Abstract
Stevia rebaudiana B. is a medicinal herb that accumulates low-caloric sweetening agents generally known as steviol glycosides among which stevioside and rebaudioside-A are predominant. These sweeteners are synthesized through the commonly known 2-C-methyl-d-erythritol-4-phosphate pathway common to steviol glycosides, gibberellic acid, pigments, and other isoprene-based secondary metabolites. The present investigation was designed to elucidate the role of steviol, an immediate precursor of sweetening agents, in the cultures of plants conducting stevioside biosynthesis in the leaf tissue. It was proposed that either the precursor or an elicitor was responsible for stimulating stevioside production in leaves. Transcript profiling of genes involved in stevioside biosynthesis was explored both in the roots and leaves of the plant and it was found that the transcript of the key enzyme kaurenoic acid hydroxylase, producing steviol, was the same in both tissue. However, when these plants were treated with the precursor steviol, most of the genes in the pathway were downregulated. Interestingly, it was found that very minute concentrations of the precursor were required to enhance the transcription level of genes of the pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stevia rebaudiana Bertoni is a medicinal plant widely used as a sweetener from ancient times in South America. The sweetening power of the herb comes from the diterpenoid glycoside generally known as steviol glycosides. The aglycon moiety in these glycosides is steviol which is glycosylated by various glycosyltransferases at the C-13 and C-19 positions to form steviol glycosides. These steviol glycosides along with other secondary metabolites like alkaloids, phenols, and flavonoids make the plant suitable for medicinal uses. Due to such metabolites, the plant and its extracts are widely used as anti-hypertensive, anti-hyperglycemic, and anti-rotavirus agents. Because of the sweetening agents the plant is also used to sweeten soft drinks, soy sauce, yogurt, and other foods in Japan, Korea, and Brazil (Tadhani and others 2007). Apart from these major glucose containing steviol glycosides, recently, Ceunen and Geuns (2013) reported several other ent-kaurene glycosides which also contain rhamnose, xylose, and fructose. Moreover, they reported that steviol glycosides are also found in Stevia phlebophylla (Mexican origin), Rubus suavissimus (Chinese blackberry), and Angelica keiskei (Japanese perennial).
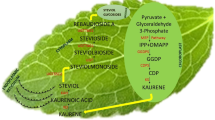
Isopentenyl diphosphate is a fundamental unit of diterpenoids and steviol glycosides and it was believed to be synthesized through the mevalonate pathway but Totte and others (2000) reported the involvement of the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway (Fig. 1) for the synthesis of steviol glycosides. This plant has contributed its large portion of secondary metabolism to steviol glycoside formation. Steviol glycosides share a common pathway with gibberellic acid (GA3) as they are terpenoid and both are derived from the plastid and thus these glycosides are synthesized mainly in the leaf (Brandle and Telmer 2007; Richman and others 1999). However, less than 0.1 % of steviol glycosides were found in roots also (Bondarev and others 2003). The steviol glycoside biosynthesis pathway separates from GA3 biosynthesis after the formation of kaurenoic acid. Gibberellins are synthesized via the action of kaurenoic acid oxidase whereas steviol (the precursor of steviol glycosides) is synthesized by a homotetramer enzyme kaurenoic acid hydroxylase having a subunit size of 39 kDa, after which further glycosylation of steviol takes place. An enzyme responsible for the production of steviolbioside has not yet been well characterized and thus reported as UGT in the pathway (Brandle and Telmer 2007), whereas some of the early genes DXS and DXR were well characterized from stevia (Totte and others 2003). Steviol glycoside content inside the leaf cell depends largely on genotype and environment. The sweetener is approved in many countries and now it is also recognized by the World Health Organization with an acceptable daily intake of steviol glycosides, expressed as steviol, at about 4 mg/kg of body weight.
