Abstract
Oxygen deprivation is a lethal stress that only a few animals can tolerate for extended periods. This study focuses on analyzing the role of DNA methylation in aiding natural anoxia tolerance in a champion vertebrate anaerobe, the red-eared slider turtle (Trachemys scripta elegans). We examined the relative expression and total enzymatic activity of four DNA methyltransferases (DNMT1, DNMT2, DNMT3a and DNMT3b), two methyl-binding domain proteins (MBD1 and MBD2), and relative genomic levels of 5-methylcytosine under control, 5 h anoxic, and 20 h anoxic conditions in liver, heart, and white skeletal muscle (n = 4, p < 0.05). In liver, protein expression of DNMT1, DNMT2, MBD1, and MBD2 rose significantly by two- to fourfold after 5 h anoxic submergence compared to normoxic-control conditions. In heart, 5 h anoxia submergence resulted in a 1.4-fold increase in DNMT3a levels and a significant decrease in MBD1 and MBD2 levels to ~30 % of control values. In white muscle, DNMT3a and DNMT3b increased threefold and MBD1 levels increased by 50 % in response to 5 h anoxia. Total DNMT activity rose by 0.6–2.0-fold in liver and white muscle and likewise global 5mC levels significantly increased in liver and white muscle under 5 and 20 h anoxia. The results demonstrate an overall increase in DNA methylation, DNMT protein expression and enzymatic activity in response to 5 and 20 h anoxia in liver and white muscle indicating a potential downregulation of gene expression via this epigenetic mechanism during oxygen deprivation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term epigenetics was first coined to refer to heritable changes in gene expression and phenotype that arise independent of changes in the primary DNA sequence (Waddington 1942; Wolffe and Matzke 1999; Bird 2002). Such epigenetic changes are coordinated via permanent (such as during tissue differentiation) or reversible (such as stress-responsive) modifications to DNA and histone proteins. Many previous studies have shown that epigenetic mechanisms play a vital role in most, if not all, cellular and physiological processes, such as gene expression (Grewal and Moazed 2003), cell cycle control (Macaluso et al. 2005), growth and development (Bestor and Tycko 1996), disease and cancer (van der Maarel 2008), aging (Calvanese et al. 2009), and genomic immunity (Barlow 1993). However, the involvement of epigenetic controls in the reversible regulation of animal adaptation to environmental stress has been understudied and is only beginning to be explored. For example, do epigenetic mechanisms such as DNA methylation play a role in metabolic rate depression, the nearly universal survival response to abiotic stresses (e.g., lack of oxygen, dehydration, temperature extremes, and food restriction)?
Hypoxia stress (low levels of oxygen) and anoxia (no oxygen) occur when the metabolic demand for oxygen exceeds the supply, and most vertebrates, particularly mammals, are highly sensitive to hypoxia/anoxia exposure. Oxygen deprivation is a particularly challenging stress to manage for intolerant species due to its dire consequences for ATP production via oxidative phosphorylation (Fraser et al. 2001). However, an obligate attachment to oxygen is not universal to all animals and many species experience natural situations where their access to oxygen can be cut off for long periods of time and yet they survive.
Among terrestrial vertebrates long-term survival without oxygen (anoxia) is most highly developed in some freshwater turtles. Turtles in the Chrysemys and Trachemys genera are champion facultative anaerobes, capable of surviving submerged underwater for >24 h at 25 °C and up to 3–5 months at 3 °C (Ultsch and Jackson 1982; Ultsch 1985). The anoxia tolerance of these species supports extended hours of breath-hold diving, and is crucial to underwater hibernation in ice-locked ponds and lakes in northern latitudes. Turtles can be blocked from surfacing to breathe by ice-locked ponds while at the same time oxygen is depleted in the water as winter progresses, thereby compromising the limited non-pulmonary oxygen uptake capacities of these species (Boutilier et al. 1997; Ultsch 2006). Red-eared sliders utilize a variety of physiological and biochemical adaptations to confer anoxia tolerance. These include maintaining high glycogen stores in liver and white muscle, use of the calciferous shell to store and buffer lactic acid, and up-regulation of cytoprotective mechanisms including antioxidant defenses and chaperone proteins (Ultsch and Jackson 1982; Hochachka et al. 1996; Lutz and Milton 2004; Storey and Storey 2007; Biggar et al. 2011). However, long-term anoxia survival is primarily achieved by suppressing, rebalancing and reprioritizing ATP use through metabolic rate depression (Storey and Storey 1990; Hochachka et al. 1996). Indeed, calorimetry has shown that the metabolic rate of submerged red-eared sliders is only about 10–20 % of the normoxic value at the same temperature (Herbert and Jackson 1985; Jackson and Heisler 1982).
Many physiological and genetic adaptations that support hypoxia/anoxia survival are interconnected with underlying epigenetic mechanisms in the form of DNA methylation and histone modifications (Krivoruchko and Storey 2010a). Epigenetic modifications are one of the essential transcriptional regulatory mechanisms in cells, and since gene transcription is a major ATP-consuming processes, typically utilizing 1–10 % of a cell’s total energy budget (Rolfe and Brown 1997), it could be expected that epigenetic mechanisms would contribute to the suppression of transcription and the induction/maintenance of a hypometabolic state. In particular, previous evidence points towards a tight link between short and long-term exposure to stresses, caloric intake, diet, and epigenetic control of certain metabolic pathways (Chiacchiera et al. 2013; Krivoruchko and Storey 2010b).
