Abstract
Strain CB756 is usually an effective competitor against indigenous bradyrhizobia for nodulation of peanut in South Africa. Recently, inoculation of peanut and cowpea with CB756 in loamy sand soils of Botswana or a sandy clay loam in South Africa proved unsuccessful, achieving <2% nodule occupancy. A survey of ‘cowpea’ bradyrhizobia from five soils in Botswana and one in South Africa showed that many were effective in ability to fix N2 on peanut and cowpea. However, 15 isolates from Good Hope, Botswana were all effective on cowpea but ineffective on peanut, three failing to nodulate the latter. Selected cowpea isolates were significantly more competitive than CB756 for nodulation of cowpea in Leonard jars, but four were unsuccessful when inoculated at Roodeplaat, South Africa. When strain CB756 and two isolates were inoculated in pots containing Roodeplaat soil, at a 4:1 inoculant to soil bradyrhizobia ratio, their average nodule occupancy was 8% on cowpea compared to 40% on peanut. Significant differences in strain nodule occupancy were not detected on either cowpea or peanut. In contrast, nodule occupancy in loamy sand from Good Hope, Botswana, inoculated at a 40:1 inoculant to soil bradyrhizobia ratio, was 22.4% on cowpea and only 6.8% on peanut. In Good Hope soil, strain CB756 was the weakest competitor on cowpea but strain differences were insignificant on peanut. Whereas the Good Hope soil population was effective on cowpea, it was ineffective on peanut. DNA fingerprinting showed that isolates from Gaborone, Francistown and Roodeplaat contained several different genotypes, whereas those from Good Hope, Rasesa and Maun were more homogeneous. The dominance at Good Hope of genotypes effective on cowpea but ineffective on peanut emphasises the value of assessing the symbiotic capabilities and structures of indigenous populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In South Africa and Botswana, legumes such as peanut (Arachis hypogaea) and the indigenous cowpea (Vigna unguiculata) are cultivated by commercial and subsistence farmers. They provide a valuable source of protein and help in maintaining soil health through biological nitrogen fixation by symbiotic bradyrhizobia (Dakora and Keya 1997; Graham and Vance 2003). Although a widely used inoculant strain, CB756, is available, many soils in South Africa contain bradyrhizobia of the ‘cowpea’ miscellany that are effective on both legumes (Strijdom 1998). The average level of effectiveness of indigenous bradyrhizobia isolated from peanut nodules was lower than CB756, although many were comparable to this strain in nitrogen-fixing ability and had similar nodulation competitiveness (Law and Strijdom 1974; Staphorst et al. 1975). The failure of peanut to respond to inoculation under dryland or irrigated conditions was attributed to the presence and size of these populations at inoculation (Van der Merwe et al. 1974; Kishinevsky et al. 1987; Strijdom et al. 1988).
A similar lack of response by cowpea to inoculation has frequently been reported in Africa and elsewhere due to the presence of effective indigenous soil populations (Bushby 1984; Danso and Owiredu 1988; Dogbe et al. 2000; Fening and Danso 2001). Cowpea bradyrhizobia isolated from Hawaiian soils exhibited appreciable specificity in nitrogen-fixing effectiveness when inoculated on heterologous legume species such as peanut (Thies et al. 1991b), and similar observations were made with cowpea bradyrhizobia from Zimbabwean soils (Mpepereki et al. 1996). Mpepereki et al. (1996) concluded that symbiotic specificity reflected inherent diversity in types within the total population and was consistent with the diverse groupings obtained amongst isolates using serological, protein profile and DNA hybridization methods.
Although cowpea and peanut cultivation is encouraged in Botswana, little is known about the nodulation capabilities of populations indigenous to this region or whether inoculation is beneficial for either crop. In the present study, indigenous isolates from five localities in Botswana and one in South Africa were compared for nitrogen-fixing effectiveness and nodulation competitiveness. Specific primer-polymerase chain reaction (SP-PCR) fingerprinting was used to assess genotype diversity at strain level (Thies et al. 2001).
Materials and methods
Field trials and nodule collection
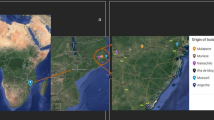
A map of the six collection sites is shown in Fig. 1. Nodules were obtained from peanut and cowpea during inoculation trials planted in December 2001 under dryland conditions at research stations at Roodeplaat, Gaborone (Sebele) and Francistown (described below). Soils at Roodeplaat and Gaborone had been cultivated but were not previously inoculated, whereas Francistown was virgin soil. Nodules from non-inoculated cowpea grown at Maun and Rasesa were obtained in July 2001 and April 2003, respectively. Soil sampled from non-inoculated cowpea plots at Good Hope Research Station in April 2003 was used to trap cowpea bradyrhizobia and was also used for a greenhouse competition experiment alongside Roodeplaat soil. A most probable number (MPN) method was used to estimate the size of soil populations in pooled soil samples (15 cm depth) from non-inoculated plots (Woomer et al. 1990). Some properties of the soils are shown in Table 1.