Biosynthetic pathway for the production of steviol glycoside in Stevia rebaudiana Bertoni. DXS deoxyxylulose-5-phosphate synthase, DXR deoxyxylulose-5-phosphate reductase, CMS 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase, CMK 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase, MCS 4-diphosphocytidyl-2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase, HDS 1-hydroxy-2-methyl-2(E)-butenyl-4-diphosphate synthase, HDR 1-hydroxy-2-methyl-2(E)-butenyl-4-diphosphate reductase, GGDPS geranylgeranyldiphosphate synthase, CDPS copalyl diphosphate synthase, KS kaurene synthase, KO kaurene oxidase, KAH Kaurenoic acid hydroxylase, UGT85C2 UDP glucosyltransferase-85C2, UGT74G1 UDP glucosyltransferase-74G1, UGT76G1 UDP glucosyltransferase-76G1
Swanson and others (1992) studied stevioside biosynthesis in callus, root, shoots, and leaf of rooted shoot cultures of S. rebaudiana and found that stevioside is not synthesized in roots, shoots, or callus; moreover, it is exclusively synthesized in leaves of rooted shoots. So two hypotheses were proposed: (1) Either the precursor of stevioside is synthesized in roots and then transported to leaves or (2) the root produces some elicitor which stimulates the production of steviol glycosides in leaf. But the exact mechanism was not well understood. Furthermore, very minute quantities of precursors showed induction of steviol glycosides thus further treatments with the same precursor may reveal the co-ordination between precursor and steviol glycoside accumulation.
Steviol glycosides are synthesized in the plant through the MEP pathway. A number of researchers have identified, cloned, and characterized genes from the MEP pathway (Brandle and Telmer 2007). One of the most interesting features about such pathways is their manipulation for metabolic engineering and production of valuable secondary metabolites flashed as intermediates throughout the pathway. Moreover, these genes are differentially expressed when they are subjected to several experimental conditions. Siahsar and others (2011) reviewed the patterns of genes and enzymes in secondary pathways of medicinal plants. They concluded that the production of such metabolites requires the detailed understanding of the biosynthesis pathway and its regulation with analysis at the transcriptome, proteome, and metabolome levels. With the help of powerful techniques for gene expression analysis, it is now feasible to check the regulation of such a pathway at the transcriptome level.
The present scenario implies that the elucidation of the secondary metabolite pathway is carried out by evaluating the up- and down-regulation, at quantitative expression levels, of the genes involved (Ghazi and others 2009). Tai and others (2009) reported that among various gene expression methods like microarray, amplified fragment length polymorphism, serial analysis of gene expression, differential display reverse transcriptase PCR, and qPCR, the most accurate and wide dynamic range of quantification can be achieved through real-time PCR (qPCR). With this aim in the center, we planned to treat a culture of S. rebaudiana with precursor steviol and to check the regulation of various genes involved in steviol biosynthesis within the leaves of treated plants.
Materials and Methods
Culture Establishment and Treatment
Micropropagation of stevia plants was carried out and cultures were maintained according to the protocol as described by Modi and others (2012). To them, different concentrations of steviol (Sigma-Aldrich), that is 0, 50, 100, 150, and 200 nM (nanomolar), mentioned as treatments T0 (control), T1, T2, T3, and T4, respectively, were fed on the same medium on which cultures were maintained. Here steviol was dissolved in a minimum amount of dimethyl sulfoxide to make the stock of 3.1 mM. Leaves were sampled at the time of subsequent subculture. Moreover, sampling of root and leaf tissue was also carried out from the control plants to find the transcript of kaurenoic acid hydroxylase (KAH) gene.
RNA Extraction and cDNA Preparation
From the leaf samples RNA extraction was carried out by the method developed by Ghawana and others (2011) with minor manipulations. Two hundred milligrams of leaf tissue were frozen in liquid nitrogen and crushed using mortar and pestle. To this 2 ml buffer (containing 3 M sodium acetate, 0.5 M EDTA, and 10 % SDS in Tris saturated phenol) and 1 ml of DEPC (diethylpyrocarbonate) treated water were added and the slurry was kept on ice for 10 min. The solution was treated with 1/4th volume of chloroform and centrifuged at 13,000 rpm at 4 °C for 10 min. The upper aqueous phase containing RNA was carefully taken and treated with 0.6th volume of isopropanol and allowed to incubate on ice for 10 min followed by centrifugation at 13,000 rpm at 4 °C for 10 min. The final precipitation was carried out using 70 % alcohol and the RNA pellet was re-suspended in 30 μl of DEPC-treated water. Extracted RNA was quantified by Nanodrop, spectrophotometer (Thermo Scientific) and 5000 ng was used as the final concentration for the preparation of cDNA using a cDNA synthesis kit (Fermentas, Thermo Scientific).
