Abstract
Recent evidence from several environments suggest that besides autotrophic ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA) are also able to perform the oxidation of NH4 + to NO2 −, although the relative importance of AOA in nitrification, compared to AOB, and their differential susceptibility to inhibitory compounds remains unclear. Experimental microcosms were set up to evaluate the effect of the addition of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) combined with a cattle effluent as organic fertilizer on the abundance and expression of ammonia oxidizers, denitrifiers, and non-target microbial populations using reverse transcription–real-time PCR, as well as on the diversity of metabolically active soil bacterial and archaeal communities by terminal restriction fragment length polymorphism. While no significant changes in soil mineral N concentrations or amoA gene copies could be detected between treatments, short-term changes in transcriptional activity revealed that DMPP impaired both bacterial and archaeal amoA mRNA, being significant at every time point for AOB and at one time point for AOA. Our findings revealed that, despite the different cellular biochemistry and metabolism existing between bacteria and archaea domains, DMPP exerts its inhibitory effect against both soil bacterial and archaeal ammonia-oxidizing transcriptional activity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The application of animal effluents to soil has the potential both to reduce the amount of fertilizer required and to increase productivity. As with many soil amendments, the use of waste materials for maintaining and improving soil productivity presents a desirable and economically attractive alternative as well as assisting in the disposal of otherwise troublesome wastes.
Up to 90 % (Dosch and Gutser 1996; Wulf et al. 2002) of the fertilizer nitrogen (N) applied to soils in the form of ammonium or ammonium-producing compounds is lost contributing to nitrate leaching as well as to the production of environmentally hazardous nitrogenous gases such as nitrous oxide (N2O) and nitric oxide (NO), both during nitrification and subsequent denitrification (Jarvis et al. 1996).
In cultivation systems, reduction of nitrification rates may be implemented by applying the exact amounts of N-fertilizer required by the crop plants (Bronson and Mosier 1994; Subbarao et al. 2006). Consequently, this leads to an increase in plant N-use efficiency through reduced N application rates. Depending on soil conditions and plant species, these beneficial effects could be increasing N-use efficiency, reducing N losses, fertilizer applications, and consequently, preventing environmental and health risk.
Nitrification inhibitors (NIs) are natural or synthetic compounds that delay the microbial oxidation of NH4 + to NO2 −, the first step of the nitrification process with nitrapyrin, dicyandiamide (DCD), and 3,4-dimethylpyrazole phosphate (DMPP), the three major commercial NIs (Subbarao et al. 2006). DMPP, however, is one of the most highly effective and widely used nitrification inhibitors (Pasda et al. 2001; Weiske et al. 2001; Irigoyen et al. 2003). DMPP has been evaluated under field (Merino et al. 2005; Li et al. 2008; Chen et al. 2010) and laboratory (Hatch et al. 2005; Barth et al. 2008; Pereira et al. 2010; Yang et al. 2013) conditions, mixed with both inorganic (Linzmeier et al. 2001; Weiske et al. 2001) or organic (Dittert et al. 2001; Macadam et al. 2003) fertilizer. It has the potential to reduce losses of N2O up to 60 % (Weiske et al. 2001; Zerulla et al. 2001; Hatch et al. 2005; Menéndez et al. 2012) even if added to slurry (Dittert et al. 2001), and to increase crop yield up to 21 % (Linzmeier et al. 2001; Pasda et al. 2001; Zerulla et al. 2001; Martínez-Alcántara et al. 2012). However, only recently, attention has been paid to the possibility of using inhibitors with organic fertilizer (Dittert et al. 2001; Macadam et al. 2003; Di and Cameron 2004; 2012), especially in Mediterranean ecosystems, where areas frequently experience temperatures above 25–30 °C and high nitrification rates. Furthermore, to our knowledge, the application of DMPP combined with cattle effluents and its effects on soil microbiota has never been studied.
Traditionally, autotrophic ammonia-oxidizing bacteria (AOB) were considered to be the exclusive contributors to ammonia oxidation (Bock and Wagner 2006). However, recent environmental metagenomics analysis revealed that ammonia monooxygenase α-subunit (amoA) genes are also present in soil ammonia-oxidizing archaea (AOA) and that in most mesophilic environments, members of the phylum Thaumarchaeota within the domain of Archaea are considerably more abundant than characterized AOB populations (Leininger et al. 2006; Spang et al. 2010).
Little information is available concerning the differential susceptibility of AOB and AOA to nitrification inhibitors. Contrasting observations have been reported for DCD (Di et al. 2010; Zhang et al. 2012) and AOA and AOB could be differentiated through the inhibition of bacteria by using the antibiotics sulfadiazine (Schauss et al. 2009), whereas, a reduced abundance of AOB, but not AOA, was observed both in rhizosphere and bulk soil following the application of DMPP (Kleineidam et al. 2011).
Evidence for ammonia oxidation by bacteria and archaea in soil has largely been based on abundance of amoA genes, rather than activity. However, the presence or high abundance of a functional gene does not mean that the function is operating (Prosser and Nicol 2008). Messenger-RNA may be an indicator of living cells or the specific activity even in a complex microbial community because its turnover is rapid in living bacterial cells, although increased steady-state mRNA level do not alone guarantee an increase in any associated metabolic activity.
Since archaeal and bacterial ecophysiology, metabolism and biochemistry might presumably be different; it will therefore be crucial to determine whether AOA are also sensitive to AOB inhibitory compounds, and whether nitrification processes can be attributed specifically to AOA. To date, there have been few studies on the effects of nitrification inhibitors on non-target soil microbiota. Given the importance of maintaining soil quality and function, it is essential that soil treatments such as nitrification inhibitors do not negatively impact on important ecosystem functional activities of non-target soil microorganisms.