The field experiments, laid out in randomised blocks, are only briefly described as the inoculation treatments were all unsuccessful. A commercial inoculant containing Bradyrhizobium strain CB756 was applied to seed using sticker (approximately 105 cells per cowpea seed and 106 cells per peanut seed) or as slurry mixed in water and applied to seed rows at either 105 or 106 cells cm−1. Non-inoculated seeds were planted as a control. Soils were fertilised with K and P at recommended levels. After 8 to 10 weeks, five plants from each treatment were sampled; those from Botswana being stored on ice for 2 to 3 days during transport. Root nodules were removed and stored in 20% glycerol in a freezer at −20°C. Strain-specific polyclonal rabbit antiserum was used to determine nodule occupancy by indirect enzyme-linked immunosorbent assay (ELISA) (Kishinevsky and Maoz 1983). Nodules from non-inoculated cowpea and peanut plants were used to isolate bradyrhizobia from each locality. As only decayed nodules were present on cowpea planted at Francistown, fresh nodules were obtained from plants used for MPN counts. Animal depredation and/or drought at the sites depleted peanut plants and at the end of the 2001/2002 season only seed yields of cowpea at Gaborone and Roodeplaat could be measured. A subsequent experiment during 2002/2003 was carried out with seed-inoculated cowpea grown under irrigation at Roodeplaat.
Isolation and detection of bradyrhizobia
Cowpea and peanut nodules were surface-sterilised, crushed in water and bradyrhizobia isolated on yeast extract-mannitol (YM) agar containing Congo red (Vincent 1970). Nitrogen-fixing effectiveness was compared with that of strain CB756 under greenhouse conditions by inoculation of seedlings grown in Leonard jar assemblies containing quartz sand and N-free Hoagland solution (Vincent 1970). Isolates from Gaborone, Francistown and Roodeplaat were tested during July/October 2002, those from Maun and Rasesa during October/November 2004 and those from Good Hope during April/June 2004. The strain/cultivar experiment was during March/May 2005. Shoot dry mass was measured after 8 to 9 weeks for cowpea and after 10 weeks for peanut.
Insertion of the marker gene for β-glucuronidase into strains by conjugational transfer of the transposon mTn5SSgusA31 and the X-glcA staining procedure for nodule occupancy identifications were as previously described (Wilson 1996; Bloem and Law 2001). Nodules of cowpea grown for 4 to 5 weeks in Leonard jars were amenable to staining but those of cowpea grown in potted soil stained poorly. Adequate staining of peanut nodules was not achieved, possibly due to reduced penetration of X-glcA in the absence of surface lenticels (Sen et al. 1986). In addition, the epidermal layer turned dark brown during aerated staining and could not be cleared by bleach. As a result, streptomycin resistance was used to identify gusA31-marked strains in the soil competition experiment (Wilson 1996). Surface-sterilised cowpea or peanut nodules were crushed in water and droplets streaked on YM agar containing streptomycin (200 μg/ml) as well as on plain YM agar. Agar plates were observed for growth after 7 days incubation at 28°C.
Strain competitiveness
Strains containing the gusA31 marker were inoculated in combination with unmarked strain CB756 onto cowpea seedlings (cv. Bechuana white) grown in Leonard jars as described by Bloem and Law (2001). The competitive index (Cx:y) was calculated according to Beattie et al. (1989) using the linear relationship: \(\log {\left( {{\text{N}}_{x} + {\text{N}}_{{both}} :{\text{N}}_{y} + {\text{N}}_{{both}} } \right)} = {\text{C}}_{{x:y}} + k{\left( {\log \,{\text{I}}_{x} :{\text{I}}_{y} } \right)}\) where Ix:Iy is the ratio of reference strain (x) and test strain (y) in inoculum and Nx + Nboth:Ny + Nboth is their ratio in nodules. Nx and Ny represent the proportion of nodules occupied by x and y, respectively, and Nboth represents the proportion of nodules occupied by both strains. If the intercept C x:y has a positive value, x is more competitive than y. If the value is negative, then y is more competitive than x. Cowpea plants were harvested 4 to 5 weeks after inoculation and nodule occupancy determined by staining.
