Abstract
The tropical Indo-West Pacific is the biogeographic region with the highest diversity of marine shallow water species, with its centre in the Indo-Malay Archipelago. However, due to its high endemism, the Red Sea is also considered as an important centre of evolution. Currently, not much is known about exchange among the Red Sea, Indian Ocean and West Pacific, as well as connectivity within the Indo-Malay Archipelago, even though such information is important to illuminate ecological and evolutionary processes that shape marine biodiversity in these regions. In addition, the inference of connectivity among populations is important for conservation. This study aims to test the hypothesis that the Indo-Malay Archipelago and the Red Sea are important centres of evolution by studying the genetic population structure of the giant clam Tridacna maxima. This study is based on a 484-bp fragment of the cytochrome c oxidase I gene from 211 individuals collected at 14 localities in the Indo-West Pacific to infer lineage diversification and gene flow as a measure for connectivity. The analysis showed a significant genetic differentiation among sample sites in the Indo-West Pacific (Φst = 0.74, P < 0.001) and across the Indo-Malay Archipelago (Φst = 0.72, P < 0.001), indicating restricted gene flow. Hierarchical AMOVA revealed the highest fixation index (Φct = 0.8, P < 0.001) when sample sites were assigned to the following regions: (1) Red Sea, (2) Indian Ocean and Java Sea, (3) Indonesian throughflow and seas in the East of Sulawesi, and (4) Western Pacific. Geological history as well as oceanography are important factors that shape the genetic structure of T. maxima in the Indo-Malay Archipelago and Red Sea. The observed deep evolutionary lineages might include cryptic species and this result supports the notion that the Indo-Malay Archipelago and the Red Sea are important centres of evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Connectivity of populations and evolution in the Indo-West Pacific (IWP)
The tropical IWP shows an incredible diversity of biota with more than 4,000 species of fishes, 6,000 species of molluscs, 800 species of echinoderms, and 500 species of hermatypic corals. The shelf waters of the IWP cover a huge area of approximately 6.6 million km2 and extend longitudinally more than half around the world from the Red Sea to the Society Islands in the Central Pacific. The Indo-Malay Archipelago, also called Coral Triangle, connecting the Philippines, Sumatra and New Guinea hosts the world’s greatest diversity of marine species (Briggs 1999). This high diversity in the Coral Triangle might be explained by several theories, e.g. centre of origin from where new species disperse (Briggs 1999), centre of overlap of the Indian and Pacific Ocean biota (Woodland 1983) and centre of accumulation of species that originated in peripheral areas (Jokiel and Martinelli 1992). However, these theories are not exclusive and one theory might not be sufficient to explain the high biodiversity in the Coral Triangle (Hoeksema 2007).
Besides this region of outstanding biodiversity, the Red Sea also should be considered as an important centre of evolution (Klausewitz 1989). Due to the high number of coral species, compared to the Indian Ocean, it is regarded as a secondary centre of diversity (Veron 2000). The Red Sea is considered as a multitaxon centre of endemism (Roberts et al. 2002), with 13.7% endemic fish species (Goren and Dor 1994).
Despite their importance as centres of marine biodiversity, not much is known about connectivity of populations within the Indo-Malay Archipelago and Red Sea, as well as exchange among the Red Sea, Indian Ocean and West Pacific (Ridgway and Sampayo 2005). However, such information is important to illuminate ecological and evolutionary processes that shape marine biodiversity in these regions. In addition, the inference of gene flow and connectivity among populations is important for the conservation of species.
Studies on the giant clam Tridacna crocea (DeBoer et al. 2008; Kochzius and Nuryanto 2008), the sponge Leucetta chagosensis (Wörheide et al. 2008), the gastropod Nerita albicilla (Crandall et al. 2008), the starfishes Linckia laevigata (Williams and Benzie 1998) and Acathaster planci (Benzie 1999), the tiger prawn Penaeus monodon (Duda and Palumbi 1999; Benzie et al. 2002; You et al. 2008), as well as the anemonefish Amphiprion ocellaris (Timm and Kochzius 2008; Timm et al. 2008) showed a genetic differentiation between populations in the Indian Ocean and West Pacific.
However, such a pattern could not be observed in other species, such as the sea urchin Diadema savignyi (Lessios et al. 2001), the gastropods Nerita plicata (Crandall et al. 2008), Echinolittorina trochoides clade D and E. reticulata (Reid et al. 2006), as well as the swordfish Xiphias gladius (Chow et al. 1997), the bigeye tuna Thunnus obesus (Alvarado Bremer et al. 1998; Chow et al. 2000), and the tasslefish Polynemus sheridani (Chenoweth and Hughes 2003).
Connectivity of populations in the Indo-Malay Archipelago
Currently, studies on the genetic population structure and gene flow of marine organisms within the Indo-Malay Archipelago investigated only a few invertebrates and fishes, showing different patterns of genetic diversification. Studied invertebrates are the boring giant clam T. crocea (DeBoer et al. 2008; Kochzius and Nuryanto 2008), the mushroom coral Heliofungia actiniformis (Knittweis et al. 2009), the giant tiger prawn P. monodon (Sugama et al. 2002), the gastropod Haliotis asinina (Imron et al. 2007), and mantis shrimps (Barber et al. 2002, 2006). Investigations on fishes include scad mackerels (Arnaud et al. 1999; Perrin and Borsa 2001; Borsa 2003; Rohfritsch and Borsa 2005), the anemonefish A. ocellaris (Nelson et al. 2000; Timm and Kochzius 2008), the jobfish Pristipomoides multidens (Ovenden et al. 2004), and seahorses (Lourie et al. 2005). Of these species, only giant clams, the anemonefish, seahorses, mantis shrimps and the gastropod H. asinina are well covered, with many populations sampled in the Coral Triangle.
Connectivity of populations in the Red Sea and Western Indian Ocean
Studies on the mud crab Scylla serrata (Fratini and Vannini 2002), the sponge L. chagosensis (Wörheide et al. 2008), and the damselfish Chromis viridis (Froukh and Kochzius 2008) have shown a genetic isolation of Red Sea populations. However, such a genetic differentiation could not be detected in the lionfish Pterois miles (Kochzius et al. 2003; Kochzius and Blohm 2005).
Within the Red Sea, only a few studies investigated gene flow in a coral (Maier et al. 2005), eulittoral bivalve (Shefer et al. 2004), and fishes (Hassan et al. 2003; Kochzius and Blohm 2005; Froukh and Kochzius 2007).
Connectivity of populations and evolution in giant clams
Even though several studies deal with population genetics of giant clams (Ayala et al. 1973; Campbell et al. 1975; Benzie and Williams 1992a, b, 1995, 1997; Macaranas et al. 1992; Kittiwattanawong 1997; Kittiwattanawong et al. 2001; Laurent et al. 2002; Juinio-Meñez et al. 2003; Ravago-Gotanco et al. 2007; Yu et al. 2000), only two studies investigated the genetic population structure of a giant clam (T. crocea) in detail on a large scale across the centre of marine biodiversity (DeBoer et al. 2008; Kochzius and Nuryanto 2008). They revealed deep divergence between genetic lineages, which might represent cryptic species. These studies showed that there is very high, but so far unknown, diversity in this rather well-studied group, which is underlined by the recent discovery of a new living species of giant clam, T. costata, in the Red Sea (Richter et al. 2008).
