Abstract
Purpose
To investigate the effect of the deleted in azoospermia (DAZ) copy cluster deletion on spermatogenesis in the South Chinese population.
Methods
In this study, the prevalence and characteristics of different DAZ copy cluster deletions and their association with spermatogenic failure were analyzed. A total of 186 infertile men with different spermatogenic impairments and 190 normozoospermic fertile men were studied. Three DAZ-specific single nucleotide variant loci and seven AZFc-specific sequence-tagged sites were examined using polymerase chain reaction (PCR)–restriction fragment length polymorphism and routine PCR.
Results
Gr/gr deletions were observed in a total of 9 of the 190 normozoospermic fertile men, and 11 gr/gr deletions were found in 186 infertile men. In addition, 3 b2/b3 deletions were identified in the infertile, but not in the fertile men. DAZ-SNV loci analysis revealed 4 DAZ copies that had 8 gr/gr-DAZ3/DAZ4 deletions and 1 gr/gr-DAZ1/DAZ2 deletion in the fertile men (8/190 vs. 1/190, p = 0.037). Analysis of DAZ deletion copies in infertile men revealed 10 gr/gr-DAZ1/DAZ2 deletions, 1 gr/gr-DAZ3/DAZ4 deletion (10/186 vs. 1/186, p = 0.011) and 3 b2/b3-DAZ1/DAZ2 deletions (13/186 vs. 1/186, p = 0.002).
Conclusions
Analysis of DAZ gene copies in AZFc microdeletions suggests that the contribution of the different deletions to male infertility varies. Removing DAZ1/DAZ2 seems to be associated with spermatogenic impairment, whereas removing DAZ3/DAZ4 seems to have little or no effect on fertility in the South Chinese population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genes on the Y chromosome are essential for male sex determination, early sexual differentiation and control of spermatogenesis [1]. Structural Y chromosome abnormalities are associated with human infertility [2, 3], and the microdeletion of the azoospermia factor (AZF) region in the Y chromosome has been discovered to be a frequent genetic cause associated with male infertility [4]. The AZFc region is particularly interesting as, approximately 80 % of AZF microdeletions occur in this region and most of them result in entire DAZ (deleted in azoospermia) gene deletion [5]. The DAZ gene has four copies, and most commonly encodes an RNA-binding protein exclusively in testicular tissue [6, 7]. Studies have demonstrated that the DAZ gene plays an important role in spermatogenesis [8, 9].
Detailed analysis of the AZFc region using new molecular non-repeating STS markers has confirmed the existence of three such microdeletions, namely gr/gr, b1/b3 and b2/b3 [10, 11]. The most prevalent partial deletion, the gr/gr deletion, is caused by recombination between amplicons g and r (g1, r1 and/or r2, with their respective homologous amplicons g2, r3 and/or r4) [12, 13].
Many studies found a positive correlation between AZFc microdeletions (gr/gr deletion and b2/b3 deletion) and spermatogenic failure [14, 15], whereas other studies could not confirm the relationship between gr/gr deletions and male infertility [16]. This disagreement may be due to different factors, such as ethnicity, country of study or geographic region [17]. The aim of the current study is to evaluate the frequency and type of DAZ copy cluster deletions on spermatogenesis in the South Chinese population. We assessed the prevalence of AZFc microdeletions (gr/gr deletion) in a population of both fertile and infertile men with severe impairment of spermatogenesis, in order to determine the contribution of these AZFc microdeletions to male infertility. A combined methodological approach that included the detection of the different DAZ gene copies by sequence nucleotide variant (SNV) analysis and AZFc-specific sequence-tagged site (STS) markers was used.
Materials and methods
Patients
The institutional ethics committee of the First Affiliated Hospital of Nanjing Medical University approved this study. The study included 376 patients who were examined in our clinic between September 2010 and January 2012. Patients included 186 men with primary infertility and 190 normozoospermic fertile men.