Comparison of strain competitiveness against indigenous bradyrhizobia in Good Hope and Roodeplaat soils used 1.8 kg quantities of non-amended soil in 150 mm diameter plastic pots. Pots were watered daily with deionised water to field capacity. Surface-sterilised imbibed seeds were incubated on water agar at 28°C to point of germination and two peanut (cv. Akwa) and two cowpea (cv. Bechuana white) seeds planted per pot. The seeds were each inoculated with 1 ml volumes of 5-day-old YM broth cultures diluted in sterile water to contain 106 viable cells per milliliter. After shoot emergence, peanut and cowpea were each reduced to one plant per pot. Plant growth, especially cowpea, was stunted because of soil compaction. Roodeplaat soil was harvested after 8 weeks and Good Hope soil after 10 weeks. At harvest, all tap-root nodules and up to 18 lateral root nodules were removed from each plant and pooled. Nodule occupancy was determined by plating on YM-streptomycin agar as described above.
SP-PCR procedures
DNA preparation, PCR amplifications and information capture methods were described previously. (Bloem et al. 2002; Botha et al. 2002). Specific primers included the nif-gene derived RPO1 sequence (Richardson et al. 1995) and arbitrary sequences CRL-7 (Mathis and McMillin 1996) and DAF9 (Niemann et al. 1997). Cluster analysis, dendrogram construction and presentation were as described in Botha et al. (2004).
Statistical analysis
The Genstat 5 (3.2) programme (NAG, UK) was used for variance analysis of completely randomised greenhouse experiments. The multiple t-distribution test procedure of Gupta and Panchapakesan (1979) was performed by the Biometry Unit of the ARC. Other statistical manipulations, including regression analysis for calculation of competition indices and comparison of regression lines, followed standard procedures (Steel and Torrie 1960; Chatterjee and Price 1977).
Results and discussion
Field trials
Field inoculation trials carried out during 2001/2002 used loamy sand soils at Gaborone and Francistown, and a sandy clay loam at Roodeplaat (Table 1). Estimated populations of cowpea-nodulating bradyrhizobia (cells g−1) were 76 at Gaborone, 467 at Francistown and 102 at Roodeplaat, whereas peanut-nodulating bradyrhizobia were either similar in size at Gaborone and Roodeplaat or four times smaller at Francistown. Strain CB756 was inoculated either as a seed coating or trickled onto seed rows as a liquid suspension at recommended and ×10 concentrations. Plants at Roodeplaat were examined for nodules after 8 weeks and those in Botswana after 10 weeks. Only decayed nodules were found on cowpea at Francistown. Serological identification by ELISA showed that nodule occupancy by CB756 was virtually undetectable (<2%) irrespective of legume, inoculation method or locality (data not shown). Only cowpea at Gaborone and Roodeplaat could be harvested at the end of season and neither site showed seed yield response to inoculation (data not shown). As hot dry conditions during December–February (145 mm at Gaborone and Roodeplaat, respectively) may have affected inoculant survival in the dryland experiments, another inoculation experiment with cowpea was carried out under irrigation at Roodeplaat during 2002/2003. The population of cowpea-nodulating bradyrhizobia at this site had increased to 570 cells g−1. Single strain seed inoculation treatments included CB756 as well as four effective indigenous isolates GC1d, GP4a, GC5d and RC3b found to be highly competitive in the greenhouse (see below). Appreciable nodule formation (20–30 nodules plant−1) occurred on plants of inoculated and non-inoculated treatments but of the five strains inoculated, only CB756 was detected at a low (2%) occupancy level in nodules sampled at 54 days (data not shown). Seed yields of the inoculated and non-inoculated treatments did not differ at harvest (data not shown).
Nitrogen-fixing effectiveness of nodule isolates
Thies et al. (1991b) did not find a significant correlation between the effectiveness of Hawaiian cowpea bradyrhizobia inoculated on cowpea and peanut, suggesting a degree of nitrogen-fixing specificity for the legume of origin. The cowpea and peanut bradyrhizobia from Gaborone, Francistown and Roodeplaat elicited growth responses on cowpea and peanut that ranged from effective to ineffective, commercial inoculant strain XS21 ranking ninth on each plant (Table 2). Comparison of the relative effectiveness of the 15 cowpea isolates (GC, FC, RC) on peanut showed a significant (P = 0.01) Spearman rank correlation coefficient (r = 0.740) suggesting an overall lack of specificity. This contrasted with an insignificant (P = 0.05) Spearman rank correlation coefficient (r = −0.079) obtained for the 14 peanut isolates (GP, FP, RP) on cowpea, suggesting that relative effectiveness of the peanut group was species dependent.
In additional tests, strain CB756 was compared for effectiveness with cowpea isolates from Rasesa, Maun and Good Hope in Botswana (Table 2). Results showed that 75% of the isolates from Rasesa and 43% of those from Maun were significantly more effective than CB756 on cowpea, whilst the remainder were similar to this strain. Of the isolates from Good Hope, 20% elicited a growth response on cowpea significantly greater than CB756 whilst 67% were less effective (Table 2).