DNA methylation is a chemically stable, reversible, and post-replicative modification of the 5th position on cytosine (5mC). Approximately 60–70 % of all CpG sites (regions of DNA in which cytosine nucleotides are located next to guanine nucleotides and separated by a single phosphate group) are methylated with the exception of relatively short regions characterized by high CpG density (called CpG islands). CpG islands are located upstream of the promoters of most genes. These CpG islands are differentially methylated in different tissues at different times, suggesting a highly dynamic transcriptional regulatory mechanism (Bird 2002). Hypermethylation of CpG islands correlates with transcriptional silencing due to: (1) direct interference of transcription factor binding at the promoter, and/or (2) through recruitment of repressive methyl-binding proteins such as MBD1 and MBD2 (Bogdanovic and Veenstra 2009). MBD1 and MBD2 bind to methylated CpG islands and recruit repressive chromatin modifiers (such as histone deacetylases) and remodeling complexes that indirectly prevent the transcriptional machinery from binding to the promoter regions to initiate transcription. MBD1 has also been shown to bind DNA and induce chromatin compaction independent of DNA methylation (Boyes and Bird 1991).
DNMTs regulate the transfer of methyl groups from S-adenosylmethionine (SAM) to cytosine residues on genomic DNA and are essential for the maintenance and de novo creation of genomic methylation patterns (Goll and Bestor 2005). DNMT1 is attributed with preferring hemimethylated DNA, is known to maintain existing methylation patterns (Goll and Bestor 2005), and is considered a replication factor. Complete inhibition of DNMT1 has been shown to kill all dividing cells and partial inhibition may cause genome instability. Therefore, DNMT1 is considered to be a vital regulator of DNA methylation patterns in the genome (Goll and Bestor 2005). DNMT3a and 3b (de novo methyltransferases) do not require hemi-methylated DNA to function and transfer methyl groups to mainly non-methylated cytosine residues (Okano et al. 1998; Ramsahoye et al. 2000). DNMT3a and b are both essential for early development and regulation of gene expression and the loss of either enzyme is fatal (Okano et al. 1999). On the other hand, DNMT2 is quite different with little to no DNA methylation activity, but instead methylate cytosines at position 38 of the anticodon loop of several tRNAs (Goll et al. 2006; Jeltsch et al.2006) and regulates folding and stability of their structures (Alexandrov et al. 2006). Schaefer et al. (2010) suggested a link between tRNA methylation by DNMT2 and the cellular stress response in which the tRNA methyltransferase activity of DNMT2 may interfere directly with the stress-induced fragmentation of various tRNAs and thereby play a role in regulating protein translation.
The present study provides the first examination of the potential role of DNA methylation in the global suppression of gene expression in response to oxygen deprivation in a vertebrate model of anoxia tolerance, the red-eared slider turtle. We investigated the expression of four DNMTs and two MBDs, measured global 5-mC levels in genomic DNA, and assayed total DNMT activity in turtle liver, heart, and white muscle in response to 5 and 20 h anoxic submergence. The results show adjustments to tissue-specific expression patterns of DNMTs and MBDs in the three organs in response to anoxia as well as significant increases in both 5-mC levels and total DNMT enzymatic activity under anoxia in liver and white muscle.
Materials and methods
Animal care and treatment
Adult female red-eared sliders (Trachemys scripta elegans), weighing 700–1500 g, were acquired from local suppliers. The animals were held at 5 ± 1 °C in large tubs filled with dechlorinated tap water for at least a week before experiments began. Control normoxic turtles were sampled from this condition. For 5 and 20 h anoxia exposures, turtles were transferred to large tubs at 5 ± 1 °C that had been previously bubbled with nitrogen gas for 1 h; 2–3 turtles were added per tub in 30 min intervals. Bubbling was continued for 1 h after the last turtle was added to a tub, then halted and restarted during sampling of the animals. A wire mesh was fitted into the top of the tubs, situated ~5 cm below the water to prevent the turtles from surfacing. All animals were killed by decapitation and tissues were immediately excised, frozen in liquid nitrogen, and stored at −80 °C.
All animals were cared for in accordance with the guidelines of the Canadian Council on Animal Care and all experimental procedures had the prior approval of the Carleton University Animal Care Committee.
Total protein extraction
For isolation of total soluble protein, samples of frozen tissues (~0.5 g) were crushed under liquid nitrogen and then homogenized 1:2.5 w:v in homogenizing buffer (20 mM Hepes pH 7.5, 200 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate) with 10 μL/mL Sigma protease inhibitor cocktail (104 mM AEBSF, 80 μM aprotinin, 4 mM bestatin, 1.4 mM E-64, 2 mM leupeptin, 1.5 mM pepstatin A) added, and a few crystals of phenylmethylsulfonyl fluoride (PMSF) added immediately before use. Samples were homogenized using a polytron homogenizer on high for 15 s and then centrifuged at 4 °C for 15 min at 10,000×g; the supernatant was saved and the pellet was discarded. Soluble protein concentrations were quantified using the BioRad protein assay (Cat# 500-0006) with bovine serum albumin as the standard. All samples were then adjusted to a constant protein concentration by adding a calculated small volume of homogenizing buffer. Aliquots of samples were then mixed 1:1 v:v with 2× SDS loading buffer (100 mM Tris-base, 4 % w:v SDS, 20 % v:v glycerol, 0.2 % w:v bromophenol blue, 10 % v:v 2-mercaptoethanol). Final sample concentrations were 3 or 5 μg/μL, depending on tissue. Proteins were denatured by placing the tubes in boiling water for 10 min and then samples were stored at −40 °C until use.
