Abstract
A study was undertaken to determine the free-living culturable diazotrophic bacteria of paddy soils from a long-term fertilizer management experiment. Long-term application of different fertilizers significantly affected the population of free-living diazotrophs. Out of 165 distinct bacterial morphotypes observed during the isolation process, only 32 were positive for both acetylene reduction assay (ARA), and nifH gene screening. The ARA activity of the isolates ranged from 1.8 to 2,844.7 nmol ethylene h−1 mg protein−1. The 16S rRNA analysis identified the isolates to be members of 13 different genera viz. Bacillus, Pseudomonas, Paenibacillus, Serratia, Ochrobactrum, Lysinibacillus, Burkholderia, Brevundimonas, Herbaspirillum, Novosphingobium, Sphingomonas, Xanthomonas, and Azorhizobium. Though partial nifH gene sequencing of diazotrophic isolates showed good consistency with that of 16S rRNA-based identification, some nifH sequences were similar to a variety of uncultured nitrogen-fixing bacteria. The diversity of free-living diazotrophic bacteria and the wide distribution of nifH sequences indicate the potential contribution of these microorganisms to nitrogen input to paddy fields.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Because of its potential economic and environmental importance, biological nitrogen fixation (BNF) is an important source of nitrogen (N) input to soil and has attracted many researchers (Demba Diallo et al. 2004; Wartiainen et al. 2008). In terrestrial ecosystems, the estimated input of BNF averages 90–130 Tg/year (Kennedy and Islam 2001) and these N inputs are important for arable soils, such as rice paddy soil (Ladha and Reddy 2003).
The N2 fixation requires the interaction of several gene products including the nitrogenase structural proteins like NifD, NifK, and nifH. The phylogeny based on nifH genes has been shown to resemble the 16S rRNA phylogeny (Zehr et al. 2003); thus nifH is an ideal phylogenetic gene marker for investigating N2-fixing organisms in natural environments. In the past, nifH gene has been successfully used to determine diversity of the diazotrophic communities (Roesch et al. 2006; Albino et al. 2006; Wakelin et al. 2007).
Paddy soils are habitats for numerous diazotrophs, which contribute significantly to soil fertility (Hashem 2001; Kennedy et al. 2004; Ariosa et al. 2005) and N requirement (Roger 1995; Ladha and Reddy 2003). Physico-chemical factors and the management practices can influence competition in soil bacterial population (Bossio et al. 1998; Ulrich and Becker 2006). Few studies have examined the influence of the fertilizer treatment on the diversity and stability of diazotrophic populations (Yevdokimov et al. 2008; Islam et al. 2009). The composition of diazotrophs was markedly influenced by inorganic N content across a range of soil types (Poly et al. 2001). However, the colony unit forming (CFU) ability of Gluconacetobacter diazotrophicus was not affected by high levels of N-fertilizations (Roesch et al. 2006). Though long-term application of chemical fertilizer amended with compost increased the average yield of rice more than chemical fertilizer applied alone (Muthukumarasamy et al. 2007), no general agreement exists on the effects of long-term inorganic or organic N-fertilizer additions on eubacterial and diazotrophic diversity of soil. Hence, identification of the major taxonomic groups of bacteria that contribute to the N input in rice ecosystem is of great relevance. Accordingly, the present study reports the isolation and characterization of the putative free-living diazotrophic bacteria from long-term fertilized paddy soil.
Materials and methods
Sampling area
Soil samples were collected from a Gangseo series (coarse loamy, mixed, nonacid, mesic family of Aquic Fluventic Eutrochrepts) paddy soils from the National Institute of Agricultural Science and Technology located at Suwon city (37°16′0″N, 127°1′0″E) under Gyeonggi Province of the Republic of Korea. This region has an average annual precipitation of 1,268.1 mm, annual evaporation of 1,102.0 mm, and a mean annual temperature of 11.6°C. The research fields were established in 1954 to evaluate the long-term effect of different fertilizer amendments on the yield of lowland rice. Rice straw was used as organic compost, prepared by fermenting straw for 5 months. Organic compost with and without nitrogen–phosphorus–potassium (NPK) fertilizer was applied to soil. Since 1986, chemical fertilizers were applied at rates of 110 kg N ha−1, 70 kg P2O5 ha−1, and 80 Kg K2O ha−1. Compost was added at 7.5, 15.0, 22.5, and 30.0 Mg ha−1 in CNPK, NPKC750, NPKC1500, NPKC2250, and NPKC3000 treatments, respectively. While CNPK received ammonium sulfate as N source, all the other treatments received urea as N source. Control treatment received neither chemical fertilizer nor compost amendments.