All patients were of Han nationality and came from the south of China. Patients with spermatogenic impairment due to causes, such as obstruction of the vas deferens, history of and/or active orchitis, hyperprolactinemia, hypogonadotropic hypogonadism, previous chemo- or radiotherapy, or a history of unilateral and bilateral cryptorchidism and varicocele were excluded. The semen analysis for sperm number, motility and morphology were performed according to World Health Organization criteria (1999) [18]. Infertile men divided into three groups on the basis of repeated semen analyses and World Health Organization criteria (1999) (Table 1). Non-obstructive azoospermia was diagnosed after bilateral testicular fine needle aspiration cytological analysis. Normozoospermia was considered when sperm concentration was more than 20 million/ml with normal sperm motility and morphology. All patients and controls exhibited a normal 46,XY karyotype and none had Y chromosome microdeletions in the AZF regions, as evaluated by multiplex PCR done on genomic DNA extracted from peripheral leukocytes with the following STSs: sY14-SRY, sY84, sY86, DBY1, sY125, sY127 and F19. This set of markers allowed identification of deletions in AZFa and AZFb.
Specific sequence-tagged site (STS) analysis of AZFc
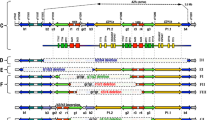
Microdeletion analysis of the AZFc region was performed by multiplex PCR using 5 STSs. The sequence-tagged site primers used were sY1161, sY1191, sY1291 and sY1206, sY1201. The ZFX gene on the X chromosome was used as an internal control to confirm PCR amplification. Five AZFc-specific STSs were used to detect entire and partial AZFc deletions, in which sY1161, sY1191, sY1291 and sY1206, sY1201 were co-amplified separately. Analysis of AZFc-specific STSs was performed on genomic DNA by PCR as first step to identifying the gr/gr deletion (Fig. 1) and b2/b3 deletion (Fig. 2).
a Analysis of the AZFc structure and results of AZFc-specific STSs and DAZ-specific SNVs. Figure a derived from the data of Kuroda-Kawaguchi [12]. The AZFc amplicon structure is drawn according to the color of Kuroda-Kawaguchi et al. The amplicon structure of the AZFc region with the complete (b2/b4) deletion is shown with an arrow. Red copies numbered r1 to r4 show the position of the four DAZ genes. Indicated above are the STSs utilized to detect deletions. b1, b2 and b3 gr/gr deletions are identified by the absence of sY1291. DAZ-specific SNV analysis distinguishes between those with a DAZ3/DAZ4 deletion (b1) and those with a DAZ1/DAZ2 deletion (b2 and b3). c The b2/b4-entire DAZ deletion identified by the positive result of sY1201 and the negative results of sY1191, sY1291 and sY1206
a Figure a derived from the data of Kuroda-Kawaguchi [12]. The AZFc amplicon structure is drawn according to the color of Kuroda-Kawaguchi et al. b Figure b derived from the data of Repping [11]. One pathway consists of an inversion involving the green and red amplicons (a “gr/rg” inversion) followed by a deletion between amplicons b2 and b3. c1 and c2 b2/b3 deletion after “gr/rg” inversion. d1 and d2 b2/b3 deletions identified by the absence of sY1291 and presence of sY1201, sY1191 and sY1206. DAZ-specific SNV analysis distinguishes between those with a DAZ1/DAZ2 deletion (d1) and those with a DAZ3/DAZ4 deletion (d2). b′ Figure b′ derived from the data of Repping [11]. The other pathway consists of an inversion between amplicons b2 and b3, followed by a deletion involving the red and green amplicons. c1′ and c2′ b2/b3 deletion after “b2/b3” inversion. d1′ and d2′ b2/b3 deletions identified by the absence of sY1291 and presence of sY1201, sY1191 and sY1206. DAZ-specific SNV analysis distinguishes between those with a DAZ3/DAZ4 deletion (d1′) and those with a DAZ1/DAZ2 deletion (d2′)
Single nucleotide variant (SNV) analysis of the DAZ gene
The partial deletion of DAZ gene copies was detected in SNV sites of the DAZ gene, including SNVII, SNVV and sY581, by PCR–restriction fragment length polymorphism. The MboI digestion fragment of SNVII distinguishes DAZ1 from other DAZ copies, the DraI digestion fragment of SNVV distinguishes DAZ1/DAZ2 from DAZ3/DAZ4, and the Sau3A digestion fragment of sY581 distinguishes DAZ1/DAZ4 from DAZ2/DAZ3. The primer sequences and PCR conditions have been described previously [11, 19].