Western immunoblotting
Samples of protein extracts containing 20–30 µg of protein were loaded onto 6–10 % SDS–polyacrylamide gels and resolved by electrophoresis for 45–90 min at 180 V in 1× Tris–glycine running buffer (75.5 g of Tris-base, 460 g glycine, 25 g SDS, ddH2O up to 2.5 L) using a BioRad Mini-Protean 3 System. Four µl aliquots of pre-stained protein molecular weight ladders (Froggabio; Cat. # PM005-0500 and PM007-0500 K) were run alongside the protein samples. Proteins were subsequently electroblotted onto 0.45 μm PVDF membranes (Millipore, Cat. #: IPVH00010) in transfer buffer (60.6 g Tris-base, 288 g glycine, 4 L methanol, 16 L ddH2O) at 160 mA for 90–120 min using a BioRad Mini-Protean Transfer cell. Subsequently, PVDF membranes were washed 3 × 5 min in 0.5× TBST (10 mM Tris, 150 mM NaCl, 0.05 % v/v Tween-20, pH 7.5) and blocked with 2.5–5 % milk for 30 min or 1 mg/ml polyvinyl alcohol (70–100 kDa) for one min. The membranes were subsequently washed 3 × 5 min in 0.5× TBST and incubated with primary antibody (diluted 1:500 for DNMT 1 and 1:1000 v:v for all remaining targets) for 24 h at 4 °C. All six antibodies used in this analysis were purchased from Genetex (DNMT1-GTX116011; DNMT2-GTX13892; DNMT3a-GTX128157; DNMT3b-GTX129127; MBD1-GTX110612; MBD2-GTX105622).
Membranes were then washed 3 × 5 min in 0.5× TBST and incubated with HRP-conjugated anti-rabbit IgG secondary antibody (Bioshop; Cat. # APA007P), diluted 1:8000 v:v in TBST, for 30–40 min at room temperature (RT) on a rocker. Proteins on the membranes were visualized using enhanced chemiluminescence and a ChemiGenius Bio-Imaging System (Syngene, Frederick, MD). Protein band densities were quantified using Gene Tools software. After immunoblotting was complete, membranes were stained with Coomassie blue (0.25 % w: v Coomassie brilliant blue stain, 7.5 % v:v acetic acid, 50 % v:v methanol) and band densities were similarly quantified using the ChemiGenius.
Preparation of nuclear extracts
Tissue samples (~0.5 g) were homogenized using a Dounce homogenizer in 1 mL of homogenization buffer [10 mM Hepes pH 7.9, 10 mM KCl, 10 mM EDTA, 1 mM dithiothreitol (DTT)]. A few crystals of PMSF and 1 μL of Sigma protease inhibitor cocktail were added just prior to homogenization. Samples were centrifuged at 10,000×g for 10 min at 4 °C and the supernatant (cytoplasmic extract) was removed. The pellet was resuspended in 150 μL of extraction buffer (20 mM HEPES, 400 mM NaCl, 1 mM EDTA, 10 % v/v glycerol, 1 mM DTT). Again, DTT and 1 μL of Sigma protease inhibitor cocktail were added just prior to addition of the buffer to the pellet. Tubes containing the samples were put on ice horizontally on a rocking platform for 1 h. Samples were then centrifuged at 10,000×g for 10 min at 4 °C. The supernatant (nuclear extract) was collected. Protein concentrations in the extracts were quantified and then extracts were treated as described above to create samples for western blotting. Final sample concentrations were 5 μg/μL. To confirm the separation of cytoplasmic and nuclear fractions samples of both fractions were immunoblotted and then probed with histone H3 antibody (diluted 1:1000 v:v; Genetex-GTX129546) to show that this nuclear protein remained in the nuclear fraction.
DNMT activity assay
Relative levels of total DNMT activity were assessed using the EpiQuick DNMT Activity/Inhibition Colorimetric Assay Ultra Kit from Epigentek (Catalog # P-3009) according to manufacturer’s instructions. In summary, 10 µg of nuclear protein extract from liver and heart, or 20 µg of nuclear protein extracts from white muscle of control, 5 h anoxic, and 20 h anoxic red-eared sliders were incubated in a 96-well microplate for 120 min at 37 °C. A blank well (containing 50 μl of assay buffer) was run per tissue type alongside a purified DNMT enzyme positive control (50 µg/ml; provided by Epigentek). After incubation, the plate was washed 3–5 times with 150 µl of 1× wash buffer. Subsequently, 50 µl of capture antibody (1000 µg/µl) was added to each well and incubated at RT for 60 min. The capture antibody was then removed, and after washing the wells, 50 µl of detection antibody (400 µg/ml) was added and incubated at RT for 30 min, followed by adding 50 µl of enhancer solution and further incubating for another 30 min at RT. Lastly, 100 µl of developer solution was added to each well and incubated at RT for 10 min away from direct light. Once the color of the positive control well had turned blue (indicating presence of sufficient methylated DNA), 100 µl of stop solution was added to stop the enzyme reaction and the plate was read using a microplate reader (Multiscan Spectrum, Thermo Labsystems) at 450 nm.
