Abstract
Closely related substrains of inbred mice often show phenotypic differences that are presumed to be caused by recent mutations. The substrains BALB/cJ and BALB/cByJ, which were separated in 1935, have been reported to show numerous highly significant behavioral and morphological differences. In an effort to identify some of the causal mutations, we phenotyped BALB/cJ and BALB/cByJ mice as well as their F1, F2, and N2 progeny for behavioral and morphological phenotypes. We also generated whole-genome sequence data for both inbred strains (~3.5× coverage) with the intention of identifying polymorphic markers to be used for linkage analysis. We observed significant differences in body weight, the weight of the heart, liver, spleen and brain, and corpus callosum length between the two substrains. We also observed that BALB/cJ animals showed greater anxiety-like behavior in the open field test, less depression-like behavior in the tail suspension test, and reduced aggression compared to BALB/cByJ mice. Some but not all of these physiological and behavioral results were inconsistent with prior publications. These inconsistencies led us to suspect that the differences were due to, or modified by, non-genetic factors. Thus, we did not perform linkage analysis. We provide a comprehensive summary of the prior literature about phenotypic differences between these substrains as well as our current findings. We conclude that many differences between these strains are unstable and therefore ill-suited to linkage analysis; the source of this instability is unclear. We discuss the broader implications of these observations for the design of future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inbred laboratory animals provide an efficient system for elucidating gene–behavior relationships because they allow for the accumulation and integration of data collected over space and time in subjects with identical genotypes. As such, inbred strains are a staple of model organism research. One limitation of inbred strains is the occurrence of new mutations. Such mutations have the potential to cause an inbred strain’s phenotype to change over time. When a colony of inbred animals are separated and maintained as two isolated breeding populations, they are considered to be substrains. This acknowledges that heterozygosity in the parent population may be differentially fixed and/or that different mutations will accumulate in each population isolate. These differences can be problematic when substrains are erroneously treated as being interchangeable, as has been the case with substrains of 129 and C57BL/6 mice (Cook et al. 2002; Bryant et al. 2008). However, the existence of closely related substrains can also been viewed as an opportunity. Since there are relatively few genetic differences between them, identification of causal alleles is somewhat analogous to the identification of ENU-induced mutations (Bull et al. 2013) or to the identification of de novo mutations in humans (Koboldt et al. 2013). Linkage mapping, in conjunction with next-generation sequencing, can be used to rapidly identify mutant alleles that give rise to marked phenotypic differences (Takahashi et al. 2008; Kumar et al. 2013).
Substrains of BALB/c inbred mice, originally derived from the Bagg albino strain, have been widely used over much of the past century. BALB/c mice are commonly used to study neuropsychiatric phenotypes; they exhibit aggressive, anxious, and stress-reactive behaviors and are sensitive to certain effects of chronic antidepressant treatment (Potter 1985; Dulawa et al. 2004; Crowley et al. 2005, 2006; Englander et al. 2005; Sankoorikal et al. 2006; Holick et al. 2008; Norcross et al. 2008; Poulter et al. 2010; Jiao et al. 2011; Mehta and Schmauss 2011; Savignac et al. 2011; Wang et al. 2011; Li et al. 2012; Vijayvargiya et al. 2013). Early reports described a highly aggressive phenotype in the BALB/cJ male as compared to BALB/cByJ males, and reported that this phenotype was transmitted in F1, F2, and N2 generations in manner that was consistent with a single recessive allele (Ciaranello et al. 1974; Kessler et al. 1977). Several other groups have examined this trait since then (Velez et al. 2010; Eppig et al. 2012).
We compiled a list of phenotypes that had been reported to be significantly different between the two substrains and examined a subset of them in BALB/cJ and BALB/cByJ mice and among their F1, F2, and N2 offspring. We also performed whole-genome re-sequencing in order to identify enough polymorphic markers to allow for linkage mapping in the F2 and N2 offspring.