Primer Design and qPCR
All the primers for endogenous genes and gene-specific primers were designed from the primer Blast tool available at the NCBI web site using the leaf EST collection developed by Brandle and others (2002). Primer sequences for all the genes involved in stevioside biosynthesis along with their in silico product size are mentioned in Table 1. The designed primers were procured from MWG, Eurofins. With these primers qRT-PCR was performed first to study the least variable housekeeping gene and subsequently regulation of target genes of the pathway were assessed. The cycling condition was set as mentioned in the Fast SYBR Green Master Mix kit (Applied Biosystems).
Relative Quantification
Relative quantification for all the targets was done with three repetitions and analyzed through the Data Assist bioinformatics tool.
Results and Discussion
Homogenous cultures of S. rebaudiana were established in control as well as treated plants. No adverse effect of treatment was observed on plant growth and plant vigor was normal (Fig. 2). Reference genes UBQ and GAPDH were found to be the most suitable among all the treated samples (Fig. 3) and analysis for relative quantification showed that mainly CMK was completely downregulated (Fig. 4) as compared to control whereas CDPS, HDR, HDS, KO, and MCS showed a differential response within treatments (Fig. 5). However, target genes CMS and GGDPS showed upregulation (Fig. 6) as compared to control (except for the second treatment in which the fold change value was slightly higher than control) and the remaining seven genes from the pathway showed upregulation in the first treatment and subsequent downregulation in other treatments (Fig. 7). Moreover, transcripts of KAH were found in both roots and leaves (Fig. 8a) but much less downregulation was observed in roots as compared to leaves (Fig. 8b). The transcripts of three predominant genes involved in glycosylation of steviol after KAH gene (UGT85C2, UGT74G1, and UGT76G1) were also reduced up to 30, 24, and 22 %, respectively, in roots as compared to leaves (Fig. 9).
Deviation plot for the selection of endogenous control (actin, ubiquitin, and GAPDH) genes. (T0: 0 nM (control); T1: 50 nM; T2: 100 nM; T3: 150 nM, and T4: 200 nM of steviol fed to micropropagated plants of Stevia rebaudiana). Values in the box at the bottom represent standard deviation of respective endogenous control
Relative quantification of CMK within treatments of precursor. Five bars represent one control as well as four treatments. RQ of untreated plants (control) is set as 1.0. (T0: 0 nM (control); T1: 50 nM; T2: 100 nM; T3: 150 nM, and T4: 200 nM of steviol fed to micropropagated plants of Stevia rebaudiana)
Relative quantification of CDPS, HDR, HDS, KO, and MCS within treatments of precursor. Five bars represent one control as well as four treatments. RQ of untreated plants (control) is set as 1.0. (T0: 0 nM (control); T1: 50 nM; T2: 100 nM; T3: 150 nM, and T4: 200 nM of steviol fed to micropropagated plants of Stevia rebaudiana)
Relative quantification of CMS and GGDPS within treatments of precursor. Five bars represent one control as well as four treatments. RQ of untreated plants (control) is set as 1.0. (T0: 0 nM (control); T1: 50 nM; T2: 100 nM; T3: 150 nM, and T4: 200 nM of steviol fed to micropropagated plants of Stevia rebaudiana)
Relative quantification of UGT74G1, UGT76G1, UGT85c2, DXR, DXS, KAH, and KS within treatments of precursor. Five bars represent one control as well as four treatments. RQ of untreated plants (control) is set as 1.0. (T0 0 nM (control); T1 50 nM; T2 100 nM; T3 150 nM, and T4 200 nM of steviol fed to micropropagated plants of Stevia rebaudiana)
Relative transcript of kaurenoic acid hydroxylase in root and leaf tissue of Stevia rebaudiana; a result of reverse transcriptase polymerase chain reaction revealing the presence of kaurenoic acid hydroxylase gene expression in leaf and root tissue and b relative quantification of the same gene through real-time PCR
Here it is noteworthy that the transcript of kaurenoic acid is also found in roots which suggests the possibility of the occurrence of steviol in roots and further glycosylation may also take place in roots because the transcript level of the three enzymes involved in glycosylation was ample.