This study aims to test the hypotheses that the Indo-Malay Archipelago is a centre of origin (Briggs 1999) and that the Red Sea is an important centre of evolution (Klausewitz 1989) by studying the genetic structure of T. maxima populations in the Indo-West Pacific (Red Sea and Indo-Malay Archipelago). Genetic lineage diversification is triggered by vicariance, which is caused by geological and oceanographic processes, such as sea level changes (Voris 2000; Siddall et al. 2002). This can be detected by molecular markers, such as the cytochrome c oxidase I gene (COI), which is suitable for studies on population genetics of giant clams (DeBoer et al. 2008; Kochzius and Nuryanto 2008).
Tridacna maxima, also called great or rugose clam, was chosen for this study because it is the geographically most widespread species of giant clam and is distributed from the Red Sea and Western Indian Ocean across the Indo-Malay Archipelago to the Tuamotu Archipelago in the central Pacific. This species grows up to 35 cm shell length and is characterised by bright and various colour patterns. Dispersal is only mediated by a short planktonic larvae duration of about 9 days (Lucas 1988), which probably leads to high genetic diversification due to reduced dispersal capability. In addition, special emphasis will be given to gene flow as a measure for connectivity between populations in the Indo-Malay Archipelago to provide baseline data for conservation.
Materials and methods
Sampling
Mantle tissue of T. maxima was collected during field trips in 2004 and 2005 at 14 sample sites across the Indo-Malay Archipelago and in the Red Sea (Fig. 1; Table 1). In order to avoid collecting complete specimens, a biopsy method was used. Small pieces of mantle tissue were cut off under water, which ensures the survival of the clam. Tissue samples were preserved in 96% ethanol and stored at 4°C in the laboratory.
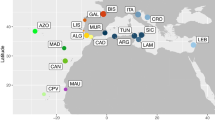
Maps of the Indo-West Pacific (a) and Indo-Malay Archipelago (b) showing samples sites (for abbreviations see Table 1). Oceanographic patterns with dominant (solid lines) and seasonally changing (dashed lines) currents (Wyrtki 1961; Gordon and Fine 1996; Gordon 2005; ITF Indonesian throughflow, SEC Southern Equatorial Current, NECC Northern Equatorial Counter Current), as well as Pleistocene maximum sea level low stand of 120 m (indicated by the light grey area; Voris 2000) are shown in map b. Pie charts represent the proportion of clades defined in the network at the different sample sites. c Network of 117 mitochondrial cytochrome c oxidase I haplotypes from 211 individuals of Tridacna maxima. Connecting lines between circles represent one mutational step. The hatches and numbers indicate additional mutational steps. The size of the circles is proportional to haplotype frequency
DNA extraction, PCR, and sequencing
Total genomic DNA was isolated using the Chelex® method, following the protocol from Walsh et al. (1991). A fragment of the cytochrome c oxidase I (COI) gene was used as a molecular marker, which was amplified using a pair of tridacnid-specific primers (Kochzius and Nuryanto 2008). PCRs in a volume of 50 μl contained 1 μl DNA template, 10 mM Tris–HCl (pH 9), 50 mM KCl, 2 mM of MgCl2, 0.2 μM of each primer, 0.2 mM of each dNTP, and 1 unit Taq polymerase. The following thermal profile was used: 94°C for 3 min, followed by 35 cycles of 1 min at 94°C, 1.5 min at 50°C and 1 min at 72°C. The final extension was carried out for 5 min at 72°C. Purification of PCR products was done with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) or Nucleosec II spin column (Macherey-Nagel, Düren, Germany) following the manufacturer’s protocol. Both strands were sequenced using the DyeDeoxy terminator chemistry and an ABI Prism 310 automated sequencer (Applied Biosystems, Darmstadt, Germany).
Genetic diversity
All sequences were edited using the programme Sequence Navigator (ver.1.0.1; Applied Biosystems) and checked manually by eye. The sequences were checked for orthology to sequences of T. crocea (Kochzius and Nuryanto 2008). No contamination with DNA from zooxanthellae was observed. In order to exclude mistakes in sequencing and to verify if a functional mitochondrial DNA sequence was obtained and not a nuclear pseudogene, the DNA sequences were translated to amino acid sequences with the programme Squint (http://www.cebl.auckland.ac.nz/index.php).
An alignment of sequences was obtained with ClustalW (Thompson et al. 1994) as implemented in the software Bioedit (ver. 7.0.4.1; Hall 1999). The programme Arlequin (ver. 2.0; Schneider et al. 2000) was used to calculate haplotype diversity h (Nei 1987) and nucleotide diversity π (Nei and Jin 1989).
Historical demography
The null hypothesis of neutral evolution of the COI gene was tested using Fu’s Fs test (Fu 1997) and Tajima’s D test (Tajima 1989) with the programme Arlequin (ver. 2.0; Schneider et al. 2000). Since significant results of these neutrality tests either indicate population hitchhiking, a bottleneck or population expansion, the sudden population expansion model (Rogers 1995) was also tested using Arlequin. These tests were run with 10,000 permutations.
Genetic population structure and gene flow
Significance of the genetic population structure was analysed with pairwise ΦST values and Analysis of Molecular Variance (AMOVA; Excoffier et al. 1992). Both analyses were performed with the programme Arlequin (ver. 2.0; Schneider et al. 2000), using a gamma shape parameter of 0.22 and the Tamura Nei substitution model. The gamma shape parameter and substitution model were obtained with the programme Modeltest (ver. 3.7; Posada and Crandall 1998). The programme Arlequin (ver. 2.0; Schneider et al. 2000) was also used to calculate a haplotype network and clades were defined based on the number of mutational steps.
Isolation-by-distance (IBD) analysis was conducted to test for a correlation between genetic (Φst-values) and geographic distances using Reduced Major Axis (RMA) regression analysis. Significance of the correlation was tested by a Mantel test with the isolation by distance web service (IBDWS 3.13), applying 30,000 permutations (Jensen et al. 2005; http://ibdws.sdsu.edu). Geographic distances represent the shortest path between two populations by sea and were measured with an electronic world atlas.
Results
Genetic diversity
A COI gene fragment of 484 bp length was obtained from 211 individuals of T. maxima collected at 14 sites across the Indo-Malay Archipelago and in the Red Sea, yielding 117 haplotypes. The sequences of these haplotypes are available at the EMBL sequence data base under the accession numbers FM244476–FM244485 and FM244513–FM244619. The sequences showed 116 (23.97%) polymorphic sites and 122 mutations. Genetic diversity within each population showed high levels of haplotype diversity, except for the populations from Karimunjava, Spermonde, and Komodo. In general, low levels of nucleotide diversity below one percent were observed, except for the population from Biak. Haplotype diversity ranges from 0.59 in the population from Karimunjava to 1.00 in Bira, nucleotide diversity from 0.23% in Komodo to 2.89% in Biak (Table 1). Overall haplotype diversity (h) was 0.94 and nucleotide diversity (π) was 2.28%.