Soil sampling and sample preparation
To sample each soil type, three 1,000 m2 plots were randomly established. In each plot, nine soil samples were collected using 10-cm long × 1.45-cm diameter soil corer at nine randomly selected points in October 2007. All samples from each plot were combined to form one composite sample and stored in a sterile polypropylene bag in coolers immediately after sampling. After removing visible root debris, field moist soil samples were sieved (2 mm) and stored at 4°C. The physico-chemical properties of soils are presented in Table 1.
Enrichment isolation and morphological characterization
Diazotrophic microorganisms were isolated using serial dilution technique on four selective N-free media viz., NFMM, LGI-P, BAz, and JNFb. NFMM medium had the following composition 10 g L−1 sucrose, 5 g L−1 malic acid, 0.1 g L−1 K2HPO4, 0.4 g L−1 KH2PO4, 0.2 g L−1 MgSO4∙7H2O, 0.1 g L−1 NaCl, 0.2 g L−1 CaCl2∙H2O, 0.01 g L−1 FeCl3, 0.002 g L−1 Na2MoO4∙2H2O, pH 7.2 (Piao et al. 2005). The LGI-P medium contained 100 g L−1 sucrose, 0.2 g L−1 K2HPO4, 0.6 g L−1 KH2PO4, 0.2 g L−1 MgSO4∙7H2O, 0.02 g L−1 CaCl2, 0.002 g L−1 Na2MoO4∙2H2O, 0.01 g L−1 FeCl3∙6H2O, 5 ml of bromothymol (0.5% solution in 0.2 M KOH), pH 5.5 (Reis et al. 1994). The BAz medium contained 2.0 g L−1 azelaic acid, 0.4 g L−1 K2HPO4, 0.4 g L−1 KH2PO4, 0.2 g L−1 MgSO4∙7H2O, 0.02 g L−1 CaCl2, 0.002 g L−1 Na2MoO4∙H2O, 0.01 g L−1 FeCl3, 0.075 g L−1 of bromothymol blue, pH 5.7 (Estrada-De Los Santos et al. 2001). JNFb contained 5 g L−1 malic acid, 0.6 g L−1 K2HPO4, 1.8 g L−1 KH2PO4, 0.2 g L−1 MgSO4∙7H2O, 0.1 g L−1 NaCl, 0.02 g L−1 CaCl2, 4.5 g L−1 KOH, 12 ml of bromothymol blue (0.5% in 0.2 N KOH), 1 ml of vitamin solution, 2 ml of micronutrient solution, 4 ml of Fe EDTA solution (1.64% w/v), pH 5.8. A 100 ml of vitamin solution, 10 mg biotin, and 20 mg pyridoxal-HCl. Micronutrients solution had 0.4 g L−1 CuSO4, 0.12 g L−1 ZnSO4∙7H2O, 1.4 g L−1 H2BO3, 1 g L−1 Na2MoO4∙2H2O, 1.5 g L−1 MnSO4∙H2O (Kirchhof et al. 1997).
Aliquots (0.1 ml) from the serially diluted samples (10−3 to 10−6) were added to four different N-free media in Petri plates and kept in an incubator at 30°C. Five days after incubation, colonies growing on N-free media were counted and grouped according to their morphological characteristics. Single colonies were picked from the Petri dishes and sub-cultured several times to obtain pure cultures. Stock cultures were made in nutrient broth containing 50% (w/v) glycerol and stored at –80°C.
Nitrogen fixation
Nitrogen fixing of the bacterial isolates was determined by acetylene reduction assay (ARA) using Gas chromatograph (DS 6200, Donam Instruments Inc., Republic of Korea) fitted with flame ionization detector and a Porapak-Q column (Park et al. 2005). All the experiments were carried out in semi-solid JNFb medium. Uninoculated media served as control. The protein concentration was determined using bovine serum albumin as standard (Lowry et al. 1951).
