5 kb)
Supplementary Fig. S8
Neighbor-Joining tree of sequences of arbuscular mycorrhizal fungi (AMF) derived from soil and roots (open and filled symbols, respectively) from three levels of intensification of land use: high intensity (diamond; HI: maize monoculture), low intensity (square; LI: extensive grassland) and zero intensity (triangle; ZI: agricultural soil left abandoned). Analysis is based on SSU rRNA gene sequences (SSU ≈ 550 bp; NS31/AM1 fragment) and involved 339 sequences (139 newly detected nucleotide sequences, 160 sequences retrieved from Pellegrino et al. (2014), 27 from the reference dataset, 12 from GenBank). The tree is rooted with Corallochytrium limacisporum. The sequences retrieved were classified in 17 AMF molecular operational taxonomic units (MOTUs) affiliated with Funneliformis sp. (12, 13), Rhizophagus sp. (5, 6, 7), Sclerocystis sp. (4), Scutellospora sp. (15), Glomus spp. (1, 2, 3, 8, 9, 10, 11, 14) and uncultured Glomeromycota (16, 17). Bootstrap values (based on 1000 replicates) are shown at the nodes. The scale bar indicates substitutions per site. Sequences obtained in the present study are shown in bold, and their accession numbers are prefixed by the internal clone identifier (clones from roots and soil are identified by a and G, respectively) and the root/soil sample number from which they were obtained. Sequences from HI and ZI land-use types are from Pellegrino et al. (2014). All new sequences were submitted to the EMBL nucleotide sequence database (accession numbers: LN555384- LN555522). The correspondence between MOTUs and the closest virtual taxa (VT) after BLAST search against the MaarjAM database (Öpik et al., 2010) is shown (JPEG 867 kb)
ESM 6
(DOCX 16.0 kb)
ESM 7
(DOCX 16.0 kb)
Supplementary Fig. S9
Community diversity of arbuscular mycorrhizal fungi (AMF) shown as relative abundances of the 17 molecular operational taxonomic units (MOTUs) retrieved in (a) the soil and (b) the roots from three levels of intensification of land use: high intensity (HI; maize monoculture), low intensity (LI; extensive grassland) and zero intensity (ZI; agricultural soil left abandoned). AMF relative abundances from HI and ZI land-use types are from Pellegrino et al. (2014) (JPEG 423 kb)
Rights and permissions
About this article
Cite this article
Ciccolini, V., Bonari, E. & Pellegrino, E. Land-use intensity and soil properties shape the composition of fungal communities in Mediterranean peaty soils drained for agricultural purposes. Biol Fertil Soils 51, 719–731 (2015). https://doi.org/10.1007/s00374-015-1013-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-015-1013-4