Materials and methods
Animals
All experiments were performed in accordance with the National Institutes of Health (NIH) guidelines for the care and use of laboratory animals and approved by the University of Chicago’s Institutional Animal Care and Use Committee. Subjects were inbred male and female BALB/cJ and BALB/cByJ mice obtained from the Jackson Laboratory (JAX; N = 13 BALB/cJ/sex; N = 12 BALB/cByJ/sex). Two BALB/cJ males were removed from the study for health reasons and were not included in aggression testing and morphological data. All mice were born on the same day ± 1 week. We also bred and tested F1 (N = 65), F2 (N = 125), and N2 (backcross to BALB/cJ, N = 112) male and female mice. F1 mice were produced using eight breeding pairs equally balanced for sex of the BALB/cJ parent. F2 crosses were produced using 11 breeding pairs of F1 mice; the F2s represented all possible combinations of F1s. Five F2 breeding pairs consisted of F1 females derived from female cJ and male cByJ males (cJxcByJ F1) crossed with F1 males derived from female cByJ and male cJ males (cByJxcJ F1). Two breeding pairs consisted of cByJxcJ F1 males crossed with cJxCByJ F1 females. Three F2 breeding pairs were produced by intercrossing male and female cJxcByJ F1s. Finally, one F2 breeding pair was produced by intercrossing male and female cByJxcJ F1s. N2 were produced using ten breeding pairs of F1xBALB/cJ mice equally balanced for sex of the BALB/cJ parent. All mice were housed in clear plastic cages with absorbent corn cob bedding in groups of 3–5 mice of the same sex and strain with food and water available ad libitum. Colony rooms were maintained on a 12:12 h light–dark cycle with lights on at 0630 h. Behavioral testing except for the resident-intruder (RI) test (see below) was conducted during the light phase between 0900 and 1600 h. For RI, the light cycle was off at 1200 h, and testing was conducted between 1230 h and 1600 h. All animals underwent the same sequence of behavioral testing: open field test (OFT), tail suspension test (TST), and RI test when applicable. BALB/cJ and BALB/cByJ mice were 78 days ± 1 week old at first testing. At least 1 week rest was given between OFT and TST, and 2 weeks rest was given before undergoing single housing prior to resident-intruder testing. For RI testing, an equal number of male intruder C57BL/6 J mice were obtained from JAX with weights <90 % of the residents. Mice were acclimated to the testing room in their home cages for 30 min prior to the start of all behavioral tests.
Open field test (OFT)
The protocol for measuring locomotor activity in an open field chamber has been previously described in detail (Bryant et al. 2009). Briefly, activity was measured using automated chambers (Versamax, AccuScan Instruments, Columbus, OH, USA) that monitored infrared beam breaks and converted data into distance traveled. Activity chambers were housed within sound-attenuating chambers with 80 lux overhead lighting and a rear wall fan providing masking of background noise. Mice were placed in chambers and allowed to freely explore for 30 min. After testing, mice were placed in their home cages and returned to the vivarium. The data reported are average values from six 5 min bins.
Tail suspension test (TST)
Mice were suspended using a small strip of adhesive tape attached to the distal third of their tail to a horizontal bar 30 cm above a soft surface. Mice were suspended for 6 min, removed and placed back in their home cages. Immobility behavior was automatically scored from saved videos by the Noldus Ethovision XT v. 5.1 software (Noldus Information Technology, Lessburg, VA, USA). The software was calibrated so that immobility was scored when the animal showed <2 % movement from one frame to the next, which corresponded behaviorally to passive suspension without voluntary movement.
Resident-intruder (RI) test
Procedures were performed strictly as in a previous manuscript (Velez et al. 2010). Prior isolation has been reported to elicit higher levels of aggression in BALB/cJ mice compared to BALB/cByJ mice in the resident-intruder paradigm (Miczek and O’Donnell 1978; Velez et al. 2010). Therefore, males were placed in individual cages 2 weeks after TST and remained individually housed for 4 weeks. During routine cage changes, only half the bedding was removed in order to maintain the cage as the resident’s territory. Intruder mice were C57BL/6 J males which were ~90 % the body weight of the residents. During isolation, mice were placed in a 12:12 light–dark cycle in which lights went off at 1200 h. Testing was conducted during the beginning of the dark cycle (between 1300 and 1700 h) when aggression levels were expected to be high. The test was conducted twice a week with 2–3 days rest in between sessions for 2 weeks, for a total of 4 sessions. RI testing was conducted under a red light only and videotaped using additional infrared illumination. After a 30 min acclimation, resident mice were placed in sound-attenuated, insulated boxes in their individual home cages. The intruder was introduced to the cage. Once the intruder had been introduced into the resident’s cage, an observer watched for an attack, defined as a bite directed at the back or flanks of the intruder. The test was concluded 5 min after the first attack or, if no attack occurred, 5 min after the intruder was placed into the cage. Residents were then returned to the vivarium in their home cages. The same intruder was paired with the same resident for each session. F2 and N2 males were not tested in RI.