PCR amplification, sequencing, and phylogeny of nifH gene
The presence of nifH gene was determined by amplifying the 390 bp fragment using a pair of specific primers, 19F (5′-GCIWTYTAYGGIAARGGIGG-3′) and 407R (5′-AAICCRCCRCAIACIACRTC-3′) (Ueda et al. 1995). The conditions of the polymerase chain reaction (PCR) were: 0.5 min at 94°C, 1 min at 50°C, and 0.5 min at 72°C with 40 cycles. The amplified products were resolved on a 1% agarose gel in 1×TBE buffer and visualized under UV light (Bio-Rad Laboratories, CA, USA). The purified PCR products were sequenced directly using an ABI 3730XL capillary DNA sequencer (50 cm capillary) with the same set of primers. The aligned sequence was compared using BLAST in GenBank to obtain closely related sequences. Phylogenetic tree was generated after performing multiple sequences alignment (CLUSTAL W version 1.8). The method of Jukes and Cantor (1969) was used to calculate evolutionary distances, phylogenetic dendrograms were constructed by neighbor-joining method, and the tree topologies were evaluated by bootstrap analysis of 1,000 dataset using MEGA version 3.10 software (Kumar et al. 2004).
16S rRNA gene amplification, sequencing, and phylogenetic analysis
The amplification of 16S rRNA gene of the bacterial genomic DNA was done using universal primers 27F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R 5′-GGTTACCTTGTTACGACTT-3′. The resultant product was sequenced directly using the fluorescent dye terminator method (ABI prism™ Bigdye™ Terminator cycle sequencing ready reaction kit V.3.1). The BLAST analysis and phylogenetic tree were constructed as described above.
Nucleotide sequence accession numbers
The nucleotide sequences of 16 S rRNA and nifH genes were deposited to GenBank under the accession numbers FJ266313–FJ266342, and FJ829453–FJ829468, respectively.
Statistical analysis
All the data were subjected to a variance analysis using SAS software (version 9.1; Cary, NC). When analysis of variance showed significant treatment effects, the Tukey’s test (P < 0.05) was applied to make comparisons between treatments.
Results
Determination of culturable diazotrophic bacteria and their purification
The diazotrophic bacterial counts of the visible colonies on four N-free media after 5 days of incubation are presented in Fig. 1. The highest diazotrophic population was observed in NPKC3000-treated soil inoculated in BAz medium (7.5 log CFU (g DW soil)−1) and the lowest (2.4 log CFU (g DW soil)−1) in JNFb medium.
Screening for nitrogenase activity and phylogeny of nifH gene
The samples varied significantly in N2-fixation rates. There were significant interactions between sample and media. The ARA ranged from 2.0 to 57.8 nmol ethylene h−1 mg protein−1 in the NFMM medium, from 1.8 to 5.2 nmol ethylene h−1 mg protein−1 in the LGI-P medium, from 23.2 to 511.2 nmol ethylene h−1 mg protein−1 in the BAz medium, and from 3.3 to 2,844.7 nmol ethylene h−1 mg protein−1 in the JNFb medium, respectively (Table 2). When the nitrogenase positive isolates were screened for the presence of nifH gene, only 32 were found positive. The maximum number of isolates were from the JNFb medium (40.63%), followed by BAz (28.13%), NFMM (21.88%), and LGI-P (9.38%) medium, respectively. When samples were compared, the highest numbers (28.13%) of nifH positive isolates were found in control plots. Some nitrogenase positive isolates were negative for the nifH gene amplification studies.
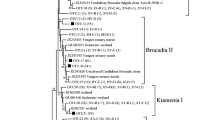
When the 32 positive nifH gene fragments were sequenced, only 16 sequences had the correct product size. The nucleotide sequences translated into amino acids and analyzed subsequently showed high diversity of NifH sequences among diazotrophic isolates (Fig. 2). The NifH sequences of Burkholderia sp. showed 100% homology to NifH of Burkholderia xenovorans retrieved from GenBank database. Similarly, Herbaspirillum sp. and Paenibacillus sp. showed 95–100% and 85–88% amino acid similarity to NifH database of Herbaspirillum seropedicae and Paenibacillus massiliensis, respectively. The sequences of Azorhizobium sp. showed 99% homology with Azorhizobium cauliodans. The NifH amino acid sequences from Sphingomonas sp., Novosphingobium sp., Methylobacterium sp., and Ochrobactrum sp. clustered with those of nitrogen-fixing uncultured bacteria forming a distant group with no close relation to the other sequences obtained in this study (Fig. 2).