Historical demography
The majority of Tajima’s D test and Fu’s Fs test for each sample site rejected the null hypothesis of neutral evolution of the COI marker. However, these could indicate population expansion, which is supported by the mismatch distribution analysis and Rogers’ test of sudden population expansion for all sample sites except Bira, which might be an artefact due to low sample size (Table 1).
Genetic population structure and gene flow
The haplotype network consists of 117 haplotypes, which can be assigned to four different clades (Fig. 1). Their proportion at each sample site is also shown in Fig. 1. At sites along the ITF (Indonesian throughflow; Sulawesi Sea, Makassar Strait, Flores Sea, Timor) and the eastern coast of Sulawesi, only clade 1 is found. A small proportion of clade 1 is also present in the Java Sea (Karimunjava) and the Western Pacific (Biak). Clades 2, 3, and 4 are separated by 8, 44, and 12 substitution from clade 1, respectively. Clade 2 could only be found in populations from the Eastern Indian Ocean (Padang) and Java Sea (Pulau Seribu, Karimunjava). Clade 3 is only found in the Western Pacific and clade 4 exclusively consists of haplotypes from the Red Sea. The most common haplotype was found in a frequency of 22.3% and was unique for the eastern region of the Indo-Malay Archipelago (from Makassar Strait eastward to New Guinea). The second most common haplotype occurred in 9.5% of the individuals and was only found in populations from the Java Sea and Eastern Indian Ocean.
The significance of the observed genetic population structure in T. maxima was verified by AMOVA and pairwise Φst-values, considering a gamma shape parameter of 0.22 and the Tamura Nei substitution model. This analysis showed a highly significant genetic structure in the Indo-West Pacific (Indo-Malay Archipelago and the Red Sea; Φst = 0.74, P < 0.001) and across the Indo-Malay Archipelago (Φst = 0.72, P < 0.001). Pairwise Φst-values showed a complex pattern of homogeneity and significant differences, most of them revealing restricted gene flow (58%) (Table 2). Significant Φst-values ranged from 0.06 to 0.91. The populations from the Red Sea (Φst = 0.76–0.91), Eastern Indian Ocean and Java Sea (Padang, Pulau Seribu, Karimunjava: Φst = 0.78–0.91), as well as Western Pacific (Biak: Φst = 0.66–0.80) were the most divergent populations, whereas most of the other populations showed panmixing.
Based on geography and pairwise analysis, a hierarchical AMOVA was carried out with different groupings (Table 3). AMOVA revealed the highest fixation index (Φct = 0.8, P < 0.001) and variation (83.8%) among groups when sample sites were assigned to the following regions: (1) Red Sea, (2) Eastern Indian Ocean (off Padang = Western Sumatra) and Java Sea (Pulau Seribu, Karimunjava), (3) Indonesian throughflow (off Manado = North Sulawesi, Sangalaki = East Borneo, Spermonde and Bira = South Sulawesi, Komodo, off Kupang = West Timor) as well as seas in the East of Sulawesi (Sembilan, off Kendari, off Luwuk, Togian Islands), and (4) Western Pacific (Biak = New Guinea).
Isolation-by-distance is indicated by a significant correlation between genetic (Φst) and geographic distances, analysed for all Indo-Malay populations (r = 0.7, P = 0.002) as well as for a subset from the central Indo-Malay Archipelago (r = 0.33, P = 0.045) (Fig. 2). The population from the Red Sea was not considered in this analysis.
Relationships between genetic and geographic distances in Tridacna maxima from the central Indo-Malay Archipelago (Bi, Ko, Ku, Lu, Ma, Sa, Se, Sp, TI; for abbreviations see Table 1) using Reduced Major Axis (RMA) regression and Mantel test
Discussion
Genetic diversity
The molecular marker used in this study shows a high level of polymorphism and is therefore suitable to study the genetic population structure of the giant clam T. maxima. High levels of polymorphism and genetic diversity were also observed in a study on T. crocea from the Indo-Malay Archipelago (DeBoer et al. 2008; Kochzius and Nuryanto 2008) and other studies on marine invertebrates (e.g. Barber et al. 2002; Luttikhuizen et al. 2003; Duran et al. 2004; Shefer et al. 2004). High haplotype diversity (≥0.85) was found in all populations, except in Karimunjava (Java Sea), Spermonde Archipelago (Makassar Strait), and Komodo (Flores Sea). High genetic diversity reflects a large population size, whereas a reduction in population size entails a reduction of genetic diversity. There are three potential factors, or a combination of them, that can lead to low haplotype diversity in these populations. It could be due to a founder effect, which is caused by the re-colonisation of the Sunda and Spermonde shelf with the rising sea level after glacials (Voris 2000). However, this explanation seems to be contradictory to the high haplotype diversity in Pulau Seribu, which is also located on the Sunda shelf. It also cannot account for the low diversity in Komodo, which lacks extensive shelf areas.
Overexploitation could also be a reason for low haplotype diversity. The shell industry in Jepara and Jakarta (Java) obtained giant clam shells from Karimunjava and Pulau Seribu, respectively. They have been utilised as material for the production of so-called ‘teraso’ tiles and other purposes, but it is not reported which giant clam species have been used in the shell industry (Pasaribu 1988; Wells 1997). Overexploitation could also be caused by fishery and marine ornamental trade, but there are no data available. Since haplotype diversity seems to be not effected in Pulau Seribu, it is difficult to estimate to which extend exploitation is responsible for the low haplotype diversity in Karimunjava. Giant clams are collected in Spermonde Archipelago (pers. obs.), but quantitative data are not available. Pollution and reef degradation is a severe problem in Spermonde Archipelago (Erdmann 1995; Edinger et al. 1998; Risk and Erdmann 2000), which might account for a reduction of tridacnid populations. In Komodo, more than 50% of the coral reefs are damaged by blast fishing, cyanide fishing, and destructive reef gleaning (Pet 1997; Fox et al. 2005). Due to these destructive fishing practices, the population of giant clams could be reduced, leading to a low genetic diversity.
Bleaching events in the Java Sea (Pulau Seribu and Karimunjava in 1998, Wilkinson 2002) might have caused a population bottleneck, because giant clams depend nutritionally on their zooxanthellae (Ishikura et al. 1999) and the loss of the zooxanthellate symbionts negatively affects their fitness (Leggat et al. 2003). In 1997/1998, some coral bleaching was observed in Spermonde Archipelago (Hopley and Suharsono 2000), but no information is available for Komodo.
Since re-colonisation, overexploitation and bleaching could have a negative influence on both populations in the Java Sea (Karimunjava and Pulau Seribu), it is difficult to explain why only the population in Karimunjava seems to be affected with regard to haplotype diversity. Populations of the giant clam T. crocea (Kochzius and Nuryanto 2008) showed low haplotype diversity in Karimunjava as well as Pulau Seribu, as one would expect.