The results in Table 2 were used to assign isolates from the different soils to arbitrary effectiveness categories relative to CB756. The percentage in each category is shown in Fig. 2. At least 50% of the isolates from each soil were in the effective (76–125%) or highly effective (≥126%) categories. The isolates from Roodeplaat appeared to be similar in effectiveness on cowpea and peanut whereas those from Francistown and Gaborone were more effective on cowpea than on peanut. The isolates from Maun and Rasesa were all in the effective or highly effective category, on cowpea (Fig. 2).
Effectiveness profiles of indigenous bradyrhizobia from Roodeplaat (RP), Francistown (F), Gaborone (G), Good Hope (GH), Rasesa (R) and Maun (BM) tested on cowpea (c) and peanut (p). Categories relative to CB756: HE highly effective (>126%); E effective (76–125%); M moderately effective (36–75%); I ineffective (≤35%)
Isolates that were highly effective on cowpea cv. Bechuana white showed similar high effectiveness on other cultivars of cowpea and the response of three cultivars to five of these strains is shown in Table 3. Strain CB756 was a relatively poor performer on cowpea, although it was usually equally or more effective on peanut (Tables 2 and 3). This contrasted with isolate GC1d which was the best performer on cowpea but was only moderately effective on peanut, whereas isolate RC3b was effective on both legumes (Tables 2 and 3). The cowpea isolates from Rasesa and Maun were not tested on peanut, except for R2m which was effective on both species (Table 3). Unexpectedly, two effective cowpea isolates from Good Hope, GHiv and GHvi were completely ineffective on peanut (Table 3). Subsequent examination showed that all the Good Hope cowpea isolates were ineffective on peanut (Fig. 2). Three (GHa, GHii and GHx) did not nodulate peanut. This contrasted with the small percentage (≤25%) of isolates from Gaborone, Francistown and Roodeplaat that were ineffective on peanut (Table 2, Fig. 2). A similar 19% average was reported for slow-growing Zimbabwean isolates by Mpepereki et al. (1996).
Nodulation competitiveness of bradyrhizobial isolates
Previous field and greenhouse trials had shown strain CB756 to be a successful competitor in South African soils for nodulation of peanut (Law and Strijdom 1974; Van der Merwe et al. 1974; Kishinevsky et al. 1987; Strijdom et al. 1988). As competitiveness on cowpea had not previously been determined, CB756 was compared with several effective indigenous isolates by inoculating mixtures onto cowpea grown in Leonard jars.
Regression analysis of data yielded linear relationships with significant correlation between the log ratio of inoculated strains and their corresponding log ratio in nodules (Table 4). In three different experiments, combinations of unmarked CB756 and marked CB756gusA31 resulted in competitive indices close to the zero value expected for equal competitiveness (0.108, −0.047, 0.012). However, competitive indices with large negative values were obtained when CB756 was inoculated in combination with gusA31-marked indigenous isolates, indicating that CB756 was the weaker competitor (Table 4). As slopes (k values) of regression lines did not differ significantly within each experimental set, line elevations could be compared by variance analysis. Significant differences were obtained between CB756gusA31 and the indigenous isolates in each set (Table 4). The first experimental set included isolates from sites (Roodeplaat, Gaborone and Francistown) used for field trials. Amongst these, the most competitive strains RC3bgusA31 and GC5dgusA31 had elevations significantly greater than those of GC1dgusA31 and GP4agusA31 (Table 4). Because of their apparent superior competitiveness, all four isolates were compared with CB756 in the irrigated field inoculation experiment planted at Roodeplaat. As noted above, however, none of the four were detected in nodules of inoculated cowpea plants.
The second experimental set included effective isolates from Maun and Rasesa (Table 1). All were significantly more competitive than CB756gusA31, whilst BM25gusA31 was significantly superior to R2mgusA31 and BM28gusA31 (Table 4).
The third set compared nodulation competitiveness of the five isolates chosen because of their effectiveness on cowpea, including two from Good Hope (Table 3). Comparison of regression line elevations indicated that all five were significantly more competitive than CB756gusA31 (Table 4). Although RC3bgusA31 was superior to GC1dgusA31 in the first set of experiments, it was less competitive in the third set. Variation in competition indices was noted by Sessitch et al. (1997) in different experiments using the same marked strains.
Competitiveness in Good Hope and Roodeplaat soils
Strains RC3b and R2m were further compared with CB756 for competitiveness on cowpea and peanut against soil populations in non-amended Roodeplaat and Good Hope soils. Identical MPN counts were obtained using cowpea and peanut, the estimated population in the Roodeplaat soil being 576 cells g−1 and that in Good Hope 58 cells g−1. Visual observation showed that Roodeplaat soil dilutions elicited an effective response on nodulated cowpea and peanut plants, whereas Good Hope soil dilutions elicited an effective response on cowpea but not peanut. Similar observations were made for non-inoculated plants in the soil experiment.