Statistical analysis
Comparisons of proportions (prevalence of the AZFc microdeletions in patients and controls) were performed by means of the Fisher’s exact test. Comparisons of the AZFc microdeletion (gr/gr deletion and b2/b3 deletion) frequencies among subgroups of the infertile males were calculated and tested with the chi-square test. Analysis was carried out using the statistical package SPSS for Windows 16.0 (SPSS Inc., Chicago, IL, USA). P values (two-sided) of less than 0.05 were regarded as statistically significant.
Results
Deletions of the AZFc region
A total of 20 deletions resemble the gr/gr deletion with the absence of sY1291 and the presence of all of the other STSs. In three cases, STS analysis identified a b2/b3 deletion with the absence of sY1191 and the presence of all other markers. No cases of b1/b3 deletion were found. There was no difference in the prevalence of gr/gr (p = 0.630) and b2/b3 (p = 0.123) deletions comparing infertile and fertile normospermic groups (Table 2). There was no difference in the prevalence of the gr/gr deletions (p = 0.985) or the b2/b3 deletions (p = 0.445) among subgroups of infertile men (Table 1).
Distribution of gr/gr and b2/b3 with DAZ copy deletion
In this study, the type of gr/gr deletions analyzed included DAZ1/DAZ2 and DAZ3/DAZ4 (Table 1). In the SNV analysis of the DAZ gene, 23 men were found to have lost sequence family variants in the SNV loci (9 in the fertile and 14 in the infertile group; p = 0.288) (Table 3). Taking specific AZFc-STS and SNV analyses together, the deletion patterns in the three groups of subjects categorized by sperm counts are shown in Table 1.
The analysis of DAZ-specific SNVs allowed us to characterize the DAZ gene copy number in men presenting partial AZFc deletions. Of the 14 men with the gr/gr deletion, 11 exhibited the doublet deletion of DAZ1/DAZ2 and 3 showed the DAZ3/DAZ4 deletion. Of the 3 men with b2/b3 deletion, all showed the doublet deletion of DAZ1/DAZ2, but not the DAZ3/DAZ4 deletion. There was a statistically significant difference between the DAZ1/DAZ2 deletion and DAZ3/DAZ4 deletion (p = 0.002). We found no difference in the gr/gr-DAZ1/DAZ2 deletions among the subgroups of infertile men (p = 0.754), and the incidence of the b2/b3-DAZ1/DAZ2 deletions in the different subgroups was not significantly different (p = 0.445). In the normozoospermic fertile population, we found 8 gr/gr-DAZ3/DAZ4 deletions and one gr/gr-DAZ1/DAZ2 deletion in the normospermic groups. There was a significant difference when comparing gr/gr-DAZ1/DAZ2 deletions versus gr/gr-DAZ3/DAZ4 deletions in the normospermic groups (p = 0.037).
Discussion
The four DAZ genes on the human Y chromosome exist in two clusters and each cluster consists of an inverted pair of DAZ genes (DAZ1/DAZ2 and DAZ3/DAZ4) [20]. Complete deletion of the AZFc region of the Y chromosome includes the loss of the DAZ gene family and is responsible for most cases of spermatogenic impairment in infertile men [7, 12]. However, the role of the gr/gr deletion and b2/b3 deletion in sperm production has been a controversy since 2003 [11], when the first partial AZFc deletion (gr/gr deletion) was identified. Both of the gr/gr and b2/b3 deletions have been reported to have variable phenotypes across human populations. At present, considerable attention is focused on the effects of smaller deletions in the AZFc subregion on spermatogenesis. Generally, there are four DAZ gene copies in AZFc. The gr/gr deletion (Fig. 1) and b2/b3 deletion (Fig. 2) commonly remove two copies of the DAZ gene and several other transcriptional units [21].