The DNMT activity was calculated by the following formula:
Genomic DNA extraction
Total genomic DNA was extracted using Zymo Research, Quick-gDNA MiniPrep kit (Catalog #: D3050) as per manufacturer’s instructions. In summary, 25 mg samples of frozen tissue were suspended in 95 µl of ddH2O, 95 µl of 2× digestion buffer, and 10 µl of proteinase K. The samples were incubated in a 55 °C bath for 3 h. Subsequently, 700 µl of genomic lysis buffer were added to each tube, thoroughly mixed and centrifuged at 10,000×g for 1 min. The supernatant was transferred to a Zymo-Spin IIC Column in a collection tube, and centrifuged for 10,000×g for 1 min. Then 200 µl of DNA pre-wash buffer was added to the same spin column but in a new collection tube, and centrifuged at 10,000×g for 1 min. Subsequently, 400 µl of g-DNA wash buffer was added to each spin column and centrifuged at 10,000×g for 1 min. Finally the spin column was transferred to a clean 1.5 ml microcentrifuge tube and 200 µl of DNA elution buffer was added and centrifuged at 15,000×g for 30 s to elute the extracted and purified genomic DNA. The concentrated gDNA product was diluted 25-fold with ddH2O and quantified using a GeneQuant Pro spectrophotometer (Pharmacia). The quality and integrity of the extracted genomic DNA was assessed by running each of the samples in a 0.6 % agarose gel at 130 V for 40–60 min.
Global DNA methylation
Relative levels of global DNA methylation (%) was assessed using the MethylFlash Methylated DNA Quantification Kit, Colorimetric (Source: Epigentek, Cat # P-1034), according to manufacturer’s instructions. This kit quantifies global DNA methylation levels colorimetrically by measuring levels of 5-methylcytosine (5-mC) in an ELISA format using genomic DNA. In summary, aliquots of 150 ng of extracted genomic DNA from liver, white muscle, and heart tissues of normoxic control, 5 h anoxic, and 20 h anoxic red-eared sliders were incubated with 80 µl of binding solution in a 96-well microplate for 90 min at 37 °C. Aliquots of 1 µl of negative control (20 µg/ml) representing unmethylated polynucleotide containing 50 % of cytosine, and 1 µl of positive control (5 ng/µl) representing methylated polynucleotide containing 50 % 5-methylcytosine were loaded into independent wells of the microplate. The wells were then incubated for 60 min at room temperature with a capture antibody (1 ng/ml). Subsequently, a 50 µl aliquot of detection antibody (0.2 µg/ml) was added and incubated at room temperature for 30 min and upon completion 50 µl of enhancer solution was added to each well and further incubated for 30 min at room temperature. Lastly, 100 µl of developer solution was added to initiate the colorimetric chemical reaction and incubated at room temperature for 10 min away from direct light. Subsequently, 100 µl stop solution was added to each well to halt the reaction and then absorbance values were read using a microplate reader (Multiscan Spectrum by Thermo Labsystems) at 450 nm.
The relative 5-mC levels were determined using the following formula:
where S is the input sample DNA in ng, P is the input positive control in ng, 2 is a factor that is used to normalize 5-mC in the positive control to 100 %, since the positive control contains only 50 % of 5-mC.
Data analysis
To adjust for minor protein loading irregularities and ensure equivalent protein loading, immunoblot band intensities were normalized against the summed intensity of a group of Coomassie-stained protein bands in the same lane that showed constant expression between control, 5 h anoxic, 20 h anoxic experimental conditions. In other words total protein analysis was used for normalization as an alternative technique to using a single housekeeping protein loading control. Target protein bands were identified by running FroggaBio standard protein molecular weight ladder and 4 µL of mammalian positive control samples (ground squirrel, Ictidomys tridecemlineatus) of liver, white muscle, and heart tissues were run alongside.
Statistical analysis used a one-way ANOVA with a Tukey post hoc test (p < 0.05) to compare three experimental conditions. Sigmaplot 11 software (Systat Softwaree Inc., San Jose, CA) was used for this analysis as well as construction of figures.
Results
DNMT and MBD protein expression in response to 5 and 20 h anoxia exposure
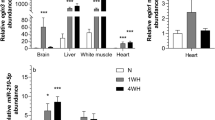
Relative protein expression levels of DNMT1, 2, 3a and 3b as well as MBD1 and 2 were assessed in liver of T. s. elegans comparing aerobic control turtles with animals given 5 and 20 h of anoxic submergence in nitrogen-gassed water (Fig. 1). DNMT1, DNMT2, MBD1, and MBD2 showed robust two- to fourfold upregulation in response to 5 h anoxic exposure as compared to normoxic-control conditions (P < 0.05). However, levels of all four proteins, as well as DNMT3a had decreased significantly at 20 h anoxia. DNMT1 and DNMT2 levels returned to near control values, DNMT3a fell to about 50 % of control, and MBD1 and MBD2 levels were reduced but still remained significantly higher (approximately twofold) than control values (P < 0.05). DNMT3b did not show any significant change in response to anoxia stress.