Nucleotide diversity in Karimunjava was rather high compared to other sample sites in this study, but the population in Komodo and Pulau Seribu showed low values. The low genetic diversity in Komodo might be due to coral reef degradation and overexploitation, even though the region is now a protected National Park (Spalding et al. 2001).
Demographic history
Neutrality tests (Tajima’s D and Fu’s Fs) rejected the null hypothesis of neutral evolution of the utilised molecular marker at half of the sample sites (Table 1). However, these tests cannot differentiate between demographic changes in population size and selection. Therefore, Rogers’ test and mismatch distribution analysis were performed to examine the null hypothesis of sudden demographic expansion (Rogers and Harpending 1992; Rogers 1995). Such a population expansion could be explained by the availability of new habitats after the rise of the sea level at the end of glacials, which enabled growth of the reduced populations (Fauvelot et al. 2003). The same signal of a sudden population expansion was found in T. crocea from the Indo-Malay Archipelago (Kochzius and Nuryanto 2008).
Genetic population structure and connectivity
The haplotypes found in T. maxima could be assigned to four distinct clades in the haplotype network. Especially, clade 3 (Western Pacific; 44 mutations to clade 1) and clade 4 (Red Sea; 12 mutations to clade 1) are very divergent, raising the question if these are cryptic species. Recently, a new species of giant clam (T. costata) was discovered in the Red Sea (Richter et al. 2008). An integrated taxonomy approach (Dayrat 2005), comparing specimens of clades 3 and 4 to additional giant clam species, using morphology, ecology, and genetics would be needed to verify if these specimens represent cryptic species.
T. maxima shows a very strong genetic population structure across the Indo-West Pacific (Indo-Malay Archipelago and Red Sea), with a Φst-value of 0.74. The genetic population structure across the Indo-Malay Archipelago also indicates highly restricted gene flow (Φst = 0.72), which is much more restricted than in T. crocea in the same region (Φst = 0. 28; Kochzius and Nuryanto 2008). Even though a Mantel test revealed a significant correlation between genetic and geographic distance in populations across the Indo-Malay Archipelago as well as in a subset of populations from the central part, the correlation was rather weak. This shows that other factors, such as oceanography and demographic history, play a major role in shaping the genetic population structure of T. maxima.
According to the pronounced genetic differences, the sample sites can be divided into four groups from West to East: (1) Red Sea, (2) Eastern Indian Ocean and Java Sea, (3) Indonesia throughflow and seas in the East of Sulawesi, and (4) Western Pacific.
Such differentiation from West to East in the Indo-Malay Archipelago was also found in the giant clam T. crocea, but these populations also showed a separation between the Eastern Indian Ocean and Java Sea (DeBoer et al. 2008; Kochzius and Nuryanto 2008). Studies on populations of fishes (Chenoweth et al. 1998; Nelson et al. 2000; Lourie et al. 2005; Timm and Kochzius 2008), a starfish (Williams and Benzie 1998), and mantis shrimp (Barber et al. 2002, 2006) also revealed such genetic breaks in the transition of the Indian and Pacific Ocean. It is hypothesised that this differentiation between populations of the Indian and Pacific Ocean is due to low sea level during glacials, when the sea level was dropped by up to 120 m. Shallow shelf areas, such as the Java Sea on the Sunda shelf, fell dry and created more or less isolated ocean basins, restricting exchange between populations (McManus 1985; Voris 2000). The genetic patterns caused by these processes still persist and additional differentiation could be induced by major prevailing currents.
In the following section, the genetic structure found in T. maxima populations is compared in detail with current patterns and results from other studies. However, comparison with other species is sometimes difficult, because different genetic markers have been applied and there are also differences in the geographic resolution of sampling.
Red Sea The population of T. maxima in the Red Sea showed a clear genetic break to all other studied populations, revealed by a distinct clade in the haplotype network and a Φst-values of up to 0.91. Such a high Φst-value may suggest that the population in the Red Sea is a cryptic species. However, as mentioned before, further analysis in an integrative taxonomy approach would be desirable to resolve this issue.
Restricted exchange between the population in the Red Sea and populations in the Indo-Malay Archipelago could be caused by two factors. On the one hand, the geographic distance of about 10,000 km restricts gene flow. On the other hand, the exchange between the Red Sea and Indian Ocean is restricted by the shallow sill of Bab El Mandab (Siddall et al. 2002). However, additional samples from the Western Indian Ocean would be desirable to reveal a detailed picture of connectivity between the Red Sea and Indian Ocean.
A genetic differentiation between populations of the Red Sea and Indian Ocean was also found in the mud crab S. serrata (Fratini and Vannini 2002) and the sponge L. chagosensis (Wörheide et al. 2008), but could not be detected in the lionfish P. miles (Kochzius and Blohm 2005).
Eastern Indian Ocean and Java Sea Low Φst-values among populations from the Eastern Indian Ocean (Padang) and Java Sea (Pulau Seribu and Karimunjava) indicate high levels of gene flow, even though the low Φst-value between Padang and Pulau Seribua was significant (Table 2). However, clade frequencies and hierarchical AMOVA suggest that the populations from the Eastern Indian Ocean and Java Sea belong to one group with high levels of connectivity. Panmixing in the populations of the Java Sea that are more than 400 km apart could be attributed to seasonally changing currents (Fig. 1). In addition, the pelagic larval duration of 9 days (Lucas 1988) seems to be sufficient for T. maxima to travel such distance mediated by currents. Although the analysis revealed panmixing of the populations in the Java Sea, both showed a different pattern of genetic differentiation to the population from Padang. The population from Pulau Seribu was statistically different from Padang, while the population from Karimunjava was not different. This is difficult to explain, because prevailing current patterns are rather expected to prevent connectivity by passive larval drift, as revealed in populations of T. crocea from the Eastern Indian Ocean and Java Sea (Kochzius and Nuryanto 2008). The lack of significant differentiation might be an analytical problem, because due to the low Φst-value, a larger sample size might be necessary to detect significance, even though a rather high number of 20 specimens from Karimunjava have been analysed. Genetic differentiation between Padang and Pulau Seribu can be explained by the current pattern in the Eastern Indian Ocean along the western coast of Sumatra and southern coast of Java (Fig. 1). Even though the main current along the western coast of Sumatra is directed to the South-East, planktonic larvae of T. maxima probably cannot enter the Java Sea through the Sunda Strait due to the prevailing outflow of water masses from the Java Sea into the Indian Ocean (Wyrtki 1961; Hendiarti et al. 2004). Due to the south-eastern current at the western coast of Sumatra, larvae from the Java Sea that enter the Indian Ocean through the Sunda Strait cannot reach the western coast of Sumatra. In addition, seasonally changing currents parallel along the southern coast of Java are deflected at Sumba in a westward direction, preventing exchange with populations in the East (Fig. 1, Wyrtki 1961).
A genetic separation of populations from the Eastern Indian Ocean was also observed in the anemonefish A. ocellaris (Nelson et al. 2000; Timm and Kochzius 2008), scad mackerel Decapterus macrosoma (Arnaud et al. 1999), the seahorse Hippocampus spinosissimus (Lourie et al. 2005), and the mantis shrimps Haptosquilla pulchella (Barber et al. 2002, 2006). However, the Indian scad mackerel D. russelli (Rohfritsch and Borsa 2005), and the seahorses H. barbouri, H. kuda, and H. trimaculatus (Lourie et al. 2005) did not show such a genetic separation of the Eastern Indian Ocean.