In the present study, the b2/b4 (entire DAZ) deletion was not detected in any of the 190 fertile males. This suggests that the b2/b4 (entire DAZ) deletion may be the major AZFc abnormality responsible for impaired spermatogenesis in Chinese men. This is supported by results from previous studies in other populations [22].
Four copy deletion haplotypes were observed in both of the groups with different spermatogenic status after subtyping the gr/gr rearrangement types with DAZ copy analysis. We found an increased prevalence of DAZ1/DAZ2 deletions in infertile men with spermatogenic impairment compared to normozoospermic fertile men (Table 1), suggesting that such mutations represent a risk factor for male infertility. In fact, association of the gr/gr-DAZ1/DAZ2 deletions and b2/b3-DAZ1/DAZ2 deletions was found not only in azoospermia and severe oligozoospermia, similar to complete AZFc deletion, but also with moderate oligozoospermia. Among the partial AZFc deletions, the vast majority of gr/gr subtypes [21, 23], we found three b2/b3 deletions, indicating that the amplicons localize where the deletion reaches the breakpoints. Clear association with spermatogenic impairment could therefore be drawn only for the gr/gr deletion. Analysis of DAZ gene copy number in these cases allowed us to identify two classes of gr/gr deletions that may have different impacts on spermatogenesis. Deletions may remove DAZ1/DAZ2 genes or DAZ3/DAZ4 genes, but only the former is found in infertile men. The gr/gr deletion removing DAZ1/DAZ2 is rarely found also in normozoospermic men, though it can be compatible with full fertility. This finding implies that the simple gr/gr deletion involving DAZ1/DAZ2 might be the most significant risk cofactor for spermatogenic impairment among partial AZFc deletions and might have important diagnostic value in a clinical setting in the South Chinese population, although study results in other populations did not support subtyping in cases of gr/gr deletion for clinical purposes.
It has been suggested that the gr/gr-DAZ1/DAZ2 deletion represents an important genetic defect leading to impaired spermatogenesis in white men [14]. In the current study, the gr/gr-DAZ1/DAZ2 deletion was observed in 0.53 % of the normozoospermic Chinese men with a significant difference between infertile and fertile males (p = 0.011), suggesting that the gr/gr-DAZ1/DAZ2 deletion on its own may result in spermatogenic impairment in the South Chinese population (Table 3). This is in contrast to previous studies in which gr/gr-DAZ1/DAZ2 deletion was found not to cause spermatogenic impairment in Chinese men [24]. The different results in different studies could be due to various reasons, including environmental factors, genetic modification or Y chromosome haplogroups in different ethnic populations.
Two rearrangement types were observed in b2/b3 deletion, and all of the b2/b3 deletion rearrangements involved the removal of DAZ1/DAZ2 in infertile men. However, no b2/b3 deletions were found in the normozoospermic fertile group. The b2/b3 deletion is rare in the South Chinese population, but our research indicates that it can have an influence on spermatogenic phenotype (p = 0.002), similar to that reported in the South Chinese Han population [25].
The biological function of the DAZ family is not fully understood [6], but expressional patterns indicate that it may play a role in spermatogenesis. Coupled with the results from the present study this suggests that the loss of DAZ1/DAZ2 is sufficient to disturb spermatogenesis.
In summary, we investigated the presence of microdeletions in South Han Chinese infertile men with severe impairment of spermatogenesis using a combined approach of DAZ-SNVs and AZFc-STS. Our results suggest that the gr/gr-DAZ3/DAZ4 deletion may not be sufficient to cause spermatogenic impairment. In addition, gr/gr and b2/b3 microdeletions, which involve the DAZ1/DAZ2 region, may be a main cause of low sperm concentration and spermatogenic failure in the South Chinese population.