These procedures differ from the aggression testing of BALB/c substrains conducted 40 years ago by Ciaranello et al. (1974); in that study, mice were single-housed mice for two rather than 4 weeks, the resident’s tail was pinched before testing, and BALB/cN mice were used as intruders.
Morphological measures
Mice were sacrificed by cervical dislocation and whole body weight was recorded to the nearest 0.1 g. The brain, heart, left lobe of the liver, left kidney, and spleen were quickly removed, cleaned of connective tissue and blotted for excess blood, and weighed to the nearest 0.1 mg. Brain was collected into 15 mL 10 % phosphate buffered formalin (Fisher Scientific). BALB/cJ and BALB/cByJ mice were sacrificed at 161 days of age ± 1 week.
Gold chloride staining for corpus callosum
Whole brains were stained using a protocol developed for staining white matter in blocks of tissue as previously described (Wahlsten et al. 2003). Briefly, individual brains were removed from fixative and bisected with a razor blade at the midsagittal plane. The brain was then immersed in 10 mL 0.2 % gold chloride in phosphate buffer at room temperature. Staining was complete after 15 min or when commissures were clearly visible with little background. The brain was removed and immediately placed in 10 mL 2.5 % sodium thiosulfate anhydrous (Fisher Scientific) for 5 min, removed, and stored in fresh 10 % formalin (Fisher Scientific). Brains were digitally scanned using an Epson Stylus NX420 scanner (Epson, Long Beach, CA, USA). Corpus callosum (CC) length was measured using arbitrary units in ImageJ (ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA) along a straight line from the tip of the genu to the tip of the splenium so as to maximize the length of the line, and normalized to body weight. Others have often used a criterion of <0.7 of a calculated expected CC value to qualify as abnormally small (Bohlen et al. 2012). As our study did not have a wide variety of brain sizes, we modified this criterion to include brains with <0.65 of the mean.
Genotyping a CNV on Chr 17
We previously characterized a ~475 kb tandem duplication on chromosome 17 (Williams et al. 2009). The duplication is located on Chr 17 from 30,174,390 to 30,651,226 Mb (build 36) and encompasses full copies of Glo1 and Dnahc8 and partial copies of Glp1r and Btbd9. While BALB/cByJ and BALB/cJ mice share the same haplotype at this location, only the BALB/cByJ substrain still has this duplication as BALB/cJ mice have undergone reversion to the non-duplicated state (see Williams et al. 2009). In order to assure ourselves that the mice were properly identified, genomic DNA was extracted from spleens of BALB/cByJ and BALB/cJ male and female mice using standard methods. PCR was conducted with a primer pair that spans the duplication boundary and only produces a product if duplication is present. The primers used were as follows: positive control amplifies in both substrains: CAGTCGTCGACAGTCATCGT and GAGCTGAAGGGATCTGCAAC, and amplifies only when duplication is present: CTCTGCCCCAGAGAACAGTC and TGATAGAGGCCACACAGCAG. Cycling conditions were as follows: initial denaturation at 95 °C, 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min; and final extension at 72 °C for 5 min. PCR products were electrophoresed on 1.5 % agarose gel.
Preliminary whole-genome sequencing
BALB/cJ and BALB/cByJ genomic libraries (one sample per substrain) were generated according to the Illumina TruSeq protocol. We sequenced these libraries on two lanes of an Illumina HiSeq 2000 using single end 100 bp reads to obtain ~110 M reads per strain. Sequencing reads for each strain were mapped to the mm9 mouse reference genome using BWA (version 0.5.9) (Li and Durbin 2009). Single nucleotide polymorphisms (SNPs) and insertions and deletions (indels) were then called for each strain compared to the mm9 reference using Samtools (Li et al. 2009).