Populations of the boring giant clam T. crocea (Kochzius and Nuryanto 2008), the mushroom coral H. actiniformis (Knittweis et al. 2009), and the mantis shrimp Gonodactylellus viridis (Barber et al. 2006) in the Java Sea were also genetically distinct, but such a pattern could not be detected in the Indian scad mackerel D. russelli (Rohfritsch and Borsa 2005) as well as the seahorses H. kuda, H. spinosissimus, and H. trimaculatus (Lourie et al. 2005). However, the genetic break observed in T. maxima in the eastern Java Sea (Fig. 1, Table 2) was also observed in the boring giant clam T. crocea (Kochzius and Nuryanto 2008), the Indian scad mackerel D. russelli (Rohfritsch and Borsa 2005), and the seahorse H. trimaculatus (Lourie et al. 2005).
Indonesian throughflow (ITF) and seas in the East of Sulawesi Pairwise comparison of populations along the ITF in the Sulawesi Sea at the north-eastern coast of Borneo (Sangalaki) and northern coast of Sulawesi (Manado), Makassar Strait (Spermonde) and Flores Sea (Bira) at the south-western tip of Sulawesi, as well as Komodo and Timor (Kupang) did not reveal significant Φst-values. These sites are well connected by the ITF, which reaches surface currents of up to 36 nautical miles per day (Wyrtki 1961), transporting about 10 million m3 of water per second (=10 Sverdrup; Godfrey 1996; Gordon and Fine 1996; Godfrey and Masumoto 1999; Gordon 2005; Susanto and Gordon 2005). Populations around Sulawesi mostly show panmixing, except the pairwise comparisons between Spermonde and Luwuk, Togian Islands, as well as Manado (Fig. 1, Table 2).
However, T. crocea showed a significant genetic differentiation between the western and eastern coast of Sulawesi (Kochzius and Nuryanto 2008), which is not present in T. maxima. Such a different pattern of connectivity in the two giant clam species around Sulawesi might be due to differences in larval ecology and dispersal capabilities. A lack of genetic separation around Sulawesi was also observed in the two mantis shrimp species H. glyptocercus and G. viridis (Barber et al. 2006). In contrast, studies on the mantis shrimp H. pulchella (Barber et al. 2002) and the mushroom coral H. actiniformis (Knittweis et al. 2009) revealed a genetic isolation of the populations from Tomini Bay, which is not present in T. maxima and T. crocea (Kochzius and Nuryanto 2008). With regard to the populations in Tomini Bay, the two giant clam species rather exhibit a similar pattern as the scad mackerel D. macarellus, which did not show genetic heterogeneity between populations in Tomini Bay and the Molluccas (Arnaud et al. 1999). Panmixing along the ITF at sites in the Sulawesi Sea and Makassar Strait was also found in the mushroom coral H. actiniformis (Knittweis et al. 2009), the anemonefish A. ocellaris (Nelson et al. 2000), the Indian scad mackerel D. russelli (Rohfritsch and Borsa 2005), as well as the mantis shrimps H. pulchella, H. glyptocercus, and G. viridis (Barber et al. 2006). These differences between species could be attributed to different dispersal capabilities of early life stages and adults.
Western Pacific The genetic isolation of the population from Biak off the north-western coast of New Guinea could be caused by the Halmahera eddy, which redirects the westward Southern Equatorial Current (SEC) into the North Equatorial Counter Current (NECC) to the East (Fig. 1, Wyrtki 1961; Gordon and Fine 1996). A similar pattern was also found in the above-mentioned three species of mantis shrimps, with a genetic break occurring in the Moluccas (Barber et al. 2006). Even though the sample site in the Western Pacific is geographically closer, the haplotypes from there (clade 3) are more divergent from the ones found in the central Indo-Malay Archipelago (clade 1) than haplotypes from the much more distant Red Sea (clade 4). This underlines the strong separation of the northern coast of New Guinea from the other regions of the Indo-Malay Archipelago.
This study revealed deep evolutionary lineages in the giant clam T. maxima, especially in the Red Sea and western Pacific coast of the Indo-Malay Archipelago. These populations are so divergent that they might constitute cryptic species, which should be evaluated in an integrative taxonomy approach (Dayrat 2005; Richter et al. 2008). The major observed genetic differentiation between the Red Sea and Eastern Indian Ocean as well as Western Pacific coasts of the Indo-Malay Archipelago is most probably due to vicariance, mainly caused by historical isolation by sea level changes. The complex pattern of connectivity in the Indo-Malay Archipelago, characterised by panmixing between some sites and restricted gene flow between others, can be attributed to prevailing current regimes and the geological history of the region. Current oceanographic conditions in the Indo-Malay Archipelago facilitate connectivity along the ITF, whereas prevailing current regimes in the Java Sea as well as the Halmahera eddy off north-western New Guinea prevent connectivity of populations in the East and West to the central Indo-Malay Archipelago.
These findings support the idea that the Red Sea is an important centre of evolution (Klausewitz 1989) and that the Indo-Malay Archipelago can be regarded as a centre of origin (Briggs 1999), which does not exclude additional mechanisms creating high biodiversity in the Coral Triangle.
Implications for conservation
International trade with non-captive bred giant clams increased from about 40,000 specimens in 1993 to about 100,000 in 2001 (Wabnitz et al. 2003). Similar to other giant clam species, wild stocks of T. maxima in the Indo-Malay Archipelago are heavily exploited for food, shells, and aquarium trade (Lucas 1994; Wells 1997). Enforcement of legislations is poor, even though giant clams are fully protected in Indonesia and collection, sale, and export of wild specimens is banned (Wells 1997). The coastal population consumed clam meat extensively and shells were used for different purposes, such as ornaments, basins, or production of tiles (Pasaribu 1988; Wells 1997).
Red Sea populations of giant clams are also subject to reef-top gathering, indicated by higher abundances in marine protected areas (MPAs) (Ashworth et al. 2004).
T. maxima is a popular species in the aquarium trade, due to its bright colouration patterns. The United States of America and European Union are the main importers for giant clams in the marine ornamental trade, with about 45,000 and 48,000 specimens in 2001, respectively (Wabnitz et al. 2003). T. maxima is classified in the IUCN Red List of Threatened Animals as ‘Lower Risk: Conservation Dependent’.
MPAs are considered as a helpful tool to ensure the sustainable use of living marine resources and to prevent overexploitation (Palumbi 2001). Therefore, MPAs are strongly recommended as a management tool for Indonesia’s marine fisheries (Mous et al. 2005). In order to ensure connectivity between MPAs, it is proposed that the spatial distribution should match the dispersal capabilities of the species to be protected (Palumbi 2003).