Thus, the aim of this study was to evaluate the short-term effect of the addition of the nitrification inhibitor DMPP combined with a cattle effluent as organic fertilizer on the abundance and expression both of direct and indirect microbial targets (ammonia oxidizers and denitrifiers), and non-target microbial populations. Furthermore, denitrification genes were monitored to determine whether the amount of nitrate can be attributed specifically to the inhibitory effect exerted by DMPP or whether an increase in denitrifier numbers and stimulation of denitrification leads to increased removal of nitrate from the soil. To determine whether amoA genes from AOA and AOB, bacterial 16S rRNA and nosZ genes are actively transcribed, total RNA from soil microcosms was isolated and relative cDNA copies of these genes after reverse transcription was quantified. Changes in diversity of metabolically active bacterial and archaeal communities were also evaluated.
Materials and methods
Soil and cattle effluent
The soil used (Casalotti soil) was collected from a Eucalyptus, short rotation, high-density plantations field managed by the research unit for intensive wood production (CRA-PLF) located in Rome (Italy) (41°54′33.55″ N, 12°21′37.83″ E). The soil is classified as luvisols (http://www.bluegoldmedia.com/post/102218.html) having a sandy loam texture. Six replicate samples of soil from the top 30 cm (about 30 kg in total) were collected in June 2009 and stored in sterile plastic bags. The soils were air-dried, homogenized by sieving (2-mm mesh size), pooled, and stored at room temperature. Main physico-chemical and some microbiological properties of the soil are reported in Table 1. The cattle effluent used was obtained from a dairy farm situated in Rome. Sampling was performed in June 2009 and sample was stored in PVC barrels at 4 °C until required. On day 0, it was sampled after thorough stirring and blending and analyzed by standard laboratory methods to assess the following physico-chemical properties: moisture (88.9 %), dry matter (11.1 %), Ntot (0.32 %), N-NH4 + (0.17 %), and TOC (5.97 %).
Experimental design
A non-disruptive laboratory incubation experiment was performed to assess potential N mineralization and nitrification, whereas short-term effect on microbial dynamics was assessed in disruptive soil microcosms fertilized with cattle effluent (E) and cattle effluent combined with DMPP (ED). An untreated soil (C) and soil + DMPP (D) control treatments were included.
Liquid formulation of DMPP (25 %, provided by EuroChem Agro Spa, Italy) was used at a final concentration of 1 % according to the organic fertilizer NH4 +-N content, as recommended by the providers (Zerulla et al. 2001). Nitrogen from cattle effluent was added at a concentration of 250 mg N kg soil−1.
Potential N mineralization and nitrification
Cattle effluent and nitrification inhibitor were added singly or as a pre-mixed solution to 50-g air-dried soil mixed with quartz sand in a 1:1 ratio (Stanford and Smith 1972). The mixture was incubated at 60 % water holding capacity in the dark at 30 °C for 12 weeks (Benedetti et al. 1994; Dell’Abate et al. 2003). The Büchner funnels used were covered by cling film which prevented water loss but allowed gas exchange. Every 3–4 days, funnels were aerated and moisture was replenished throughout the duration of the experiment. NO3 −-N, NO2 −-N, and NH4 +-N produced during the incubation were monitored at 1, 2, 4, 8, and 12 weeks. The soils were eluted with 900 ml 0.01-M CaSO4 solution, and then with 100-ml N-free solution [0.002-M CaSO4, 0.005-M Ca(H2PO4)2, 0.0025-M K2SO4, 0.002-M MgSO4] in order to reintegrate nutrient elements. Nitrogen forms were determined colorimetrically in the eluate by continuous flux analyser (AutoAnalyzer Technicon II), according to Wall et al. (1975) for NH4 +-N and Kamshake et al. (1967) for (NO3 −+NO2 −)-N. Cumulative net nitrification and net N mineralization were expressed as (NO3 −+NO2 −)-N (milligram per N kilogram dry soil) and (NH4 ++NO3 −+NO2 −)-N (milligram per N kilogram dry soil) respectively. Each treatment was replicated three times.
Microcosm study of microbial analysis
Microcosms containing 1.1 kg of homogenized soil were set up to examine microbial community structure and functioning in Casalotti soil after cattle effluent and DMPP addition. Microcosms were incubated at 60 % water holding capacity in the dark at 30 °C for 28 days, and covered by cling film which prevented water loss but allowed gas exchange. Every 3 days, moisture was replenished throughout the experiments’ duration. Each treatment was replicated three times. Ten-gram subsamples were taken at days 1, 4, and 7, and immediately frozen at −80 °C and stored for molecular analysis until required.
Nucleic acid extraction
RNA and DNA were extracted from 2 g of each soil sample and slurry using a RNA PowerSoil® Total RNA Isolation Kit and RNA PowerSoil® DNA Elution Accessory Kit, respectively (Mo Bio, Carlsbad, CA, USA) following slight modification to the manufacturer’s instructions, whereby the 15-min shaking on a flat bed vortex was replaced by a 30-s bead beading step (5.5 m s−1, Fastprep). DNA and RNA concentrations were determined using the Qubit quantification platform with Quant-iT™ dsDNA high sensitivity Assay Kit and Quant-iT™ RNA Assay Kit (Invitrogen, NZ). DNA and RNA were diluted to 5 ng/μL and stored in −80 °C freezer for the following molecular applications.
Quantitative real-time PCR
The abundances of bacterial 16S rRNA, bacterial amoA, archaeal amoA, nirK, nirS, and nosZ genes in all the soil samples were quantified using real-time PCR (Clark et al. 2012). Quantitative real-time PCR (qPCR) was performed using primers described in Table 2 and a Stratagene Mx3000P QPCR thermal cycler (Agilent Technologies) in a 25-μL reaction mixture consisting of 20-ng DNA template, 12.5-μL QuantiTect SYBR green master mix (QIAGEN), and the following primer concentrations: 1 μM for bacterial 16S rRNA and archaeal amoA, 1.5 μM for bacterial amoA, 5 μM for nirK and nosZ, and 7.5 μM for nirS. Real-time amplification was performed using the following programs developed: for bacterial 16S rRNA gene: initial 15 min at 95 °C; 30 cycles of 15 s at 95 °C for denaturing, 30 s at 58 °C for annealing, 30 s at 78 °C, and 15 s at 84 °C (both readout) for extension; for bacterial amoA gene: initial 15 min at 95 °C; 45 cycles of 15 s at 95 °C for denaturing, 30 s at 58 °C for annealing, 45 s at 72 °C, and 15 s at 84 °C (both readout) for extension; and for archaeal amoA gene: initial 15 min at 95 °C; 40 cycles of 15 s at 95 °C for denaturing, 30 s at 58 °C for annealing, 45 s at 72 °C, and 15 s at 78 °C (both readout) for extension; for nirK, nirS, and nosZ genes see Clark et al. (2012).