Phylogenetic tree based on partial NifH amino acid sequences of diazotrophic isolates obtained from paddy soils under long-term fertilization treatments. The database accession numbers are indicated after the bacterial names. Bootstrap values (from 1,000 replicates) higher than 50% are shown at the nodes. The sequences obtained in this study are shown in bold
16S rRNA gene amplification, sequencing, and phylogenetic analysis
The 16S rRNA analysis revealed that the bacterial isolates of paddy soils belong mainly to Proteobacteria and the subgroup Alphaproteobacteria was dominant and most diverse, having four different families with nine isolates belonging to the genera Ochrobactrum, Brevundimonas, Novosphingobium, Sphingomonas, and Azhorhizobium (Table 2, Fig. 3). On the other hand, the Betaproteobacteria subgroup though was dominant, it had only two genera. The subgroup Gammaproteobacteria composed of three genera belonged to three families. Two Bacillus sp. (RFNB2 and RFNB6), two Paenibacillus sp. (RFNB4 and RFNB5), and one Lysinibacillus sp. (RFNB10) made the group Firmicutes. Overall, majority of the isolates (46.66%) belonged to Herbaspirillum, Serratia, and Burkholderia sp. (Fig. 3).
Phylogenetic tree of 16S rRNA gene sequences showing the relationships among the diazotrophic bacteria isolated from paddy soils and the related genera. Bootstrap values (from 1,000 replicates) higher than 50% are shown at the nodes. The sequences obtained in this study are shown in bold. The numbers in the parenthesis are the nucleotide sequence accession numbers in the GenBank. Scale bar presents 0.02 nucleotide substitutions
Discussion
Soil management improves soil quality through creating durable soil structure and suitable microhabitats for high N2-fixation activity (Chotte et al. 2002). In this study, different soils maintained under various fertilizer management practices were selected to find out the influence of inputs on the nifH gene pool and diazotrophic bacteria.
Irrespective of the media used, plate count analysis revealed that the numbers of culturable diazotrophic bacterial population were significantly higher in NPKC2250 amended soils than other treatments and control soil. At the same time, the NPK-treated soil recorded significantly lower diazotrophic populations in the used media with the exception of the JNFb medium then the control (Fig. 1). Our results are in agreement with an earlier report (Muthukumarasamy et al. 2007), where a higher diazotrophic population was observed in rhizosphere soils receiving both N and compost.
When a total of 165 diazotrophic bacterial isolates were screened for acetylene reduction, higher ARA was observed in bacterial isolates picked from BAz and JNFb media, indicating the carbon source was important for the growth of the selective microbes. Out of this, only 32 were found positive for the presence of nifH gene. Our results are in agreement with earlier studies (Kuklinsky-Sobral et al. 2004, Chowdhury et al. 2007) where nifH gene could not be amplified because of the variability of this gene (Zehr et al. 2003). It has been reported that no direct correlation between the presence of nifH and the ARA activity of the bacterial strains (Dean and Jacobson 1992). Among the four N-free media used, the maximum number of nifH positive isolates was obtained in JNFb used to select Azospirillum or Herbaspirillum species followed by BAz medium used for selecting Burkholderia. Various authors have reported the occurrence of these bacteria in soil (Perin et al. 2006; Soares et al. 2006). A higher number of nifH gene positive isolates was obtained from control plots than the chemical fertilizer and organic amended treatments.
Partial nifH gene sequencing showed good consistency with 16S rRNA identification and revealed high diversity of nifH gene among diazotrophs of paddy soils thus confirming the large nifH gene diversity among isolates isolated from sweet potato (Reiter et al. 2003) and rice (Knauth et al. 2005). As expected, nifH sequences from Burkholderia sp., Herbaspirillum sp., and Azorhizobium sp., were closely related with the type I nifH genes (Zehr et al. 2003), codifying molybdenum nitrogenases, and isolated from cyanobacteria and proteobacteria. The sequences from Sphingomonas sp., Novosphingobium sp., Methylobacterium sp., and Ochrobactrum sp. were moderately similar to a bacteriochlorophyllide reductase gene (bchL), a nifH homolog, and probably belonging to the so-called type IV nitrogenases, which have been described as a highly divergent and loosely coherent group of nif-like genes including sequences from the Archaea and homologous chlorophyllide reductase genes (Zehr et al. 2003).