Even though 51 MPAs with an area of about 58,000 km2 in Indonesia include coral reefs (Spalding et al. 2001), these MPAs cover only about 1% of the total marine area of the Indonesian Archipelago. Due to the strong genetic population structure of T. maxima, this network seems to be not sufficient to protect species with limited larval dispersal. Since exchange between the three defined groups in the Indo-Malay Archipelago is limited, they should be managed separately and patterns of connectivity should be considered in the spatial arrangement of MPAs. In order to draw a generalised picture of connectivity among populations in the Indo-Malay Archipelago for designing a functional network of MPAs, further studies on other invertebrates and fishes are needed. However, the results of this study and others cited herein already provide preliminary information about regions that should be considered as separate management units by regional authorities.
References
Alvarado Bremer JR, Stequert B, Robertson NW, Ely B (1998) Genetic evidence for inter-oceanic subdivision of bigeye tuna (Thunnus obesus Lowe) populations. Mar Biol 132:547–557
Arnaud S, Bonhomme F, Borsa P (1999) Mitochondrial DNA analysis of the genetic relationships among populations of scads mackerel (Decapterus macarellus, D. macrosoma, and D. russelli) in South-East Asia. Mar Biol 135:699–707
Ashworth JS, Ormond RFG, Sturrock HT (2004) Effects of reef-top gathering and fishing on invertebrate abundance across take and no-take zones. J Exp Mar Biol Ecol 303:221–242
Ayala FJ, Hedgecock D, Zumwalt GS, Valentine JW (1973) Genetic variation in Tridacna maxima, an ecological analog of some unsuccessful evolutionary lineages. Evolution 27:177–191
Barber PH, Palumbi SR, Erdmann MV, Moosa MK (2002) Sharp genetic breaks among populations of Haptosquilla pulchella (Stomatopoda) indicate limits to larval transport:patterns, causes and consequences. Mol Ecol 11:659–674
Barber PH, Erdmann MV, Palumbi SR (2006) Comparative phylogeography of three codistributed stomatopods:origins and timing of regional lineage diversification in the coral triangle. Evolution 60:1825–1839
Benzie JAH (1999) Major genetic differences between crown-of-thorns starfish (Acanthaster planci) populations in the Indian and Pacific Oceans. Evolution 53:1782–1795
Benzie JAH, Williams ST (1992a) Genetic structure of giant clam (Tridacna maxima) populations from reefs in the Western Coral Sea. Coral Reefs 11:135–141
Benzie JAH, Williams ST (1992b) No genetic differentiation of giant clam (Tridacna gigas) populations in the Great Barrier Reef, Australia. Mar Biol 112:1–5
Benzie JAH, Williams ST (1995) Gene flow among giant clam (Tridacna gigas) populations in Pacific does not parallel ocean circulation. Mar Biol 123:781–787
Benzie JAH, Williams ST (1997) Genetic structure of giant clam (Tridacna maxima) populations in the west Pacific is not consistent with dispersal by present-day ocean currents. Evolution 51:768–783
Benzie JAH, Ballment E, Forbes AT, Demetriades NT, Sugama K, Haryanti Moria S (2002) Mitochondrial DNA variation in Indo-Pacific populations of the tiger prawn, Peneus monodon. Mol Ecol 11:2553–2569
Borsa P (2003) Genetic structure of round scad mackerel Decapterus macrosoma (Carangidae) in the Indo-Malay Archipelago. Mar Biol 142:575–581
Briggs JC (1999) Coincident biogeographic patterns: Indo-West Pacific Ocean. Evolution 53:326–335
Campbell CA, Valentine JW, Ayala FJ (1975) High genetic variability in a population of Tridacna maxima from the Great Barrier Reef. Mar Biol 33:341–345
Chenoweth SF, Hughes JM (2003) Oceanic interchange and nonequilibrium population structure in the estuarine dependent Indo-Pacific tasselfish, Polynemus sheridani. Mol Ecol 12:2387–2397
Chenoweth SF, Hughes JM, Keenan CP, Lavery S (1998) When oceans meet:a teleost shows secondary integration at an Indian-Pacific interface. Proc R Soc Lond B Biol Sci 265:415–420
Chow S, Okamoto H, Uozumi Y, Takeuchi Y, Takeyama H (1997) Genetic stock structure of the swordfish (Xiphias gladius) inferred by PCR-RFLP analysis of the mitochondrial DNA control region. Mar Biol 127:359–367
Chow S, Okamoto H, Miyabe N, Hiramatsu K, Barut N (2000) Genetic divergence between Atlantic and Indo-Pacific stocks of bigeye tuna (Thunnus obesus) and admixture around South Africa. Mol Ecol 9:221–227
Crandall ED, Frey MA, Grosberg RK, Barber PH (2008) Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Mol Ecol 17:611–626
Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85:407–415
De Boer TS, Subia MD, Ambariyanto Erdmann MV, Kovitvongsa K, Barber PH (2008) Phylogeography and limited genetic connectivity in the endangered giant boring clam, Tridacna crocea, across the Coral Triangle. Conserv Biol 22:1255–1266
Duda TF, Palumbi SR (1999) Population structure of the black tiger prawn, Penaeus monodon, among western Indian Ocean and western Pacific populations. Mar Biol 134:705–710
Duran S, Palacin C, Becerro MA, Turon X, Giribet G (2004) Genetic diversity and population structure of the commercially harvested sea urchin Paracentrotus lividus (Echinodermata, Echinoidea). Mol Ecol 13:3317–3328
Edinger EN, Jompa J, Limmon GV, Widjatmoko W, Risk MJ (1998) Reef degradation and coral biodiversity in indonesia:Effects of land-based pollution, destructive fishing practices and changes over time. Mar Pollut Bull 36:617–630
Erdmann M (1995) An ABC guide to coral reef fisheries in southwest Sulawesi, Indonesia. Naga, ICLARM Quarterly 18(2):4–6
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes:application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fauvelot C, Bernardi G, Planes S (2003) Reduction in the mitochondrial DNA diversity of coral reef fish provides evidence of population bottlenecks resulting from Holocene sea-level change. Evolution 57:1571–1583
Fox HE, Mous PS, Pet JS, Muljadi AH, Caldwell RL (2005) Experimental assessment of coral reef rehabilitation following blast fishing. Conserv Biol 19:98–107
Fratini S, Vannini M (2002) Genetic differentiation in the mud crab Scylla serrata (Decapoda:Portunidae) within the Indian Ocean. J Exp Mar Biol Ecol 272:103–116
Froukh T, Kochzius M (2007) Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Mol Ecol 16:1359–1367
Froukh T, Kochzius M (2008) Species boundaries and evolutionary lineages in the blue green damselfishes Chromis viridis and C. atripectoralis (Pomacentridae). J Fish Biol 72:451–457
Fu FX (1997) Statistical test of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Godfrey JS (1996) The effect of the Indonesian throughflow on ocean circulation and heat exchange with the atmosphere:A review. J Geophys Res 101:12217–12238