A melting curve analysis was performed to confirm PCR product specificity after amplification by measuring fluorescence continuously as the temperature increased from 50 to 99 °C. The specificity of the PCR was further evaluated by running randomly selected samples on a 1.5 % (w/v) agarose gel. The corresponding real-time PCR efficiency for each of these genes was estimated based on serial dilutions of the common DNA mixture described below. Triplicate qPCR was performed for each target gene for all samples. The copy numbers of these genes per gram of dry soil was calculated from the copy number of each gene per nanogram of DNA multiplied by the amount of DNA contained in each gram of dry soil.
Standards were generated from PCR products that had been obtained from soil DNA extracts, gel purified, quantified using a Qubit™ fluorometer (Invitrogen, NZ) and diluted accordingly. This was to minimize the bias inherent in the usual approach, where one or only a few genes are used to standardize complex communities (Töwe et al. 2010). Problems arise because the degenerate primers have only partial homology to the gene variants used to generate the standard curve, and there are differences in the amplification efficiencies of the range of sequences internal to the primers in the environmentally extracted DNA samples.
The number of copies of each gene was calculated using the following equation: gene copy number = (nanogram*number/mole)/(base pairs*nanogram/gram*gram mole base pairs) (http://www.uri.edu/research/gsc/resources/cndna.html). Standards were diluted accordingly to give a concentration range from 0 to 107 (109 for bacterial 16S rRNA) gene copies per microliter. All DNA preparations were checked for the absence of inhibitors prior to PCR and all results were analyzed using LinRegPCR program version 11.1 (Ramakers et al. 2003; Ruijter et al. 2009) to confirm the efficiency of amplification and the absence of inhibition.
Reverse transcription–real-time PCR
Quantitative real-time reverse transcription (RT-PCR) assays were used to specifically measure the bacterial 16S, bacterial amoA, archaeal amoA, and nosZ abundance mRNA. The mRNA concentrations of the four genes investigated were quantified with one-step RT-PCR assays. RNA was treated with TURBO DNA-free™ Kit (Applied Biosystems, USA). Standard PCRs were performed on all TURBO DNA-free treated RNAs to confirm that no PCR products were produced and hence, all traces of contaminating DNA had been removed. RT-PCR assays were subsequently performed using SensiFAST™ SYBR Lo-ROX One-Step Kit (Bioline, UK) in a 40-μL mix containing 20 μL of 2X SensiFAST SYBR Lo-ROX One-step Mix, 0.4 μL of reverse transcriptase, 0.8 μL of RiboSafe RNase Inhibitor, 0.2-ng DNA for bacterial 16S rRNA gene, 20-ng DNA for nosZ gene, and 50-ng DNA for bacterial and archaeal amoA genes, and the same primers concentrations used in real-time amplification assays described before.
The RT-PCR reaction programs were the same as those used in real-time amplification assays.
cDNA synthesis
RT was carried out on half of the total RNA extracted using QuantiTect Reverse Transcription Kit (QIAGEN). Ten-microliters RNA were added to 2 μL gDNA wipeout buffer to a final volume of 14 μL, in order to eliminate genomic DNA. The suspension was heated for 2 min at 42 °C and then placed immediately on ice. An RT mix of 1-μL reverse transcriptase containing RNase inhibitor, 5X Quantiscript RT buffer containing Mg2+ and dNTPs, and 1-μL RT primer mix, to a final volume of 20 μL, was added. RNA free of contaminating DNA obtained previously was added on ice to each tube containing reverse transcription master mix. The suspension was incubated for 15 min at 42 °C, and then heated for 3 min at 95 °C to stop the reaction. The absence of DNA contamination was confirmed by PCR amplification of non reverse transcribed RNA samples.
RT-T-RFLP analysis
The samples collected from soil microcosms described previously were analyzed by terminal restriction fragment length polymorphism (T-RFLP) on metabolic active microbial populations. The bacterial and archaeal 16S rRNA genes were amplified in multiplex with VIC- and NED-labeled (on forward primer) primers, respectively, 63f/1087f and Ar344F/Ar927R, details in Table 2.
Following PCR amplification (MacDonald et al. 2011) products were visualized with ethidium bromide staining on a 1 % (w/v) agarose gel using UV radiation and purified using Wizard® Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s instructions. Purified PCR products (about 500 ng) were digested with 20 U of Msp I (Promega, UK) restriction enzyme as previously described (Macdonald et al. 2007). Terminal restriction fragments (T-RFs) were resolved on an ABI PRISM 3130xl genetic analyzer (Applied Biosystems, UK).
Data analysis
Excel and SPSS 11 software was used for the analysis of potential N mineralization and nitrification data, as well as qPCR and RT-qPCR data, and comparisons of means were conducted using one-way ANOVA and post hoc (Duncan) test at a level of p < 0.05.
Bacterial and archaeal T-RFLP profiles were generated from the raw data using GeneMarker (version 1.7; Softgenetics, USA). Fragment analysis was performed between 50 and 500 base pair (bp), which was within the line range of the internal size standard (LIZ 500, Applied Biosystems). A baseline threshold of 50 fluorescence units was used to minimize the effect of artifacts. To account for T-RF drift, fragments that differed by less than 0.5 bp were aligned as the same peak, i.e., binning (Dunbar et al. 2001). The relative abundance of a T-RF in a profile was calculated as a proportion of the total peak height of all the T-RFs in a profile. Any peak that was <0.5 % of the total fluorescence unit was removed from the data before statistical analysis.