Strains CB756, RC3b and R2m, each containing the gusA31marker, were inoculated onto cowpea and peanut planted in the same pot. As 4 × 106 cells were inoculated onto 1.8 kg soil, the ratio of inoculated cells to the total indigenous population in each pot at planting was about 4:1 for Roodeplaat soil and 40:1 for Good Hope soil. Similar numbers of nodules per replicate plant were obtained from cowpea and peanut, respectively, averaging 23 and 16 for Roodeplaat and 18 and 24 for Good Hope. Streptomycin-resistance, used to detect the gusA31-marked inoculant strains, was not detected amongst isolates from non-inoculated control plants from either soil.
Although strain CB756 was significantly less competitive than RC3b and R2m in Leonard jar experiments (Table 4), nodule occupancy differences in Roodeplaat soil were not significant nor were strain × legume interactions evident (Table 5). In Good Hope soil, a strain × legume interaction was detected in which nodule occupancy on cowpea by RC3b (35.6%) was significantly greater than that of CB756 (10.7%) whilst strain R2m was intermediate (Table 5). The strains did not differ in competitiveness on peanut, however, nor did mean values for strains differ when data for the two legumes was combined (Table 5).
Although peanut-nodulating bradyrhizobia appeared to be equal in number to cowpea-nodulating bradyrhizobia in each soil, nodule occupancy of the inoculated strains showed a highly significant soil × legume interaction (Table 5). Whereas, their mean nodule occupancy was significantly higher on peanut (40%) than cowpea (8%) in Roodeplaat soil, this value was significantly lower on peanut (6.8%) than cowpea (22.4%) in Good Hope soil. As the effect was similar for each of the three inoculant strains, the result suggested that the two indigenous bradyrhizobial populations differed in competitiveness on each legume. The Roodeplaat population appeared to present less of a competitive barrier to inoculation of peanut than cowpea whilst the situation was reversed in Good Hope soil. Different soils and their associated populations have been shown to influence the competitive success of inoculant strains (Berg et al. 1988; Botha et al. 2004).
Mean inoculant nodule occupancy was significantly lower (14.6%) in Good Hope soil than in Roodeplaat soil (24%), although the inoculant to soil population ratio of the former was tenfold higher. This suggests that overall competitiveness of the Good Hope soil population, which is dominated by bradyrhizobia that are ineffective on peanut (Fig. 2), was higher than that of the effective Roodeplaat population. The leaves of inoculated peanut plants in the low nitrogen Good Hope soil were as yellow as non-inoculated plants at harvest; the small proportion of nodules formed by effective inoculant strains having no visible stimulatory effect (data not shown).
Analysis of soil isolates by PCR fingerprinting
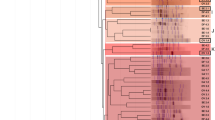
Genomic DNA of isolates from each of the six localities was compared by SP-PCR using primers CRL7, DAF9 and RPO1. Data from individual primer comparisons were then combined in the dendrogram shown in Fig. 3. Fifteen clusters were observed with similarities ≥80%, each of which contained isolates from the same locality. Their distribution agreed with previous findings that individual clones have high sampling site endemicity and that different clones may occur at the same sampling site (Vinuesa et al. 2005). For example, four different clones were evident amongst the fifteen isolates from Good Hope even though all had the same characteristic of ineffectiveness on peanut whilst being effective on cowpea (Fig. 3). One of the four clones contained the three cowpea isolates GHii, GHx and GHa that were unable to nodulate peanut, whilst the fourth isolate (GHe) in the clone produced many tiny brown ineffective nodules.