The search for potential free-living diazotrophs and their diversity in nature is still a fascinating ongoing research and there is much work to be done to harness the whole potential of diverse diazotrophic bacterial communities in soil and their interaction with plants. The results of this study provide evidence for the presence of 32 distinct free-living diazotrophic bacteria, representing 12 different families, associated with paddy fields. The growth of different bacterial isolates on four different N-free media, nifH, and 16S rRNA phylogenetic analyses indicate that diazotrophic bacteria are diverse. The knowledge on the diversity of diazotrophic bacteria is required not only for understanding their ecological importance in the paddy soils, but also for their utilization in sustainable agricultural as inoculants of rice.
References
Albino U, Saridakis DP, Ferreira MC, Hungria M, Vinuesa P, Andrade G (2006) High diversity of diazotrophic bacteria associated with the carnivorous plant Drosera villosa var. villosa growing in oligotrophic habitats in Brazil. Plant Soil 287:199–207
Ariosa Y, Carrasco D, Leganés F, Quesada A, Fernández-Valiente E (2005) Development of cyanobacterial blooms in Valencian rice fields. Biol Fertil Soils 41:129–133
Bossio DA, Scow KM, Gunapala N, Graham KJ (1998) Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb Ecol 36:1–12
Chotte J, Schwartzmann A, Bally R, Monrozier LJ (2002) Changes in bacterial communities and Azospirillum diversity in soil fractions of a tropical soil under 3 or 19 years of natural fallow. Soil Biol Biochem 34:1083–1092
Chowdhury SP, Schmid M, Hartmann A, Tripathi AK (2007) Identification of diazotrophs in the culturable bacterial community associated with roots of Lasiurus sindicus, a perennial grass of Thar desert, India. Microb Ecol 54:82–90
Dean DR, Jacobson MR (1992) Biochemical genetics of nitrogenase. In: Stacy G, Burris RH, Evans HJ (eds) Biological nitrogen fixation. Chapman and Hall, New York, pp 763–834
Demba Diallo M, Willems A, Vloemans N, Cousin S, Vandekerckhove TT, de Lajudie P, Neyra M, Vyverman W, Gillis M, Van der Gucht K (2004) Polymerase chain reaction denaturing gradient gel electrophoresis analysis of the N2-fixing bacterial diversity in soil under Acacia tortilis ssp. raddiana and Balanites aegyptiaca in the dryland part of Senegal. Environ Microbiol 6:400–415
Estrada-De Los Santos P, Bustillos-Cristales R, Caballero-Mellado J (2001) Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol 67:2790–2798
Hashem A (2001) Problems and prospects of cyanobacterial biofertilizer for rice cultivation. Aust J Plant Physiol 28:881–888
Islam MR, Trivedi P, Palaniappan P, Reddy MS, Sa TM (2009) Evaluating the effect of fertilizer application on soil microbial community structure in rice based cropping system using Fatty acid methyl esters (FAME) analysis. World J Microbiol Biotechnol 25:1115–1117
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kennedy IR, Islam N (2001) The current and potential contribution of a symbiotic nitrogen fixation to nitrogen requirements on farms: a review. Aust J Exp Agr 41:447–457
Kennedy IR, Choudhury ATMA, Kecske’s ML (2004) Non symbiotic bacterial diazotrophs in crop-farming systems: can their potential for plant growth promotion be better exploited? Soil Biol Biochem 36:1229–1244
Kirchhof G, Reis VM, Baldani JI, Eckert B, Döbereiner J, Hartmann A (1997) Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 194:45–55
Knauth S, Hurek T, Brar D, Reinhold-Hurek B (2005) Influence of different Oryza cultivars on expression of nifH gene pools in roots of rice. Environ Microbiol 7:1725–1735
Kuklinsky-Sobral J, Araujo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL (2004) Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol 6:1244–1251
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Ladha JK, Reddy PM (2003) Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant Soil 252:151–167