Statistics
BALB/cJ and BALB/cByJ data were analyzed independently from F1, F2, and N2 data as they were tested in different cohorts. Data were analyzed first by two-way ANOVA with sex and strain as factors. If there was no significant effect of sex, then data were pooled and analyzed by two-tailed t test or one-way ANOVA as appropriate. Resident-intruder data were analyzed by repeated measures two-way ANOVA with session and strain as factors. Post hoc comparisons were made using Bonferroni adjustment. Significance was set at p < 0.05.
Results
Genotyping at a CNV on Chromosome 17
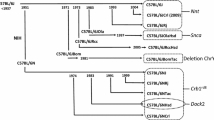
The BALB/cJ and BALB/cByJ substrains were separated in 1935 (JAX Notes 1990) (Fig. 1a). In a prior study, we obtained BALB/c DNA samples taken from BALB/c animals between 1982 and 2006 to evaluate their haplotypes at a CNV duplication on Chr 17. We found that the BALB/cByJ and BALB/cJ substrains both belong to a common duplication-containing haplotype, yet BALB/cJ did not contain the duplication. This haplotype structure suggests that non-allelic homologous recombination (unequal crossing over) occurred in BALB/cJ, explaining the reversion to a non-duplicated state (Williams et al. 2009). Here, we used this CNV duplication on Chr 17 as a genetic marker to confirm genotypes of all BALB/cJ and BALB/cByJ mice at the end of the study (Fig. 1b).
a The BALB/cJ and BALB/cByJ substrains were separated in 1935 and were maintained at JAX and NIH, respectively. BALB/cByJ obtained its name from Dr. D.W. Bailey who acquired NIH BALB/c mice in 1961. b PCR from genomic DNA of male animals that were phenotyped (see Fig. 2–4) showing the presence (cByJ) or absence (cJ) of a duplication on Chr 17 that has previously been reported. Genotyping of females gave equivalent results (not shown).
Open field test (OFT)
There was no difference in total locomotor activity in the open field test between the substrains [t(47) = 1.87, p > 0.05] (Fig. 2a), and there was no interaction between substrain and activity during the six individual time bins [F(5, 235) = 2.1; p > 0.05]. There were no differences among F1, F2, and N2 offspring in total distance traveled (Fig. 2a). However, BALB/cJ mice exhibited fewer entries to the center region compared to BALB/cByJ mice [t(45) = 2.83, p < 0.01] (Fig. 2b); and BALB/cJ also traveled significantly less distance than BALB/cByJ in the center region of the field [t(46) = 2.84, p < 0.01] (Fig. 2c). There were no differences among F1, F2, and N2 mice in total distance traveled or in number of entries into the center region (Fig. 2e, f). F2 mice traveled more distance in the center region than F1 mice [F(2, 293) = 5.5, p < 0.01] (Fig. 2g).
a–c Behavior in the open field and d tail suspension tests for BALB/cJ (cJ) and BALB/cByJ (cByJ) male and female mice (N=23-24/strain). e–h Right panels contains data from the same tests in F1, F2, and N2 mice (N=65, 125, 112, respectively). Data are mean +/- s.e.m. *p<0.05, **p<0.01. M, male. F, female.
Tail suspension test (TST)
BALB/cJ and BALB/cByJ mice are favored for environmental, pharmacological, and developmental studies of depression-like behavior. Prior reports about depression-like behavior have disagreed about the differences between these two substrains (Supplementary Table 3). In the present study, females exhibited more immobility than males in both substrains [F(1, 39) = 5.2, p < 0.05] and in the F1, F2, and N2 crosses [F(1, 271) = 15.1, p < 0.001]. BALB/cByJ mice were more immobile than BALB/cJ [F(1, 39) = 4.2, p < 0.05] (Fig. 2d). F2 mice exhibited more immobility than either F1 or N2 mice [F(2, 271) = 18.1, p < 0.001] (Fig. 2h).