Effect of 5 and 20 h anoxic submergence on total protein levels of DNMT1, DNMT2, DNMT3a, DNMT3b, MBD1, and MBD2 in liver of T. s. elegans as determined by Western immunoblotting. Data are mean ± SEM, and n = 3–4 independent trials on tissue samples from different animals. a Significantly different from the corresponding control (P < 0.05). b Significantly different values from the 5 h anoxic value
In heart, short-term anoxic submergence (5 h) resulted in an approximately 1.4-fold increase in DNMT3a levels and significant decreases in MBD1 and MBD2 to about 30 % of control values (P < 0.05) (Fig. 2). With longer anoxia exposure (20 h), these changes were reversed for DNMT3a and MBD2. However, two proteins showed strong increases after 20 h anoxia: DNMT3b levels increased by 2.5-fold over control values and MBD1 levels increased to about 1.6-fold over control (or about fourfold higher than 5 h values).
Effect of 5 and 20 h anoxic submergence on total protein levels of DNMT1, DNMT2, DNMT3a, and DNMT3b, MBD1, and MBD2 in heart of T. s. elegans as determined by Western immunoblotting. Other information as in Fig. 1
The pattern in white skeletal muscle (neck retractor) included approximately threefold increases in DNMT3a and DNMT3b in response to 5 h anoxic submergence as well as a 50 % increase in MBD1 levels (P < 0.05) (Fig. 3). With prolonged anoxia (20 h) DNMT3a levels fell strongly to only about 30 % of controls. In contrast, DNMT3b levels continued to increase to about 3.8-fold higher than controls and MBD1 levels also remained elevated. DNMT1, DNMT2, and MBD2 in skeletal muscle were largely unaffected by anoxia exposure.
Effect of 5 and 20 h anoxic submergence on total protein levels of DNMT1, DNMT2, DNMT3a, DNMT3b, MBD1, and MBD2 in white muscle of T. s. elegans as determined by Western immunoblotting. Other information as in Fig. 1
DNMT activity in liver, heart, and white skeletal muscle in response to anoxia
Total DNMT activities were measured as the change in optical density per h per mg nuclear protein in liver, heart, and white skeletal muscle. Activity was highest in liver with mean values of 57.3 ± 4.4, 99.8 ± 7.4 and 109.0 ± 5.6 OD/h/mg nuclear protein for aerobic controls, 5 h anoxia, and 20 h anoxia, respectively. Heart showed intermediate activity levels of 41.8 ± 2.1, 29.6 ± 3.2 and 43.6 ± 0.9 OD/h/mg, respectively, whereas white skeletal muscle had the lowest overall DNMT activities of 5.96 ± 0.6, 14.0 ± 0.5 and 11.6 ± 0.7 OD/h/mg, respectively, for controls, 5 h anoxia and 20 h anoxia. Relative changes in DNMT activities are shown in Fig. 4. In liver, activity increased significantly by 1.7 and 1.9-fold over control values after 5 and 20 h of anoxia exposure (P < 0.05). Activity also increased significantly in white muscle by 2.4 and 2.0-fold, respectively (P < 0.05). In heart, however, there was little effect of anoxia on total DNMT activity.
Total DNMT enzyme activity (activity was measured as OD/h/mg nuclear protein) in liver, heart, and white muscle of T. s. elegans as determined by EpiQuick DNMT Activity/Inhibition Colorimetric Assay Ultra Kit from Epigentek. Other information as in Fig. 1
5-mC levels in genomic DNA of red-eared slider liver, heart, and white skeletal muscle in response to anoxia
Relative changes in global DNA methylation levels (in terms of total 5-methylcytosine content) in response to anoxic submergence are shown in Fig. 5. Compared with a relative mean level of 0.77 ± 0.03 for liver control, 5mC levels significantly increased to 1.07 ± 0.02 during 5 h anoxia and to 0.98 ± 0.04 during 20 h anoxia. White muscle showed similar increase in relative 5mC levels with 0.86 ± 0.03 for normoxic control, 1.30 ± 0.02 for 5 h anoxia, and 1.11 ± 0.02 for 20 h anoxia. Heart showed no change in relative 5mC content.
Relative levels of global DNA methylation (5-mC content) in liver, heart, and white muscle of T. s. elegans as determined by MethylFlash Methylated DNA colorimetric quantification kit from Epigentek. Other information as in Fig. 1
Discussion
Most organisms can withstand unfavorable environmental conditions such as high or low temperature, dehydration, or lack of food for short periods of time. However, due to the importance of oxygen-based metabolism for generating ATP, oxygen deprivation is a lethal stress that only a handful of vertebrates can endure for extended periods of time. Freshwater turtles belonging to the genera Trachemys (sliders) and Chrysemys (painted turtles) represent the extreme of anoxia tolerance among terrestrial vertebrates and can withstand as much as three months of anoxic submergence at 3 °C and recover with little or no metabolic damage (Ultsch and Jackson 1982; Ultsch 1985, 2006; Hermes-Lima and Zenteno-Savin 2002; Jackson 2002).