UPGMA-dendrogram derived from combined PCR fingerprints of isolates and strains of bradyrhizobia generated using the CRL7, DAF9 and RPO1 primers. Strains with similarity levels ≥60% were manually assigned to specific groups. Brackets indicate the corresponding seven clusters. Scale shows percentage similarity
Seven clusters with similarity ≥60% could be delineated in the dendrogram (Fig. 3). Whilst some clusters were heterogeneous, containing isolates from distant localities, others were quite homogeneous with regard to origin of locality. From the top, cluster 1 contained one Roodeplaat, two Francistown and three Rasesa isolates. Cluster 2 contained three highly effective Gaborone isolates, the eleven ineffective Good Hope isolates discussed above and the effective and competitive Roodeplaat isolate RC3b. Cluster 3 contained a single Roodeplaat isolate, the inoculant strain CB756 (originally isolated in Zimbabwe), the Bradyrhizobium elkanii type strain LMG 6134 and the clone of four Good Hope isolates noted in the previous paragraph. Cluster 4 contained a mixed selection of five Gaborone, four Francistown, five Roodeplaat and one Rasesa isolates. Cluster 5 contained two closely related Roodeplaat isolates. Cluster 6 contained all thirteen Maun isolates and Cluster 7 contained eight isolates from Rasesa. The remaining isolates from Francistown (FP4a), Rasesa (R5), Maun (BM1) and Roodeplaat (RC3e) grouped apart from the seven clusters as did the Bradyrhizobium japonicum type strain USDA6 (Fig. 3). The dendrogram indicated that genetic diversity within sites such as Gaborone, Francistown and Roodeplaat was as great as that between the sites. The diversity within the groups from Rasesa, Maun and Good Hope, many of which were highly effective on cowpea, appeared lower than that of isolates from the other soils (Figs. 2 and 3). The latter three groups may contain naturally dominant genotypes for which a legume such as cowpea may show preference. McInnes et al. (2004) suggested that such genotypes could be used in inoculation strategies to improve nitrogen fixation in the field.
McInnes et al. (2004) further noted that genetic diversity of rhizobial populations was often studied to characterise more effective or better adapted strains for particular applications. Clearly, symbiotic responses are more likely to be linked to symbiotic gene relatedness than other measures of genetic relatedness (e.g. Urtz and Elkan 1996); thus, genetic fingerprinting of the entire genome may not be the most appropriate tool for grouping and selecting effective strains. Nevertheless, comparison of the relative effectiveness of such groupings may be helpful in making initial screening decisions. For example, using cowpea data, the mean percent effectiveness (±standard deviation) relative to CB756 of bradyrhizobial isolates in each of the seven clusters shown in Fig. 3 ranked in the following order:Cluster 7 (140 ± 18) > cluster 6 (127 ± 18) > cluster 5 (113 ± 11) > cluster 1 (101 ± 21) > cluster 2 (99 ± 22) > cluster 3 (92 ± 30) > cluster 4 (77 ± 37).
According to this ranking, genotypes belonging to clusters 7 and 6 might be more likely to contain strains with superior nitrogen-fixing properties than genotypes belonging to clusters 3 and 4.
Although SP-PCR and related techniques are not suitable for taxonomic classification at species or higher level (Vinuesa et al. 2005), the similarity of the isolates to LMG6134 (B. elkanii) and USDA6 (B. japonicum) suggests that they are closely related to the two Bradyrhizobium type strains (Fig. 3). Phylogenetic studies of their symbiotic and other genes will be reported elsewhere.
Conclusions
The soils sampled in Botswana and South Africa each contained appreciable resident populations of bradyrhizobia that were generally effective in nitrogen fixation with cowpea and peanut. The same implications for inoculation practices thus appeared to apply to these soils as previously noted for other southern African soils by Strijdom (1998). In our field experiments, unexpectedly, poor nodulation by strain CB756 (nodule occupancy =2%) compared to that of indigenous bradyrhizobia was observed even though soil populations were at moderate levels (76–570 cells g−1) where appreciable inoculation success might have been expected (Weaver and Frederick 1974; Brockwell et al. 1995). For example, the ratio of CB756 to soil bradyrhizobia would have been about 1:20 for the seed inoculation treatment at Roodeplaat in 2001, using the criterion of Brockwell et al. (1995). The inoculation failures at Gaborone, Francistown and Roodeplaat contrast with the 50% or more nodule occupancy previously achieved on peanut with CB756 and other strains using similar seed or liquid inoculation methods (Law and Strijdom 1974; Van der Merwe et al. 1974; Kishinevsky et al. 1987). Poor survival after inoculation due to hot, dry environmental conditions may have been a contributing factor. In addition, the earlier experiments generally used sandy to sandy loam soils, often with low pH and these soils may have been more favourable for survival of strain CB756 than the higher pH, loamy sand or sandy clay loam soils used in the present study (Table 1). In Australia, strain CB756 failed to grow in heavy clay soils and showed poor competitiveness against indigenous populations for nodulation of two Vigna species (Bushby 1984). Improved nodulation occupancy levels were achieved in the greenhouse when higher inoculum to soil bradyrhizobia ratios of 4:1 for Roodeplaat and 40:1 for Good Hope soils were used. However, even these ratios did not achieve the 66% nodule occupancy level regarded as crucial for impact in the field (Thies et al. 1991a). Strain CB756 was significantly less competitive on cowpea in Leonard jars than all the indigenous isolates with which it was compared (Table 4). However, differences in strain competitiveness were generally less evident in the soil experiment (Table 5). The finding that an appreciable number of indigenous isolates were significantly more effective on cowpea than CB756 suggests potential for improvement of this inoculant.