Godfrey JS, Masumoto (1999) Diagnosing the mean strength of the Indonesian Throughflow in an ocean general circulation model. J Geophys Res 104:7889–7895
Gordon AL (2005) Oceanography of the Indonesian seas and their throughflow. Oceanography 18:14–27
Gordon AL, Fine RA (1996) Pathways of water between the Pacific and Indian oceans in the Indonesian seas. Nature 379:146–149
Goren M, Dor M (1994) An updated checklist of the fishes of the Red Sea–CLOFRES II. Israel Academy of Sciences and Humanities, Jerusalem
Hall TA (1999) BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hassan M, Harmelin-Vivien M, Bonhomme F (2003) Lessepsian invasion without bottleneck:examples of two rabbitfish species (Siganus rivulatus and Siganus luridus). J Exp Mar Biol Ecol 291:219–232
Hendiarti N, Siegel H, Ohde T (2004) Investigation of different coastal processes in Indonesian waters using SeaWiFS data. Deep-Sea Res Pt II 51:85–97
Hoeksema BW (2007) Delineation of the Indo-Malayan centre of maximum marine biodiversity: the coral triangle. In: Renema W (ed) Biogeography, time and place: distributions, barriers and islands, Top Geobiol 29: 117–178
Hopley D, Suharsono (2000) The Status of coral reefs in eastern Indonesia. Global Coral Reef Monitoring Network (GCRMN)
Imron Jeffrey B, Hale P, Degnan BM, Degnan SM (2007) Pleistocene isolation and recent gene flow in Haliotis asinina, an Indo-Pacific vetigastropod with limited dispersal capacity. Mol Ecol 16:289–304
Ishikura M, Adachi K, Maruyama T (1999) Zooxanthellae release glucose in the tissue of a giant clam, Tridacna crocea. Mar Biol 133:665–673
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance web service. BMC Genet 6:13
Jokiel P, Martinelli FJ (1992) The vortex model of coral reef biogeography. J Biogeogr 19:449–458
Juinio-Meñez MA, Magsino RM, Ravago-Gotanco R, Yu ET (2003) Genetic structure of Linckia laevigata and Tridacna crocea populations in the Palawan shelf and shoal reefs. Mar Biol 142:717–726
Kittiwattanawong K (1997) Genetic structure of giant clam, Tridacna maxima in the Andaman Sea, Thailand. Spec Publ Phuket Mar Biol Cent 17:109–114
Kittiwattanawong K, Nugranad J, Srisawat T (2001) High genetic divergence of Tridacna squamosa living at the west and east coasts of Thailand. Spec Publ Phuket Mar Biol Cent 25:343–347
Klausewitz W (1989) Evolutionary history and zoogeography of the Red Sea ichthyofauna. Fauna of Saudi Arabia 10:310–337
Knittweis L, Krämer WE, Timm J, Kochzius M (2009) Genetic structure of Heliofungia actiniformis (Scleractinia:Fungiidae) populations in the Indo-Malay Archipelago:implications for live coral trade management efforts. Conserv Genet 10:241–249
Kochzius M, Blohm D (2005) Genetic population structure of the lionfish Pterois miles (Scorpaenidae, Pteroinae) in the Gulf of Aqaba and northern Red Sea. Gene 347:295–301
Kochzius M, Nuryanto A (2008) Strong genetic population structure in the boring giant clam Tridacna crocea across the Indo-Malay Archipelago:implications related to evolutionary processes and connectivity. Mol Ecol 17:3775–3787
Kochzius M, Söller R, Khalaf MA, Blohm D (2003) Molecular phylogeny of the lionfish genera Dendrochirus and Pterois (Scorpaenidae, Pteroinae) based on mitochondrial DNA sequences. Mol Phylogenet Evol 28:396–403
Laurent V, Planes S, Salvat B (2002) High variability of genetic pattern in giant clam (Tridacna maxima) populations within French Polynesia. Biol J Linn Soc 77:221–231
Leggat W, Buck BH, Grice A, Yellowlees D (2003) The impact of bleaching on the metabolic contribution of dinoflagellate symbionts to their giant clam host. Plant Cell Environ 26:1951–1961
Lessios HA, Kessing BD, Pearse JS (2001) Population structure and speciation in tropical seas:global phylogeography of the sea urchin Diadema. Evolution 55:955–975
Lourie SA, Green DM, Vincent ACJ (2005) Dispersal, habitat differences, and comparative phylogeography of Southeast Asian seahorses (Syngnathidae:Hippocampus). Mol Ecol 14:1073–1094
Lucas JS (1988) Giant clams: description, distribution and life history. In: Copland JW, Lucas JS (eds) Giant clams in Asia and the Pacific. Australian Centre for International Agricultural Research, Canberra, pp 21–32
Lucas JS (1994) The biology, exploitation and mariculture of giant clams (Tridacnidae). Rev Fish Sci 2:181–223
Luttikhuizen PC, Drent J, Baker AJ (2003) Disjunct distribution of highly diverged mitochondrial lineage clade and population subdivision in a marine bivalve with pelagic larval dispersal. Mol Ecol 12:2215–2229
Macaranas JM, Ablan CA, Pante MJR, Benzie JAH, Williams STH (1992) Genetic structure of giant clam (Tridacna derasa) populations from reefs in the Indo-Pacific. Mar Biol 113:231–238
Maier E, Tollrian R, Rinkevich B, Nürnberger B (2005) Isolation by distance in the scleractinian coral Seriatopora hystrix from the Red Sea. Mar Biol 147:1109–1120
McManus JW (1985) Marine speciation, tectonics and sea-level changes in Southeast Asia. Proc 5th Int Coral Reef Symp 4:133–138
Mous PJ, Pet JS, Arifin Z, Djohani R, Erdmann MV, Halim A, Knight H, Pet-Soede L, Wiadnya G (2005) Policy needs to improve marine capture fisheries management and to define a role for marine protected areas in Indonesia. Fish Manag Ecol 12:259–268
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Jin L (1989) Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol 6:290–300
Nelson JS, Hoddell RJ, Chou LM, Chan WK, Phang VPE (2000) Phylogeographic structure of false clownfish, Amphiprion ocellaris, explained by sea level changes on the Sunda shelf. Mar Biol 137:727–736
Ovenden JR, Salini JS, O’Connor, Street R (2004) Pronounced genetic population structure in a potentially vagile fish species (Pristipomoides multidens, Teleostei, Perciformes, Lutjanidae) from the East Indies triangle. Mol Ecol 13:1991–1999
Palumbi SR (2001) The ecology of marine protected areas. In: Bertness MD, Gaines SM, Hixon ME (eds) Marine Community Ecology. Sinauer Associates, Sunderland, pp 509–530
Palumbi SR (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13:S146–S158
Pasaribu BP (1988) Status of giant clams in Indonesia. In: Copland JW, Lucas JS (eds) Giant clams in Asia and the Pacific. Australian Centre for International Agricultural Research, Canberra, pp 44–46
Perrin C, Borsa P (2001) Mitochondrial DNA analysis of the geographic structure of Indian scad mackerel in the Indo-Malay Archipelago. J Fish Biol 59:1421–1426
Pet J (1997) Destructive fishing methods in and around Komodo National Park. SPC Live Reef Fish Information Bulletin 2:20–24
Posada D, Crandall KA (1998) Modeltest:testing the model of DNA substitution. Bioinformatics 14:817–818