Red-eared sliders (T. s. elegans), the experimental model of this paper, use an array of physiological responses at the onset of hypoxia in an attempt to compensate for the drop in oxygen availability, such as increasing lung ventilation, releasing more red blood cells from the spleen, alterations to hemoglobin affinity for oxygen, an overall increase in cardiac output, and resorting to alternative modes of oxygen uptake across the epithelia of cloacal and/or buccopharyngeal cavities (Lutz and Storey 1997; Jackson 2002). However, if blood oxygen levels continue to decline and are not quickly remediated, these physiological responses fall short in meeting tissue ATP demands and then biochemical mechanisms are implemented to deal with long-term anoxia. These include: (1) a significant reduction in metabolic rate (i.e., ATP demand) to as low as 10–20 % of the corresponding aerobic rate at the same temperature by suppressing the activities of many metabolic processes (e.g., ion-motive ATPases, cell cycle, protein translation, gene expression) (Storey 2007), and (2) a switch to dependence on glycolytic ATP production supported by high glycogen reserves in liver, and (3) acid buffering by release of biocarbonate from the shell and lactate storage into the calciferous shell (Jackson and Heisler 1982; Jackson et al. 2000) that allows turtles to tolerate lactate buildup as high as 150–200 mM in plasma.
Various studies have examined transcriptional regulation during anoxia-induced hypometabolism in turtles from the point of view of altered expression of transcription factors (e.g., NF-κB, ChREBP, FoxO, HIF-1) and genes under their control (Biggar et al. 2011; Krivoruchko and Storey, 2010b, 2013, 2014). Global controls on transcriptional activity via changes in histone acetylation and HDAC expression have also been reported (Krivoruchko and Storey 2010a). However, the use of DNA methylation as a means of anoxia-induced transcriptional suppression in turtles has not previously been investigated. The present study examined genomic 5-methyl cytosine levels, DNMT enzymatic activity, and the protein expression levels of DNMTs and MBDs to show that changes in DNA methylation patterns are a previously unrecognized response to anoxia in turtle organs and represent a new regulatory mechanism to be considered in stress-induced metabolic rate depression.
In multicellular eukaryotes, DNA methylation is restricted to cytosine residues on genomic DNA and is often associated with a rigid chromatin state (heterochromatin) and inhibition of gene expression (Bird and Wolffe 1999; Klose and Bird 2006). There are two main mechanisms by which DNA methylation represses gene expression: (1) covalent modifications to the fifth position of cytosine directly prevent the association and binding of the transcriptional unit to promoter sequences through steric hindrance (Watt and Malloy 1998), and (2) 5-mC attracts methyl-CpG-binding proteins (MBD1-4) to the methylated promoter sequences and thereby indirectly represses gene expression (Boyes and Bird 1991). Furthermore, MBDs recruit and target chromatin remodeling co-repressors to methylated regions on the DNA and silence gene expression (Jones et al. 1998; Ng et al. 1999).
Relative genomic 5-methyl cytosine levels increased by 1.3–1.5-fold in liver and white muscle of red-eared sliders in response to 5 and 20 h anoxic submergence (Fig. 5). Correlated with this, protein expression levels of MBD1 and MBD2 were observed to increase two- to threefold in liver and MBD1 increased by 1.5-fold in white skeletal muscle (Figs. 1, 3). These results are consistent with the creation of an overall repressive chromatin state and probable down-regulation of the expression of many genes under anoxic conditions in liver and white muscle.
DNA methylation is mediated by DNA methyltransferases (DNMTs) with four isozymes known in vertebrates: DNMT 1, 2, 3a and 3b. Changes in DNMT protein expression and activity added further support for an overall suppression of transcriptional activity during turtle anoxia. Total DNMT activity increased by 1.7–2.4-fold in liver and white muscle, correlated with the increase in 5-mC levels in these two tissues. The changes in total DNMT activity were well supported by the up-regulation of DNMT protein expression in all three organs although organ-specific expression patterns of the DNMT isozymes were evident. Liver showed a robust increase in DNMT1 and DNMT2 expression levels in response to short-term anoxia exposure but no increase in DNMT3a or 3b (Fig. 1), whereas white muscle showed three- to fourfold increase in DNMT3a and 3b expression with no increase in DNMT1 or 2 (Fig. 3). The lack of change in DNMT1 expression in white muscle may be a reflection of the post-mitotic, non-proliferative state of muscle (Biggar and Storey 2012). The neck retractor muscle is a fast-twitch, mainly glycolytic muscle with low numbers of mitochondria and used mainly for rapid movements of the neck. Since DNMT1 has been shown to localize to DNA replication foci during the S phase of the cell cycle and is known to selectively methylate hemimethylated CpG dinucleotides to copy pre-existing methylation patterns onto newly synthesized DNA (Sharif et al. 2007; Avvakumov et al. 2008), it is perhaps not surprising that there was no significant response to anoxia by this enzyme in white muscle where little, if any, cell cycle activity would be expected, especially under anoxia. Overall, white muscle seems to exclusively utilize the de novo methyltransferases DNMT3a and DNMT3b to methylate genomic DNA in response to anoxic exposure.