Euclidian similarity matrices were generated on relative abundance T-RFLP data that had been log transformed prior to principal components analysis (PCA). PCA scores were plotted in two dimensions: principal components 1 (PC1) and PC2.
Results in the tables, text, and figures are given as means ± SD.
Results
Potential N mineralization and nitrification
The amount of N mineralized (NH4 +, NO2 − ,and NO3 −)-N and nitrification during the incubation period after addition of organic fertilizer to the soil with or without DMPP are reported in Fig. 1. DMPP had no significant effect on total N mineralization throughout the course of the experiment (Fig. 1b), but it reduced nitrification starting from the second week of incubation: at 14-days nitrification was reduced by 86.4 %, and at 28 days by 77.4 % in ED treatment with respect to E (p < 0.05) (Fig. 1a). After 8 and 12 weeks, net nitrification from each treatment was not significantly different and the inhibition effect declined. After 12 weeks of incubation, 155.04 ± 15.35 and 138.18 ± 10.78 mg N kg−1 dry soil were mineralized (E and ED, respectively) (Fig. 1b). No differences could be detected between C and D treatments and the amount both of N mineralized and nitrification was consistently lower than amended treatments (data not shown).
Weekly net nitrification (a) and cumulative net N mineralization (b) as milligram of nitrites and nitrates or total mineral N leached per kilogram dry soil. Significant effects of the inhibitor are indicated by asterisks, Duncan’s test, p < 0.05. Treatments: cattle effluent as organic fertilizer (E) combined or not with DMPP (ED)
Abundance of N-cycle organism genes
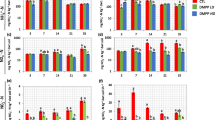
Figure 2 shows the changes in the bacterial 16S rRNA, bacterial amoA, archaeal amoA, nirK, nirS, and nosZ gene copy numbers quantified using qPCR assays. The bacterial 16S rRNA gene copy numbers during the first 7 days of incubation range from 4.19 × 108 for D at day 1 to 2.23 × 109 g−1 soil for E at day 7 (Fig. 2a). These differences were not significant across sampling times (p < 0.05); however, bacterial 16S rRNA copy numbers in soil were slightly higher in microcosms amended with cattle effluent.
Quantification of a bacterial 16S, b bacterial amoA, c archaeal amoA d nosZ, e nirK, and f nirS gene copies in untreated soil (C) and in soils treated with DMPP (D), cattle effluent (E), and cattle effluent + DMPP (ED) after 1, 4, and 7 days of microcosms incubation. Comparisons of means were conducted using one-way ANOVA and Duncan test at a level of p < 0.05
For both untreated soils and treatments, there was no significant change in bacterial or archaeal amoA gene copy numbers throughout the course of the experiment, which varied from 2.38 × 105 for C at day 1 to 1.75 × 106 g−1 soil for ED at day 1, and from 1.71 × 105 for ED at day 7 to 5.74 × 105 g−1 soil for E at day 1, respectively (Fig. 2b–c). Bacterial and archaeal amoA gene copy numbers did not differ (p < 0.05) among treatments or as a function of sampling time, and their abundances were generally comparable.
The relative abundance of nirK, nirS, and nosZ denitrification genes showed a similar pattern: nosZ gene copies, for example, varied from 3.17 × 106 (D, day 7) to 4.05 × 107 g−1 soil (E, day 7) (Fig. 2d). At days 4 and 7 DMPP significantly reduced nosZ gene copy numbers (p < 0.05). As for bacterial 16S rRNA gene, nosZ gene quantification revealed increasing copy number in both microcosms containing organic fertilizer (Fig. 2d). Furthermore, we found the following gene copy numbers concentrations per gram bovine effluent: 1.21 × 109 for bacterial 16S, 4.76 × 105 for bacterial amoA, 1.90 × 106 for nirK, 1.012 × 106 for nirS, and 2.52 × 106 for nosZ. Archaeal amoA gene copy numbers were below limit of detection.
Transcriptional activity of N-cycle organism genes
Figure 3 shows changes in the bacterial 16S rRNA, bacterial and archaeal amoA, and nosZ mRNA levels quantified using RT-qPCR assays. In contrast to gene copy number, gene expression showed significant differences in mRNA levels for all the genes detected at 1, 4, and 7 days of microcosms incubation. Bacterial amoA mRNA levels were lower in DMPP + cattle effluent treatment in comparison to cattle effluent-only treatment starting from day 1 until 7 (p < 0.05), indicating a clear inhibitory effect of DMPP on transcriptional activity (Fig. 3b). Increasing levels of AOA amoA mRNA were measured in E which ranged from 1.05 × 103 at day 1 to 1.84 × 104 g−1 soil at day 7 (Fig. 3c). A trend towards lower archaeal amoA mRNA levels in ED when compared to E at each time point was observed, although it was only significant (p < 0.05) at day 7.
Quantification of a bacterial 16S, b bacterial amoA, c archaeal amoA, and d nosZ mRNA levels in untreated soil (C) and in soils treated with DMPP (D), cattle effluent (E), and cattle effluent + DMPP (ED) after 1, 4, and 7 days of microcosms incubation. Comparisons of means were conducted using one-way ANOVA and Duncan test at a level of p < 0.05
Inhibition of denitrifiers (nosZ mRNA levels) was also found: an approximately 10-fold decrease in nosZ gene transcripts relative to ED when compared to E soils was observed at day 1, and about a 5-fold decrease in numbers at day 7 (Fig. 3d), but no significant decrease at day 4 could be detected.
However, lower 16S rRNA levels were found in ED treatment in comparison to the pure E treatment; these were significant after 24 h of application (Fig. 3a).