Ravago-Gotanco RG, Magsino RM, Juinio-Menez MA (2007) Influence of the North Equatorial Current on the population genetic structure of Tridacna crocea (Mollusca:Tridacnidae) along the eastern Philippine seaboard. Mar Ecol Prog Ser 336:161–168
Reid DG, Lal K, Mackenzie-Dodds J, Kaligis F, Littlewood DTJ, Williams ST (2006) Comparative phylogeography and species boundaries in Echinolittorina snails in the central Indo-West Pacific. J Biogeogr 33:990–1006
Richter C, Roa-Quiaoit H, Jantzen C, Al-Zibdah M, Kochzius M (2008) Collapse of a new living species of giant clam in the Red Sea. Curr Biol 18:1349–1354
Ridgway T, Sampayo EM (2005) Population genetic status of the Western Indian Ocean:what do we know? Western Indian Ocean J Mar Sci 4:1–9
Risk MJ, Erdmann MV (2000) Isotopic composition of Nitrogen in stomatopod (Crustacea) tissues as an indicator of human sewage impacts on Indonesian coral reefs. Mar Pollut Bull 40:50–58
Roberts CM, McClean CJ, Veron JEN, Hawkins JP, Allen GR, McAllister DE, Mittermeier CG, Schueler FW, Spalding M, Wells F, Vynne C, Werner TB (2002) Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295:1280–1284
Rogers AR (1995) Genetic evidence for Pleistocene population expansion. Evolution 49:608–615
Rogers AL, Harpending H (1992) Population growth makes in the distribution of pairwise genetics differences. Mol Biol Evol 9:552–569
Rohfritsch A, Borsa P (2005) Genetic structure of Indian scad mackerel Decapterus russelli:Pleistocene vicariance and secondary contact in the Central Indo-West Pacific Seas. Heredity 95:315–326
Schneider S, Roessli D, Excoffier L (2000) Arlequin, version 2.000. University of Geneva, Geneva
Shefer S, Abelson A, Mokady O, Geffen E (2004) Red to Mediterranean Sea bioinvasion:natural drift through the Suez Canal, or anthropogenic transport? Mol Ecol 12:2333–2343
Siddall M, Smeed DA, Matthiesen S, Rohling EJ (2002) Modelling the seasonal cycle of the exchange flow in Bab el Mandab (Red Sea). Deep-Sea Res Pt I 49:1551–1569
Spalding MD, Ravilious C, Green EP (2001) World atlas of coral reefs. UNEP World Conservation Monitoring Centre, University of California Press, Berkeley
Sugama K, Haryanti, Benzie JAH, Ballment E (2002) Genetic variation and population structure of the giant tiger prawn, Penaeus monodon, in Indonesia. Aquaculture 205:37–48
Susanto RD, Gordon AL (2005) Velocity and transport of the Makassar Strait throughflow. J Geophysl Res 110:C01005
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Thompson JG, Higgins DG, Gibson TJ (1994) CLUSTAL W:improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Timm J, Kochzius M (2008) Geological history and oceanography of the Indo-Malay Archipelago shape the genetic population structure in the False Clown Anemonefish (Amphiprion ocellaris). Mol Ecol 17:3999–4014
Timm J, Figiel M, Kochzius M (2008) Contrasting patterns in species boundaries and evolution of anemonefishes (Amphiprioninae, Pomacentridae) in the centre of marine biodiversity. Mol Phylogenet Evol 49:268–276
Veron JEN (2000) Corals of the world. Australian Institute of Marine Science and CRR Qld Pty Ltd
Voris HK (2000) Maps of Pleistocene sea levels in Southeast Asia:shorelines, river systems and time duration. J Biogeogr 27:1153–1167
Wabnitz C, Taylor M, Green E, Razak T (2003) From ocean to aquarium. UNEP-WCMC, Cambridge, UK
Walsh PS, Metzger DA, Higushi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506–513
Wells S (1997) Giant clams:status, trade and mariculture, and the role of CITES management. IUCN, Gland, Switzerland and Cambridge, UK
Wilkinson C (2002) Status of coral reefs of the world:2002. Australian Institute of Marine Science
Williams ST, Benzie JAH (1998) Evidence of a biogeographic break between populations of a high dispersal starfish:congruent regions within the Indo-West Pacific defined by color morphs, mtDNA, and allozyme data. Evolution 52:87–99
Woodland DJ (1983) Zoogeography of the Siganidae (Pisces): an interpretation of distribution and richness pattern. Bull Mar Sci 33:713–717
Wörheide G, Epp LS, Macis L (2008) Deep genetic divergences among Indo-Pacific populations of the coral reef sponge Leucetta chagosensis (Leucettidae):Founder effects, vicariance, or both? BMC Evol Biol 8:24
Wyrtki K (1961) Physical oceanography of the Southeast Asian waters. University of California, La Jolla, USA
You E-M, Chiu T-S, Liu K-F, Tassanakajon A, Klinbunga S, Triwitayakorn K, de la Peña LD, Li Y, Yu H-T (2008) Microsatellite and mitochondrial haplotype diversity reveal population differentiation in the tiger shrimp (Peneus monodon) in the Indo-Pacific region. Anim Genet [doi:10.1111/j.1365-2052.2008.01724.x]
Yu ET, Juinio-Meñez MA, Monje VD (2000) Sequence variation in the ribosomal DNA internal transcribed spacer of Tridacna crocea. Mar Biotechnol 2:511–516
Acknowledgement
We would like to thank the institutions and individuals that have made our study possible: German Federal Ministry of Education and Research (BMBF, Grant no. 03F0390B), which funded the project ‘Molecular Genetics as a Tool for the Management of Marine Ornamentals in Sulawesi (Indonesia)’ in the framework of the joint German-Indonesian project SPICE (Science for the Protection of Indonesian Coastal Marine Ecosystems); German Academic Exchange Service (DAAD) for supporting A. Nuryanto; Centre for Tropical Marine Ecology (Bremen, Germany) for project co-ordinated, especially C. Richter; colleagues from the University of Bremen, especially J. Timm for help during field work; colleagues from Universitas Hasanuddin (Makassar, Indonesia) for logistical support in Spermonde, especially J. Jompa; anonymous reviewers for constructive comments. The SPICE project is conducted and permitted under the governmental agreement between the German Federal Ministry of Education and Research (BMBF) and the Indonesian Ministry for Research and Technology (RISTEK), Indonesian Institute of Sciences (LIPI), Indonesian Ministry of Maritime Affairs and Fisheries (DKP), and Agency for the Assessment and Application of Technology (BPPT). This work was carried out in co-operation with Hassanuddin University (UNHAS, Makassar, Indonesia), Agricultural University Bogor (IPB, Bogor, Indonesia), and Jenderal Soedirman University (Purwokerto, Indonesia).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Biology Editor Dr. Ruth Gates
Rights and permissions
About this article
Cite this article
Nuryanto, A., Kochzius, M. Highly restricted gene flow and deep evolutionary lineages in the giant clam Tridacna maxima . Coral Reefs 28, 607–619 (2009). https://doi.org/10.1007/s00338-009-0483-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-009-0483-y