On the other hand, liver is a proliferative tissue, the largest organ in the turtle body, vital to the biosynthetic needs of the animal, and contains the majority of the glycogen reserves needed to sustain energy metabolism under anoxia. Turtle liver hepatocytes (from C. picta bellii) have been shown to respond robustly to anoxia with a 90 % reduction in metabolic rate compared to normoxic hepatocytes (Buck et al. 1993). Limiting energy expenditure is important for long-term anoxia survival and the present data indicate that liver is utilizing the actions of DNMT1, MBD1, and MBD2 and possibly DNMT2 to implement methylation. DNMT1 and genomic 5-mC marks are known to interact with and recruit chromatin modifiers such as histone deacetylase 1 (HDAC1) to increase chromosome condensation and suppress gene expression (Jones et al. 1998). Interestingly, Krivoruchko and Storey (2010a) showed a significant increase in HDAC1 transcripts and protein levels in the liver of red-eared sliders after 5 h anoxia along with a 25 % decrease in histone H3 acetylation at Lys 9 and 23 (indicative of increased chromatin compaction under anoxia).
In contrast, heart showed less evidence of genome silencing via DNA methylation during anoxia with no significant change in 5-mC levels and a significant increase for only MBD1 at 20 h. In addition, cardiac muscle showed no significant increase in total DNMT enzymatic activity and restricted changes in DNMT protein levels (Figs. 2, 4). Heart has an important role to play under anoxia to continue circulation of remaining oxygen to key tissues (e.g., brain) for as long as possible, as well as to distribute anaerobic fuel (glucose) and remove anaerobic end product (lactate) from organs over the duration of the anoxic period. The heart of diving turtles exhibits tenfold higher cardiac glycogen levels than in other terrestrial vertebrates and approximately fivefold higher than in diving mammals such as the seal and cardiac glycogen can be used as a source of hexose units during long-term anoxia (Beall and Privitera 1973; Storey 1975). In addition, a study on lactate dehydrogenase (LDH) kinetics have shown that anoxic-tolerant turtle heart LDH can function more efficiently by increasing the affinity of LDH for pyruvate during anaerobic conditions (Beall and Privitera 1973). Hence, heart may not experience a strong repression of gene expression compared to liver and white skeletal muscle during anaerobic conditions in the turtle and may continue to function.
Overall, the present study provides the first evidence that DNA methylation and its regulators, DNA methyltransferases and methyl-binding proteins, are differentially regulated during 5 and 20 h of anoxic exposure in a tissue-specific manner. An overall stringency in chromatin compaction and a significant down-regulation in gene expression in liver and white muscles correlates with a global state of metabolic arrest that supports long-term anoxia survival in turtles. DNA methylation could potentially be used as a gene regulatory mechanism not only during the early entrance phase in the hypometabolic response to anoxia but it could also be used in the maintenance phase in which global suppression of transcription and translation is stringently enforced. One of the most important feats at a time of low/no oxygen availability is to conserve ATP as much as possible, and red-eared sliders are potentially using DNA methylation and its regulator proteins to facilitate the strong suppression of gene expression when oxygen is limiting.
References
Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21:87–96
Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455:822–825
Barlow DP (1993) Methylation and imprinting: from host defense to gene regulation? Science 260:309–310
Beall R, Privitera C (1973) Effects of cold exposure on cardiac metabolism of the turtle Chrysemys picta. Am J Physiol 224:435–441
Bestor TH, Tycko B (1996) Creation of genomic methylation patterns. Nat Genet 12:363–367
Biggar KK, Storey KB (2012) Evidence of cell cycle suppression and microRNA regulation of cyclin D1 during anoxia exposure in turtles. Cell Cycle 9:1705–1713
Biggar KK, Groom AG, Storey KB (2011) Hypometabolism and turtles: physiological and molecular strategies of anoxic survival. In: Nowakowska A, Caputa M (eds) Hypometabolism: strategies of survival in vertebrates and invertebrates. Research Signpost, Kerala, pp 57–94
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16(6):21
Bird AP, Wolffe AP (1999) Methylation-induced repression belts, braces, and chromatin. Cell 99:451–454
Bogdanovic O, Veenstra JC (2009) DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118:549–565
Boutilier R, Donohoe P, Tattersall G, West T (1997) Hypometabolic homeostasis in overwintering aquatic amphibians. J Exp Biol 200:387–400
Boyes J, Bird A (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123–1134
Buck LT, Land SC, Hochachka PW (1993) Anoxia-tolerant hepatocytes: model system for study of reversible metabolic suppression. Am J Physiol 265:49–56
Calvanese V, Lara E, Kahn A, Fraga MF (2009) The role of epigenetics in aging and age-related diseases. Ageing Res Rev 8:268–276
Chiacchiera F, Piunti A, Pasini D (2013) Epigenetic methylations and their connections with metabolism. Cell Mol Life Sci 70:1495–1508
Fraser K, Houlihan D, Lutz DP, Leone-Kabler S, Manuel L, Brechin J (2001) Complete suppression of protein synthesis during anoxia with no post-anoxia protein synthesis debt in the red-eared slider turtle Trachemys scripta elegans. J Exp Biol 204:4353–4360