Structural changes of metabolically active microbial communities
The structural differentiation of metabolically active microbial communities during the first 7 days of microcosm incubation was analyzed by RT-T-RFLP. Figure 4 shows the number of bacterial (Fig. 4a) and archaeal (Fig. 4b) T-RFs, respectively, found in soil microcosms after 1, 4, and 7 days of incubation. Bacterial T-RFs were more impacted by treatments when compared to their archaeal counterparts: at day 1 bacterial T-RFs in only DMPP microcosms were significantly lower than the other treatments, and after 4 and 7 days, a significant difference between E and ED microcosms was detected. In line with the greater level of stability of the archaeal community, the number of archaeal 16S T-RFs fluctuated less than those of bacteria throughout the present experiment, and no statistical difference could be detected.
PCA was done on species abundance data: the analysis compared two sets of descriptors, T-RFs versus treatments and reduced the multidimensional relationship between them to two principal components. In all cases on the first principal component loadings (PC1) a strong laddering occurred (data not shown), and this was due to the fact that a major fraction of T-RFs were not affected by any of the microcosm treatments; furthermore, most of the time PC1 explained a greater percentage of variance, ranging from 44 to 50 % and from 75 to 96 % for bacterial and archaeal population, respectively (Fig. 5). However, significant differences in metabolically active microbial community structure were evident across treatments. At day 1, the active bacterial community structure was more similar in the two microcosms where cattle effluent was applied, regardless of the presence of the nitrification inhibitor. The first two components explained 66 % of the total variance; microcosms with DMPP were negatively affected by PC2, and C and D microcosms were negatively affected by PC3 (which explained 17 % of the total variance) whereas PC4, which explained 12 % of the total variance, was able to discriminate E (positively) and ED (negatively) treatments (data not shown). Archaeal community structure after 1 day of incubation was relatively stable: C, D, and ED microcosms didn’t show any differences; only cattle effluent microcosms were affected differently from the others by PC2, although it explained only 20 % of the total variance observed.
Principal component analysis of metabolically active soil bacterial (a, b, c) and archaeal (d, e, f) communities based on RT-T-RFLP analysis of 16S rRNA gene sequences after 1, 4, and 7 days of microcosms incubation. Treatments: black circle untreated soil (C); white circle soil + DMPP (D); black triangle soil + cattle effluent (E); white triangle soil + cattle effluent + DMPP (ED)
After 4 days of incubation, metabolically active bacterial community structure of E and ED was affected differently from their controls without cattle effluent and the inhibitor, and differently from each other on PC2. Both at day 1 and 4, the archaeal population was stable across treatments, except for E treatments at day 4 which were slightly separated by PC2 explaining only 3 % of the total variance.
After 7 days of microcosm incubation, PCA on archaeal community structure discriminated in a similar way to that seen in day 4, ED being more separated from C and D when compared to day 4.
Discussion
Mineralization of organic N and the concentration of ammonium in soil have a direct effect on nitrification and plant-available N. Most of the applied N-fertilizer to soil in the form of NH4 + is oxidized to NO3 − by ammonia-oxidizing microorganisms (Subbarao et al. 2006). However, the application of DMPP delays this oxidation over time resulting in limited nitrate availability and consequently, reducing the risk of losses by leaching. In our incubation study at constant soil temperature, DMPP strongly inhibited nitrification at 14 and 28 days after the application of cattle effluent as organic fertilizer. From day 56 until the end of the incubation period, the inhibitory effect of DMPP declined, suggesting that DMPP has undergone biological decomposition since it is non-volatile (Zerulla et al. 2001). The effectiveness of DMPP as nitrification inhibitor in soil has been widely demonstrated including when added to slurry (Dittert et al. 2001; Macadam et al. 2003; Hatch et al. 2005; Merino et al. 2005) and animal urine (Di and Cameron 2004; 2012); however, these studies were all conducted at temperatures lower than 30 °C. Zerulla et al. (2001) reported that nitrification for ammonium sulfate nitrate at 20 °C was completed within 7–21 days, compared to 40 days with DMPP. In Mediterranean regions where temperatures can be significantly higher than those registered in Central and Northern Europe, it is crucial to assess whether and to what extent warm temperatures may affect DMPP performance as a nitrification inhibitor, in order to guarantee an adequate N supply to crop plants throughout the entire vegetative cycle (Grisi et al. 1998).
Furthermore, we found no significant differences in total mineral N leached between the two treatments (Fig. 1b), hence the reduction in nitrate concentration found in the presence of DMPP can be attributed to inhibition of nitrification rather than stimulation of denitrification.
The number of ammonia oxidizers in certain environments is relatively low (about 104 to 106 cells gram per soil), making real-time PCR, which is highly sensitive, as a convenient method for enumeration of these microorganisms. Real-time PCR can also be used to determine a broad range of starting molecule template concentrations spanning at least five orders of magnitude (Heid et al. 1996). Thus, it is also suitable for quantifying soil microorganisms with population dynamics radically different than those of ammonia oxidizers. However, molecular fingerprinting techniques target the most dominant microbial community members (Muyzer and Smalla 1998) and can be biased due to the formation of heteroduplexes and PCR selection and drift (Suzuki and Giovannoni 1996; Head et al. 1998). Therefore, it might be possible that the contribution of less abundant species would not be reproducibly detected using this approach. Hence, both archaeal and bacterial community structures were profiled by T-RFLP of 16S rRNA (rather than DNA) to increase sensitivity and potentially target the predominantly active members of the microbial community (Felske and Akkermans 1998). Great sensitivity will arise through the high copy number of 16S rRNA per cell and growth and activity may be detected through increase in ribosome content prior to cell division.