This result reveals the production of steviol glycoside in roots and supports the result obtained by Bondarev and others (2003). Seven genes from the pathway showed upregulation in the first treatment and downregulation in subsequent treatments. In the present investigation, we hypothesized that the steviol supplied at increasing concentrations may induce glycosylation but the result revealed that the plant promotes glycosylation only at very low supplementation of the immediate precursor. Thus, a very low quantity of precursor stimulates steviol glycosides but it is just required to induce that, because further application of precursor was unable to stimulate the enhancement. Some of the genes responded differentially to the treatments thus there may be no direct impact of precursor supply on their expression levels. Analysis of these genes in roots and leaves suggested that synthesis of steviol glycosides also occurred in roots but at much lower levels than that observed in leaves. Thus, the immediate precursor of steviol glycosides (steviol) may be synthesized in roots (because transcripts of kaurenoic acid hydroxylase and the genes involved in glycosylation were also found in the roots). This is in agreement with the result obtained by Swanson and others (1992) and there is much less chance for synthesis of any elicitor molecule which could be further transported in the leaf to stimulate as suggested.
References
Bondarev NI, Sukhanova MA, Reshetnyak OV, Nosov AM (2003) Steviol glycosides content in different organs of Stevia rebaudiana and its dynamics during ontogeny. Biol Plant 47:261–264
Brandle JE, Telmer PG (2007) Steviol glycosides biosynthesis. Phytochemistry 68:1855–1863
Brandle JE, Richman A, Swanson AK, Chapman BP (2002) Leaf ESTs from Stevia rebaudiana: a resource for gene discoveries in diterpene synthesis. Plant Mol Biol 50:613–622
Ceunen S, Geuns JMC (2013) Steviol glycosides: chemical diversity, metabolism, and function. J Nat Prod 76:1201–1228
Ghawana S, Paul A, Kumar H, Kumar A, Singh H, Bhardwaj PK, Rani A, Singh RS, Raizada J, Singh K, Kumar S (2011) An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res Notes 4:85–89
Ghazi YA, Bourot S, Arioli T, Dennis ES, Llewellyn D (2009) Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant Cell Physiol 50:1364–1381
Modi AR, Patil G, Kumar N, Singh AS, Subhash N (2012) A simple and efficient in vitro mass multiplication procedure for Stevia rebaudiana Bertoni and analysis of genetic fidelity of in vitro raised plants through RAPD. Sugar Tech 14:391–397
Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthesis pathway. Plant J 19:411–421
Siahsar B, Raissi AS, Tavassoli A, Rahimi M (2011) Pattern of gene and enzyme in secondary pathways of medicinal plants. J Med Plants Res 5:5953–5957
Swanson SM, Mahady GB, Beecher CWW (1992) Stevioside biosynthesis by callus, root, shoot and rooted-shoot cultures in vitro. Plant Cell Tiss Org Cult 28:151–157
Tadhani MB, Patel VH, Subhash R (2007) In vitro antioxidant activity of Stevia rebaudiana leaves and callus. J Food Comp Anal 27:323–329
Tai HH, Conn G, Davidson C, Platt HW (2009) Arbitrary multi-gene references for normalization of real time PCR gene expression data. Plant Mol Biol Rep 27:315–320
Totte N, Charon L, Rohmer M, Compernolle F, Baboeuf I, Geuns JMC (2000) Biosynthesis of the diterpenoid steviol, an ent-kaurene derivative from Stevia rebaudiana Bertoni, via the methylerythritol phosphate pathway. Tetrahedron Lett 41:6407–6410
Totte N, Ende WV, Damme EJMV, Compernolle F, Baboeuf I, Geuns JMC (2003) Cloning and heterologous expression of early genes in gibberellin and steviol biosynthesis via the methylerythritol phosphate pathway in Stevia rebaudiana. Can J Bot 81:517–522
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Modi, A.R., Raj, S., Kanani, P. et al. Analysis of Differentially Expressed Genes Involved in Stevioside Biosynthesis in Cultures of Stevia Rebaudiana Bertoni Treated with Steviol as an Immediate Precursor. J Plant Growth Regul 33, 481–488 (2014). https://doi.org/10.1007/s00344-013-9396-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-013-9396-7