Goll MG, Bestor TG (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398
Grewal SIS, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301:798–802
Herbert CV, Jackson DC (1985) Temperature effects on the responses to prolonged submergence in the turtle Chrysemys picta bellii. II. Metabolic rate, blood acid-base and ionic changes, and cardiovascular function in aerated and anoxic water. Physiol Zool 58:670–681
Hermes-Lima M, Zenteno-Savin T (2002) Animal response to drastic changes in oxygen availability and physiological oxidative stress. Comp Biochem Physiol C: Toxicol Pharmacol 133:537–556
Hochachka PW, Buck LT, Doll CJ, Land SC (1996) Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA 93:9493–9498
Jackson DC (2002) Hibernating without oxygen: physiological adaptations of the painted turtle. J Physiol 543:731–737
Jackson DC, Heisler N (1982) Plasma ion balance of submerged anoxic turtles at 3 °C: the role of calcium lactate formation. Respir Physiol 49:159–174
Jackson DC, Crocker CE, Ultsch GR (2000) Bone and shell contribution to lactic acid buffering of submerged turtles Chrysemys picta bellii at 3 °C. Am J Physiol Regul Integr Comp Physiol 278:1564–1571
Jeltsch A, Nellen W, Lyko F (2006) Two substrates are better than one: dual specificities for DNMT2 methyltransferases. Trends Biochem Sci 31:306–308
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylas to repress transcription. Nat Ganet 19:187–191
Klose RJ, Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31:89–97
Krivoruchko A, Storey KB (2010a) Epigenetics in anoxia tolerance: a role for histone deacetylases. Mol Cell Biochem 342:151–161
Krivoruchko A, Storey KB (2010b) Molecular mechanisms of turtle anoxia tolerance: a role for NF-kappaB. Gene 450:63–69
Krivoruchko A, Storey KB (2013) Anoxia-responsive regulation of the FoxO transcription factors in freshwater turtles, Trachemys scripta elegans. Biochim Biophys Acta 1830:4990–4998
Krivoruchko A, Storey KB (2014) Activation of the carbohydrate response element binding protein (ChREBP) in response to anoxia in the turtle (Trachemys scripta elegans). Biochim Biophys Acta 1840:3000–3005
Lutz PL, Milton SL (2004) Negotiating brain anoxia survival in the turtle. J Exp Biol 207:3141–3147
Lutz PL, Storey KB (1997) Adaptations to variations in oxygen tension by vertebrates and invertebrates. In: Dantzler WH (ed) Handbook of physiology, section 13, Comparative physiology, vol 2. Oxford University Press, Oxford, pp 1479–1522
Macaluso M, Montanari M, Cinti C, Giordano A (2005) Modulation of cell cycle components by epigenetic and genetic events. Semin Oncol 32:452–457
Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempest P, Reignberg D, Bird A (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet 1:58–61
Okano M, Xie S, Li E (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19:219–220
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257
Ramsahoye BH, Biniszkiewics D, Lyko F, Clark V, Bird AP, Jaenisch R (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 97:5237–5242
Rolfe DF, Brown GC (1997) Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev 77:731–758
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24:1590–1595
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting DNMT1 to methylated DNA. Nature 7171:908–912
Storey KB (1975) Purification and properties of turtle heart creatine kinase: role for the enzyme in glycolytic control. Int J Biochem 6:53–59
Storey KB (2007) Anoxia tolerance in turtles: metabolic regulation and gene expression. Comp Biochem Physiol A 147:263–276
Storey KB, Storey JM (1990) Metabolic rate depression and biochemical adaptation in anaerobiosis, hibernation and estivation. Q Rev Biol 65:145–174
Storey KB, Storey JM (2007) Tribute to P.L. Lutz: putting life on ‘pause’—molecular regulation of hypometabolism. J Exp Biol 210:1700–1714
Ultsch GR (1985) The viability of nearctic freshwater turtles submerged in anoxia and normoxia at 3 and 10 °C. Comp Biochem Physiol A 3:607–611
Ultsch GR (2006) The ecology of overwintering among turtles: where turtles overwinter and its consequences. Biol Rev 81:339–367
Ultsch GR, Jackson DC (1982) Long-term submergence at 3 °C of the turtle, Chrysemys picta bellii, in normoxic and severely hypoxic water: survival, gas exchange and acid-base status. J Exp Biol 96:11–28
Van der Maarel SM (2008) Epigenetic mechanisms in health and disease. Ann Rheum Dis 67:97–100
Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150:563–565
Watt F, Molloy PL (1998) Cytosine methylation prevents binding to DNA of HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev 2:1136–1143
Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286:481–486
Acknowledgements
Thanks to J.M. Storey for editorial review of the manuscript. This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (Grant #: 6793). KBS holds the Canada Research Chair in Molecular Physiology and SW holds a postgraduate Queen Elizabeth II Graduate Scholarship in Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Heldmaier.
Rights and permissions
About this article
Cite this article
Wijenayake, S., Storey, K.B. The role of DNA methylation during anoxia tolerance in a freshwater turtle (Trachemys scripta elegans). J Comp Physiol B 186, 333–342 (2016). https://doi.org/10.1007/s00360-016-0960-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00360-016-0960-x