References
Disteche CM, Brown L, Saal H et al (1986) Molecular detection of a translocation (Y; 15) in a 45, X male. Hum Genet 74:372–377
Kim JW, Park SY, Ryu HM et al (2012) Molecular and clinical characteristics of 26 cases with structural Y chromosome aberrations. Cytogenet Genome Res 136:270–277
Knebel S, Pasantes JJ, Thi DA et al (2011) Heterogeneity of pericentric inversions of the human Y chromosome. Cytogenet Genome Res 132:219–226
Vogt PH, Edelmann A, Kirsch S et al (1996) Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 5:933–943
Simon M, Bakker E, Krausz C (2004) EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions. State of the art 2004. Int J Androl 27:240–249
Kim B, Lee Y, Kim Y et al (2009) Polymorphic expression of DAZ proteins in the human testis. Hum Reprod 24:1507–1515
Writzl K, Zorn B, Peterlin B (2005) Copy number of DAZ genes in infertile men. Fertil Steril 84:1522–1525
Fernandes AT, Fernandes S, Goncalves R et al (2006) DAZ gene copies: evidence of Y chromosome evolution. Mol Hum Reprod 12:519–523
Reynold N, Cooke HJ (2005) Role of the DAZ genes in male infertility. Reprod Biomed Online 10:72–80
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequences classes. Nature 423:825–837
Repping S, Skaletsk H, Brown L et al (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35:247–251
Kuroda-Kawaguchi T, Skaletsky H, Brown LG et al (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in the infertile men. Nat Genet 29:279–286
Hucklenbroich K, Gromoll J, Heinrich M et al (2005) Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod 20:191–197
Ferlin A, Tessari A, Ganz F et al (2005) Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 42:209–213
Visser L, Westerveld GH, Korver CM et al (2009) Y chromosome gr/gr deletions are a risk factor for low semen quality. Hum Reprod 24:2667–2673
Stouffs K, Tournaye H, Van der Elst J et al (2008) Do we need to search for gr/gr deletions in infertile men in a clinical setting? Hum Reprod 23:1193–1199
Stouffs K, Lissens W, Tournaye H et al (2011) What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update 17:197–209
World Health Organization (1999) Laboratory manual for the examination of human semen and sperm-cervical mucus interaction. Cambridge University Press, Cambridge
Repping S, van Daalen SK, Korver CM et al (2004) A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics 83:1046–1052
Saxena R, de Vries JW, Repping S et al (2000) Four DAZ gene in two clusters found in the AZFc region of the human Y chromosome. Genomics 67:256–267
Shahid M, Dhillon VS, Khalil HS et al (2011) Associations of Y-chromosome subdeletion gr/gr with the prevalence of Y-chromosome haplogroups in infertile patients. Eur J Hum Genet 19:23–29
Zhang F, Lu C, Li Z et al (2007) Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet 44:437–444
Yang Y, Ma M, Li L et al (2010) Differential effect of specific gr/gr deletion subtypes on spermatogenesis in the Chinese Han population. Int J Androl 33:745–754
Yang Y, Xiao CY, Zhou CA et al (2006) DAZ1/DAZ2 cluster deletion mediated by gr/gr recombination per se may not be sufficient for spermatogenesis impairment: a study of Chinese normozoospermic men. Asian J Androl 8:183–187
Wu B, Lu NX, Xia YK et al (2007) A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod 22:1107–1113
Acknowledgments
This research was supported by the project fund by the Development of Innovative Research Team in the First Affiliated Hospital of Nanjing Medical University and Key Medical Disciplines of Jiangsu Province.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Quan Li and Di Qiao contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, Q., Qiao, D., Song, Nh. et al. Association of DAZ1/DAZ2 deletion with spermatogenic impairment and male infertility in the South Chinese population. World J Urol 31, 1403–1409 (2013). https://doi.org/10.1007/s00345-013-1058-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-013-1058-7