While no significant changes in soil mineral N concentrations (Fig. 1a) or amoA gene copies (Fig. 2b–c) could be detected between the two fertilizer treatments after 7 days of incubation, short-term changes in transcriptional activity revealed that DMPP impaired both bacterial and archaeal amoA mRNA (Fig. 3b–c), being significant at every time point for AOB and at only one time point for AOA. Recent evidence suggests that archaea may play a significant role in ammonium oxidation in soil (Leininger et al. 2006; Prosser and Nicol 2008). Schauss et al. (2009) showed AOA were potentially responsible for up to half of nitrification activity in manured soil. Other quantitative studies (Francis et al. 2005; Hai et al. 2009) revealed that the AOA are more stable and less responsive to environmental differences than AOB; the high numbers in various ecosystems and at greater soil depths indicate that AOA are adapted to a broad range of growth conditions and might therefore have a more versatile metabolism than AOB, perhaps being able to grow mixotrophically. In the effluent archaeal amoA gene copy numbers were below the limit of detection, whereas bacterial amoA gene copy numbers were found at a concentration of 4.76 × 105 g−1 bovine effluent. Therefore, although soil archaeal amoA transcription was reduced following the addiction of DMPP, it is not possible to speculate how any archaea in the effluent were affected, nor if DMPP differentially affected organisms in the original soil and in the effluent itself. However, since DMPP significantly inhibited soil archaeal amoA transcription, some soil bacterial amoA transcription might have been affected. Although several studies have identified archaeal amoA genes in soils, and transcriptional activity has been demonstrated (Treusch et al. 2005; Leininger et al. 2006; Nicol et al. 2008; Mertens et al. 2009), the actual role of AOA in N-cycling remains unclear and may vary between soil types and conditions. Recent studies (O’Callaghan et al. 2010; Kleineidam et al. 2011) revealed a pronounced impact of nitrification inhibitory compounds on AOB but not AOA gene copy numbers, and that a qPCR analysis of amoA gene transcripts is needed to confirm their role in N-cycling. However, in this study, the efficacy of a nitrification inhibitor at the transcriptional level has been demonstrated. In particular, our findings revealed that, despite the different cellular biochemistry and metabolism existing between the domains Bacteria and Archaea, DMPP exerts its inhibitory effect against both bacterial and archaeal ammonia-oxidizing transcriptional activity. However, even though transcriptional activity is a measure of ammonia monooxygenase (AMO) production potential in soil, changes in mRNA amoA abundance may not directly reflect protein production and activity, since AMO seems to be strongly protected from degradation. Pinck et al. (2001) found that cells starved of ammonia for 1 year still contained high amounts of AMO, although they showed far less ammonia oxidation activity than growing cells. Hence, the amount of AMO may not directly correlate with the activity of ammonia oxidation.
It is known that N fertilization can change the structure and activity of denitrifying community, and subsequently, affect the N2O emission (Dambreville et al. 2006; Kramer et al. 2006). The ratio N2O/N2 rises if NO3 − is abundant in the soil because the latter is preferred to N2O as an electron acceptor (Schlegel 1992). Hence, if nitrate production is inhibited by DMPP, the denitrification pathway will be affected when the inhibitor is added. In our study, denitrification genes nirK, nirS, and nosZ, showed a similar DNA-based qPCR pattern over time at least for two sampling time points. Because of its importance as the final step in denitrification, nosZ was chosen to determine the effect of DMPP on transcription: transcript abundance decreased significantly in microcosms with DMPP and cattle effluent when compared to effluent-only treatment after 1 and 7 days but not after 4 days of incubation (Fig. 3d). The same trend was also found in bacterial 16S rRNA transcripts, suggesting a short-term impact of DMPP on the non-target bacterial community.
Animal effluents contain consistent amounts of microorganisms, and both Nitrosospira and Nitrosomonas-like 16S rDNA have been detected in swine manure (Ceccherini et al. 1998). It is known that microorganisms enact transcriptional changes in response to shifts in resource availability (Brandt et al. 2004), so that the fluctuations in viable populations following the addition of cattle effluent as nutrient sources were probably reflected in transcriptional activity. No significant difference could be detected between days 0 and 1 (data not shown); however, the slight differences between treatments observed in bacterial 16S and denitrifiers qPCR data could be explained both by the introduction of specific groups at high abundance in the effluent and by rapid initial growth, due also to the addition of labile carbon.
These results were reflected in active soil microbial community structure data, where relatively small shifts, both in bacterial and archaeal active communities, were detected, even though the number of archaeal 16S T-RFs fluctuated less than those of bacteria throughout the experiment (Fig. 4), confirming that there is minimal metabolic response of archaeal community structure to treatments. O’Callaghan et al. (2010) found significant short-term shifts in active soil bacterial community structure following application of urine and urine + nitrification inhibitor DCD, as revealed by RT-DGGE; however, by day 56, general bacterial communities were similar in all soil treatments and untreated soils.
Taken together, our results indicate clearly that the nitrification inhibitor DMPP exerts its effect on ammonia oxidizers’ transcriptional activity in our experimental conditions. However, our results revealed a moderate short-term effect on denitrifiers and general bacterial mRNA levels both from soil and cattle effluent. The mode of DMPP action is unclear and it is possible that in addition to inhibiting transcription of certain bacterial and archaeal genes including amoA, the compound acts as a readily available C and N substrate for other groups. For future investigations, it would be of interest to monitor amoA mRNA changes in active soil microbial community structure over longer incubation periods and in field experiments in order to see whether or not the diversity found between treatments will be maintained.