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with folin phenol reagent. J Biol Chem 193:265–275
Muthukumarasamy R, Kang UG, Park KD, Jeon WT, Park CY, Cho YS, Kwon SW, Song J, Roh DH, Revathi G (2007) Enumeration, isolation and identification of diazotrophs from Korean wetland rice varieties grown with long-term application of N and compost and their short-term inoculation effect on rice plants. J Appl Microbiol 102:981–991
Park M, Kim C, Yang J, Lee H, Shin W, Kim S, Sa TM (2005) Isolation and characterization of diazotrophic growth promoting bacteria from rhizosphere of agricultural crops of Korea. Microbiol Res 160:127–133
Perin L, Martinez-Agular L, Castro-Gonzalez R, Estrada-de los Santos P, Cabellos-Avelar T, Guedes HV, Reis VM, Caballero-Mellado J (2006) Diazotrophic Burkhoderia species associated with field-grown maize and sugarcane. Appl Environ Microbiol 72:3103–3110
Piao Z, Cui Z, Yin B, Hu J, Zhou C, Xie G, Su B, Yin S (2005) Changes in acetylene reduction activities and effects of inoculated rhizosphere nitrogen-fixing bacteria on rice. Biol Fertil Soils 41:371–378
Poly F, Ranjard L, Nazaret S, Gourbiere F, Monrozier LJ (2001) Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262
Reis VM, Olivares FI, Döbereiner J (1994) Improved methodology for isolation of Acetobacter diazotrophicus and confirmation of its endophytic habitat. World J Microbiol Biotechnol 10:101–104
Reiter B, Bürgmann H, Burg K, Sessitsch A (2003) Endophytic nifH gene diversity in African sweet potato. Can J Microbiol 49:549–555
Roesch LFW, Olivares FL, Passaglia LMP, Selbach PA, Saccol de Sa EL, Oliveria de Camargo FA (2006) Characterization of diazotrophic bacteria associated with maize: effect of plant genotype, ontogeny and nitrogen supply. World J Microbiol Biotechnol 22:967–974
Roger PA (1995) Biological N2-fixation and its management in wetland rice cultivation. Fertil Res 42:261–276
Soares RA, Roesch LFW, Zanatta G, Oliveria de Camargo FA, Passaglia LMP (2006) Occurrence and distribution of nitrogen fixing bacterial community associated with oat (Avena sativa) assessed by molecular and microbiological techniques. Appl Soil Ecol 33:221–234
Ueda T, Suga Y, Yahiro N, Matsuguchi T (1995) Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol 177:1414–1417
Ulrich A, Becker R (2006) Soil parent material is a key determinant of the bacterial community structure in arable soils. FEMS Microbiol Ecol 56:430–443
Wakelin SA, Colloff MJ, Harvey PR, Marshner P, Gregg AL, Roger SL (2007) The effect of stubble retention and nitrogen application on soil microbial community structure and functional gene abundance under irrigated maize. FEMS Microbiol Ecol 59:661–670
Wartiainen I, Eriksson T, Zheng W, Rasmussen U (2008) Variation in the active diazotrophic community in rice paddy—nifH PCR-DGGE analysis of rhizosphere and bulk soil. Appl Soil Ecol 39:65–75
Yevdokimov I, Gattinger A, Buegger F, Munch JC, Schloter M (2008) Changes in microbial community structure in soil as a result of different amounts of nitrogen fertilizer. Biol Fertil Soils 44:1103–1106
Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5:539–554
Acknowledgments
We gratefully acknowledge Dr. Wen Xin Chen, China Agricultural University, PR China for their help in analyzing the data. Md. Rashedul Islam thanks the Korea Research Foundation (KRF) for awarding a Ph.D. fellowship. We thank H. P. Deka Boruah and R. Anandham for critical reading of this manuscript. We also wish to thank Tahera Sultana for helpful suggestions while doing molecular part of this work. The work was partially supported by the Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Islam, R., Trivedi, P., Madhaiyan, M. et al. Isolation, enumeration, and characterization of diazotrophic bacteria from paddy soil sample under long-term fertilizer management experiment. Biol Fertil Soils 46, 261–269 (2010). https://doi.org/10.1007/s00374-009-0425-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-009-0425-4