As South African soils were found to contain effective bradyrhizobia, inoculation of cowpea and peanut was previously regarded of little benefit except as a means of establishing an effective strain in the nodulating population (Strijdom et al. 1988; Strijdom 1998). In such soils, management practices that encourage effective nodulation by indigenous populations or enrichment of effective and competitive strain genotypes may be of more value than inoculation (Thies et al. 1995; van Kessel and Hartley 2000). The Good Hope population in Botswana was, however, dominated by bradyrhizobia that were effective on cowpea but ineffective on peanut (Table 3; Figs. 2 and 3). Transforming this population by inoculation and enrichment of strains effective on both legumes might be difficult, considering its competitiveness on peanut (Table 5). However, recent evidence shows that physiological sanctions placed by the host plant on ineffective nodules favour effective nodules and their rhizobia (Denison and Kiers 2004). This aspect, in conjunction with crop management aimed at increasing nitrogen demand (van Kessel and Hartley 2000), could facilitate population transformation at Good Hope.
Although the Good Hope population may be atypical for southern Africa, its existence shows that indigenous bradyrhizobia are not always inherently capable of adequate nitrogen fixation on a crop such as peanut. The dominance at Good Hope of genotypes effective on cowpea but ineffective on peanut, emphasises the value of determining the symbiotic and structural characteristics of soil populations and their suitability for an introduced crop.
References
Beattie GA, Clayton MK, Handelsman J (1989) Quantitative comparison of the laboratory and field competitiveness of Rhizobium leguminasarum biovar phaseoli. Appl Environ Microbiol 55:2755–2761
Berg RK, Lonachan TE, Zablotowicz RM, Lieberman MT (1988) Nodule occupancy by introduced Bradyrhizobium japonicum in Iowa soils. Agron J 80:876–881
Bloem JF, Law IJ (2001) Determination of competitive abilities of Bradyrhizobium japonicum strains in soils from soybean production regions in South Africa. Biol Fertil Soils 33:181–189
Bloem JF, Botha WJ, Law IJ, Steyn PL (2002) Colony variation in Sinorhizobium meliloti inoculant strain U 45. Microbiol Res 157:283–292
Botha WJ, Bloem JF, Law IJ (2002) Bradyrhizobium sp. (Lupinus) in the winter rainfall region of South Africa. Biol Fertil Soils 36:335–343
Botha WJ, Jaftha JB, Bloem JF, Habig JH, Law IJ (2004) Effect of soil bradyrhizobia on the success of soybean inoculant strain CB 1809. Microbiol Res 159:219–231
Brockwell J, Bottomley PJ, Thies JE (1995) Manipulation of rhizobia microflora for improving legume productivity and soil fertility: a critical assessment. Plant Soil 174:143–180
Bushby HVA (1984) Colonization of rhizospheres and nodulation of two Vigna species by rhizobia inoculated onto seed: influence of soil. Soil Biol Biochem 16:635–641
Chatterjee S, Price B (1977) Regression analysis by example. Wiley, New York
Dakora FD, Keya SO (1997) Contribution of legume nitrogen fixation to sustainable agriculture in Sub-Saharan Africa. Soil Biol Biochem 29:809–817
Danso SKA, Owiredu JD (1988) Competitiveness of introduced and indigenous cowpea Bradyrhizobium strains for nodule formation on cowpeas [Vigna unguiculata (L) Walp.] in three soils. Soil Biol Biochem 20:305–310
Denison RF, Kiers ET (2004) Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett 237:187–193
Denison RF, Kiers ET (2004) Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett 237:187–193
Fening JO, Danso SKA (2001) Response of cowpea to inoculation with indigenous Bradyrhizobium strains. Trop Sci 41:172–176
Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131:872–877
Gupta SS, Panchapakesan S (1979) Multiple decision procedures: theory and methodology of selecting and ranking populations. Wiley, New York
Kishinevsky B, Maoz A (1983) ELISA identification of Rhizobium strains by use of enzyme-labelled protein A. Curr Microbiol 9:45–49
Kishinevsky B, Strijdom BW, Otto CJ, Lochner HH, Kriel MM (1987) Response to inoculation of groundnuts grown under irrigation in soil containing indigenous rhizobia. S Afr J Plant Soil 4:75–78
Law IJ, Strijdom BW (1974) Nitrogen-fixing and competitive abilities of a Rhizobium strain used in inoculants for Arachis hypogaea. Phytophylactica 6:221–228
Mathis JN, McMillin DE (1996) Detection of genetic variation in Bradyrhizobium japonicum USDA 110 variants using DNA fingerprints generated with GC rich arbitrary PCR primers. Plant Soil 186:81–85