References
Barth G, von Tucher S, Schmidhalter U (2008) Effectiveness of 3,4-dimethylpyrazole phosphate as nitrification inhibitor in soil as influenced by inhibitor concentration, application form, and soil matric potential. Pedosphere 18:378–385
Benedetti A, Alianiello F, Dell’Abate MT (1994) A modified Stanford and Smith method for the study of the mineralization of nitrogen from organic materials. In: Neetson JJ, Hassink J (eds) Nitrogen mineralization in agricultural soils. AB-DLO, Haren
Bock E, Wagner M (2006) Oxidation of inorganic nitrogen compounds as an energy source. In: Dworkin M (ed) The Prokaryotes. Springer, New York, pp 457–495
Brandt BW, Kelpin FDL, van Leeuwen IMM, Kooijman SALM (2004) Modelling microbial adaptation to changing availability of substrates. Water Res 38:1003–1013
Bronson KF, Mosier AR (1994) Suppression of methane oxidation in aerobic soil by nitrogen fertilizers, nitrification inhibitors, and urease inhibitors. Biol Fertil Soils 17:263–268
Ceccherini MT, Castaldini M, Piovanelli C, Hastings RC, McCarthy AJ (1998) Effects of swine manure fertilization on autotrophic ammonia-oxidizing bacteria in soil. Appl Soil Ecol 7:149–157
Chen D, Helen CS, Islam A, Edis R (2010) Influence of nitrification inhibitors on nitrification and nitrous oxide (N2O) emission from a clay loam soil fertilized with urea. Soil Biol Biochem 42:660–664
Clark IM, Buchkina N, Jhurreea D, Goulding KWT, Hirsch PR (2012) Impacts of nitrogen application rates on the activity and diversity of denitrifying bacteria in the Broadbalk wheat experiment. Phil Trans R Soc B 367:1235–1244
Dambreville C, Hallet S, Nguyen C, Morvan T, Germon JC, Philippot L (2006) Structure and activity of the denitrifying community in a maize-cropped field fertilized with composted pig manure or ammonium nitrate. FEMS Microbiol Ecol 56:119–131
Dell’Abate MT, Benedetti A, Trinchera A, Galluzzo D (2003) Nitrogen and carbon mineralisation of leather meal in soil as affected by particle size of fertiliser and microbiological activity of soil. Biol Fertil Soils 37:124–129
Di HJ, Cameron KC (2004) Effects of temperature and application rate of a nitrification inhibitor, dicyandiamide (DCD), on nitrification rate and microbial biomass in a grazed pasture soil. Aust J Soil Res 42:927–932
Di HJ, Cameron KC (2012) How does the application of different nitrification inhibitors affect nitrous oxide emissions and nitrate leaching from cow urine in grazed pastures? Soil Use Manag 28:54–61
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2010) Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72:386–394
Dittert K, Bol R, King B, Chadwick D, Hatch D (2001) Use of a novel nitrification inhibitor to reduce nitrous oxide emission from 15 N-labelled dairy slurry injected into soil. Rapid Commun Mass Spectrom 15:1291–1296
Dosch P, Gutser R (1996) Reducing N losses (NH3, N2O, N2) and immobilization from slurry through optimized application techniques. Fert Res 43:165–171
Dunbar J, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and a new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67:190–197
Felske A, Akkermans ADL (1998) Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soil. Microb Ecol 36:31–36
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Grisi B, Grace C, Brookes PC, Benedetti A, Dell’Abate MT (1998) Temperature effect on organic matter and microbial biomass dynamics in temperate and tropical soils. Soil Biol Biochem 30:1309–1315
Hai B, Diallo NH, Sall S, Haesler F, Schauss K (2009) Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol 75:4993–5000
Hatch D, Trindade H, Cardenas L, Carneiro J, Hawkins J, Scholefield D, Chadwick D (2005) Laboratory study of the effects of two nitrification inhibitors on greenhouse gas emissions from slurry-treated arable soil: impact of diurnal temperature cycle. Biol Fertil Soils 41:225–232
Hauben L, Vauterin L, Swings J, Moore ERB (1997) Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int J Sys Bacteriol 47:328–335
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol 35:1–21
Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994
Henry S, Baudoinb E, López-Gutiérrez JC, Fabrice M-L, Braumanb A, Philippot L (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods 59:327–335
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72:5181–5189
Irigoyen I, Muro J, Azpilikueta M, Aparicio-Tejo P, Lamsfus C (2003) Ammonium oxidation kinetics in the presence of nitrification inhibitors DCD and DMPP at various temperatures. Aust J Soil Res 41:1177–1183
Isermayer H (1952) Estimation of soil respiration in closed jars. In: Alef K, Nannipieri P (eds) Method in applied soil microbiology and biochemistry. Academy, London, pp 214–216
Jarvis SC, Stockdale EA, Shepherd MA, Powlson DS (1996) Nitrogen mineralization in temperate agricultural soils: processes and measurement. Adv Agron 57:187–235
Jurgens G, Lindstrom K, Saano A (1997) Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol 63:3233–3241
Kamshake LJ, Hannah SA, Comen JM (1967) Automated analysis for nitrate by hydrazine reduction. Water Resour 1:205–216
Kleineidam K, Košmrlj K, Kublik S, Palmer I, Pfab H, Ruser R, Fiedler S, Schloter M (2011) Influence of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) on ammonia-oxidizing bacteria and archaea in rhizosphere and bulk soil. Chemosphere 84:182–186
Kramer SB, Reganold JP, Glover JD, Bohannan BJ, Mooney HA (2006) Reduced nitrate leaching and enhanced denitrifier activity and efficiency in organically fertilized soils. Proc Natl Acad Sci U S A 103:4522–4527
Leininger S, Urich T, Sclotter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soil. Nature 442:806–809
Li H, Liang X, Chen Y, Lian Y, Tian G, Ni W (2008) Effect of nitrification inhibitor DMPP on nitrogen leaching, nitrifying organisms, and enzyme activities in a rice-oilseed rape cropping system. J Environ Sci 20:149–155
Linzmeier W, Gutser R, Schmidhalter U (2001) Nitrous oxide emission from soil and from a nitrogen-15-labelled fertilizer with the new nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP). Biol Fertil Soils 34:103–108
Macadam XMB, del Prado A, Merino P, Estavillo JM, Pinto M, Gonzales-Murua C (2003) Dicyandiamide and 3,4-dimethyl pyrazole phosphate decrease N2O emissions from grassland but dicyandiamide produces deleterious effects on clover. J Plant Physiol 160:1517–1523
Macdonald CA, Singh BK, Peck JA, van Schaik AP, Hunter LC, Horswell J, Campbell CD, Speir TW (2007) Long-term exposure to Zn-spiked sewage sludge alters soil community structure. Soil Biol Biochem 39:2576–2586
Macdonald CA, Clark IM, Zhaoa FG, Hirsch PR, Singh BK, McGrath SP (2011) Long-term impacts of zinc and copper enriched sewage sludge additions on bacterial, archaeal and fungal communities in arable and grassland soils. Soil Biol Biochem 43:932–941
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium specific PCR primers that amplify genes coding for bacterial 16SrRNA. Appl Environ Microbiol 64:795–799
Martínez-Alcántara B, Quiñones A, Polo C, Primo-Millo E, Legaz F (2012) Use of nitrification inhibitor DMPP to improve nitrogen uptake efficiency in citrus trees. J Agric Sci 5:1–18
Menéndez S, Barrena I, Setien I, González-Murua C, Estavillo JM (2012) Efficiency of nitrification inhibitor DMPP to reduce nitrous oxide emissions under different temperature and moisture conditions. Soil Biol Biochem 53:82–89
Merino P, Menéndez S, Pinto M, Gonzàlez-Murua C, Estavillo JM (2005) 3,4-dimethyl pyrazole phosphate reduces nitrous oxide emissions from grassland after slurry application. Soil Use Manag 21:53–57
Mertens J, Broos K, Wakelin SA, Kowalchuk GA, Springael D, Smolders E (2009) Bacteria, not archaea, restore nitrification in a zinc-contaminated soil. ISME J 3:916–923
Michotey V, Mejean V, Bonin P (2000) Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl Environ Microbiol 66:1564–1571
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
O’Callaghan M, Gerard EM, Carter PE, Lardner R, Sarathchandra U, Burch G, Ghani A, Bell N (2010) Effect of the nitrification inhibitor dicyandiamide (DCD) on microbial communities in a pasture soil amended with bovine urine. Soil Biol Biochem 42:1425–1436
Pasda G, Hähndel R, Zerulla W (2001) Effect of fertilizers with the new nitrification inhibitor DMPP (3,4-dimethylpyrazole phosphate) on yield and quality of agricultural and horticultural crops. Biol Fertil Soils 34:85–97
Pereira J, Fanguiero D, Chadwick DR, Misselbrook TH, Coutinho J, Trindade H (2010) Effect of cattle slurry pre-treatment by separation and addition of nitrification inhibitors on gaseous emissions and N dynamics: a laboratory study. Chemosphere 79:620–627
Pinck C, Coeur C, Potier P, Bock E (2001) Polyclonal antibodies recognizing the AmoB protein of ammonia oxidizers of the beta-subclass of the class Proteobacteria. Appl Environ Microbiol 67:118–124
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption–free analysis of quantitative real-time polymerse chain reaction (PCR) data. Neurosci Lett 339:62–66
Raskin L, Stromley JM, Rittmann BE, Stahl DA (1994) Group-specific 16S RNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60:1232–1240
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker Q, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:1–12
Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, Sharma S, Wilke BM, Matthies M, Smalla K, Munch JC, Amelung W, Kaupenjohann M, Schloter M, Schleper C (2009) Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11:446–456
Schlegel HG (1992) Allgemeine mikrobiologie, 7th edn. Thieme Verlag, Stuttgart
Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C (2010) Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18:331–340
Springer U, Klee J (1954) Pr_fung der Leistungsf_higkeit von einigen wichtigeren Verfahren zur Bestimmung des Kohlemstoffs mittels Chromschwefels_ure sowie Vorschlag einer neuen Schnellmethode. Z Pflanzenernaehr Dueng Bodenkd 64:1–7
Stanford G, Smith SJ (1972) Nitrogen mineralization potentials of soils. Soil Sci Soc Am Proc 36:465–472
Subbarao GV, Ito O, Sahrawat K, Berry WL, Nakahara K, Ishikawa T, Watanabe T, Suenaga K, Rondon M, Rao IM (2006) Scope and strategies for regulation of nitrification in agricultural systems-challenges and opportunities. Crit Rev Plant Sci 25:303–335
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Throbäck IN, Enwall K, Jarvis Ǻ, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417
Töwe S, Kleineidam K, Schloter M (2010) Differences in amplification efficiency of standard curves in quantitative real-time PCR assays and consequences for gene quantification in environmental samples. J Microbiol Methods 82:338–341
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C (2005) Novel genes for nitrite reductase and Amo related proteins indicate a role of uncultivated mesophilic Crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring microbial biomass C. Soil Biol Biochem 19:703–707
Violante P (2000) Metodi di Analisi Chimica del Suolo. Angeli, Milan
Wall L, Gehrke CW, Neuner JE, Lathey RD, Rexnord PR (1975) Cereal protein nitrogen: evolution and comparison of four different methods. Assoc Off Anal Chem 58:811–817
Weiske A, Benckiser G, Herbert T, Ottow J (2001) Influence of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) in comparison to dicyandiamide (DCD) on nitrous oxide emissions, carbon dioxide fluxes and methane oxidation during 3 years of repeated application in field experiments. Biol Fertil Soils 34:109–117
Wulf S, Maeting M, Clemens J (2002) Application technique and slurry co-fermentation effects on ammonia nitrous oxide, and methane emissions after spreading: II. Greenhouse gas emissions. J Environ Qual 31:1795–1801
Yang J, Li X, Xu L, Hu F, Li H, Liu M (2013) Influence of the nitrification inhibitor DMPP on the community composition of ammonia-oxidizing bacteria at microsites with increasing distance from the fertilizer zone. Biol Fertil Soils 49:23–30
Zerulla W, Barth T, Dressel J, Erhardt K, von Locquenghien KH, Pasda G, Rädle M, Wissemeier AH (2001) 3,4-Dimethylpyrazole phosphate (DMPP)—a new nitrification inhibitor for agriculture and horticulture. Biol Fertil Soils 34:79–84
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. The ISME J 6:1032–1045
Acknowledgments
A.F. is funded by a Ph.D. fellowship from EuroChem Agro Spa, Italy, and Timac Agro Italia. The authors would like to thank Dr. Giovanni Mughini and the CRA-PLF (Rome) for providing soil samples. The authors have no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Florio, A., Clark, I.M., Hirsch, P.R. et al. Effects of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) on abundance and activity of ammonia oxidizers in soil. Biol Fertil Soils 50, 795–807 (2014). https://doi.org/10.1007/s00374-014-0897-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-014-0897-8