McInnes A, Thies JE, Abbott LK, Howieson JG (2004) Structure and diversity among rhizobial strains, populations and communities—a review. Soil Biol Biochem 36:1295–1308
Mpepereki S, Wollum AG, Makonese F (1996) Diversity in symbiotic specificity of cowpea rhizobia indigenous to Zimbabwean soils. Plant Soil 186:167–171
Niemann S, Pühler A, Tichy H-V, Simon R, Selbitschka W (1997) Evaluation of the resolving power of three different DNA fingerprinting methods to discriminate among isolates of a natural Rhizobium meliloti population. J Appl Microbiol 82:477–484
Richardson AE, Viccars LA, Watson JM, Gibson AH (1995) Differentiation of Rhizobium strains using the polymerase chain reaction with random and directed primers. Soil Biol Biochem 27:515–524
Sen D, Weaver RW, Bal AK (1986) Structure and organization of effective peanut and cowpea root nodules induced by rhizobial strain 32H1. J Exp Bot 37:356–363
Sessitch A, Jjemba PK, Hardarson G, Akkermans ADL, Wilson KJ (1997) Measurement of the competitive index of Rhizobium tropici strain CIAT899 derivatives marked with the gusA gene. Soil Biol Biochem 29:1099–1110
Staphorst JL, Strijdom BW, Otto JF (1975) Nitrogen-fixing ability of rhizobia which nodulate groundnuts in South African soils. Phytophylactica 7:133–136
Steel RGD, Torrie JH (1960) Principles and procedures of statistics. McGraw–Hill, New York
Strijdom BW (1998) South African studies on biological nitrogen-fixing systems and the exploitation of the nodule bacterium-legume symbiosis. S Afr J Sci 94:11–23
Strijdom BW, Otto CJ, Lochner HH (1988) Effects of inoculant strains applied over two seasons on nodulation of groundnuts by indigenous rhizobia. S Afr J Sci 84:115–118
Thies JR, Bohlool BB, Singleton PW (1991a) Influence of the size of indigenous rhizobial populations on establishment and symbiotic performance of introduced rhizobia on field-grown legumes. Appl Environ Microbiol 57:19–28
Thies JR, Bohlool BB, Singleton PW (1991b) Subgroups of the cowpea miscellany: symbiotic specificity within Bradyrhizobium spp. for Vigna unguiculata, Phaseolus lunatus, Arachis hypogaea, and Macroptilium atropurpureum. Appl Environ Microbiol 57:1540–1545
Thies JE, Woomer PL, Singleton PW (1995) Enrichment of Bradyrhizobium spp populations in soil due to cropping of the homologous host legume. Soil Biol Biochem 27:633–636
Thies JE, Holmes EM, Vachot A (2001) Application of molecular techniques to studies in Rhizobium ecology: a review. Aust J Exp Agric 41:299–319
Urtz BE, Elkan GH (1996) Genetic diversity among Bradyrhizobium isolates that effectively nodulate peanut (Arachis hypogaea). Can J Microbiol 42:1121–1130
Van der Merwe SP, Strijdom BW, Uys CJ (1974) Groundnut response to seed inoculation under extensive agricultural practices in South African soils. Phytophylactica 6:295–302
van Kessel C, Hartley C (2000) Agricultural management of grain legumes: has it led to an increase in nitrogen fixation? Field Crops Res 65:165–181
Vincent JM (1970) A manual for the practical study of root-nodule bacteria, I.B.P. Handbook No. 15. Blackwell, Oxford
Vinuesa P, Silva C, Werner D, Martínez-Romero E (2005) Population genetics and phylogenetic inference in bacterial molecular systematics: the role of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54
Weaver RW, Frederick LR (1974) Effect of inoculum rate on competitive nodulation of Glycine max L. Merrill. I Greenhouse studies. Agron J 66:229–232
Wilson KJ (1996) GusA as a marker gene to track microbes. In: Ackermans ADL, van Elsas JD, de Bruijn F-J (eds) Molecular Microbial Ecology Manual 6.1.5. Kluwer, Dordrecht, pp 1–25
Woomer P, Bennet J, Yost R (1990) Overcoming the inflexibility of most-probable-number procedures. Agron J 82:349–353
Acknowledgements
We are grateful to the staff of the Department of Agricultural Research, Botswana for their help and support. South African researchers were funded through the National Research Foundation by a grant from the Regional S&T Programme of the South African Department of Science and Technology. The excellent technical assistance of Martie M. Kriel is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Law, I.J., Botha, W.F., Majaule, U.C. et al. Symbiotic and genomic diversity of ‘cowpea’ bradyrhizobia from soils in Botswana and South Africa. Biol Fertil Soils 43, 653–663 (2007). https://doi.org/10.1007/s00374-006-0145-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-006-0145-y