Resident-intruder (RI) test
BALB/cJ male mice have been used to study aggression since the 1970s. They have been reported to exhibit high levels of attack behavior compared to BALB/cByJ males (Ciaranello et al. 1974; Kessler et al. 1977; Couppis et al. 2008; Velez et al. 2010; Dow et al. 2011). Here, we used the RI paradigm to measure aggression. Both the BALB/cJ and BALB/cByJ males (the residents) showed decreased latencies to attack the intruder over the sessions 1–4 [F(3, 63) = 4.2, p < 0.01] (Fig. 3a), and the number of attacks on the intruder increased over sessions 1–4 [F(3, 63) = 4.1, p < 0.05)] (Fig. 3b). However, surprisingly, BALB/cByJ males had a lower attack latency than BALB/cJ males [strain: F(1, 21) = 9.6, p < 0.01)] and a higher number of attacks compared to BALB/cJ males [strain: F(1, 21) = 9.6, p < 0.01)] (Fig. 3a, b). This was opposite to the difference that has been reported by many prior publications (Ciaranello et al. 1974; Kessler et al. 1977; Couppis et al. 2008), including a relatively recent study by our lab (Velez et al. 2010).
Corpus callosum (CC) length
Brain size and CC defects have been hypothesized to be related to antisocial behavior of BALB/c mice (Fairless et al. 2008; Li et al. 2012). Here, we found that BALB/cJ mice had significantly shorter CCs normalized to brain weight (BALB/cJ: 2.9 ± 0.24; BALB/cByJ: 3.2 ± 0.35), [t(43) = 3.1, p < 0.01] (Fig. 4). One BALB/cJ and two BALB/cByJ corpus callosa were abnormally small (60.2, 59.3, and 39.7 % of the mean, respectively; Fig. 4b). No differences were observed in the normalized CC area between the strains (data not shown).
a Representative male brains showing typical and abnormally small corpus callosa from BALB/cJ and BALB/cByJ adult mice. b BALB/cJ mice had shorter CCs compared to BALB/cByJ mice. Whole brains were bisected along the mid-sagittal plane and stained with 0.2% gold chloride solution to visualize white matter fibers. Corpus callosum length was defined from the tip of splenium to the tip of genu (arrows).
Organ weights
There were no differences between strains in total body weight. We found significant strain differences in brain, liver, male heart, and female spleen (Table 1), where BALB/cByJ mice had heavier organ weights with the exception of the brain, for which the BALB/cByJ were lighter than BABL/cJ.
Comparison to prior results
The behavioral results from the present study are summarized and compared to prior studies (Supplementary Tables 1–4). Findings for heart, liver, spleen, body, brain, and corpus callosum length were consistent with past reports (Supplementary Table 5).
Preliminary whole-genome sequencing
Roughly, 100 M reads were uniquely mapped per substrain, resulting in coverage of ~3.5x per substrain (Supplementary Table 6). Approximately, 450 K SNPs and 300 K indels were called per substrain (Supplementary Table 7). Called SNPs tended to be of low quality, likely due to the low read depth. The empirical cumulative distribution function (CDF) for read depth and PHRED quality per called SNP showed that ~60 % of SNPs per strain were supported by three or fewer reads and had a PHRED quality score <50 (Supplementary Fig. 1A, B). To obtain a high-quality set of SNPs for comparison between strains, SNPs were filtered for read depths of at least ten reads and PHRED quality scores of at least 100. This filtering yielded roughly 1500 SNPs per strain (included in this manuscript as supplementary files), of which ~300 were common to both substrains. The majority (>90 %) of these SNPs were not found within the Wellcome Trust SNP reference panel for 17 inbred lines (Supplementary Table 8) (Keane et al. 2011; Yalcin et al. 2011). These preliminary data are available upon request, and might be useful for identifying a panel of SNPs that could be used for linkage mapping in these strains.
Discussion
We examined a number of previously reported substrain differences between the BALB/cJ and BALB/cByJ substrains. We also took initial steps toward mapping the causal loci, including using next-generation sequencing to identify polymorphic markers and producing F1, F2, and N2 offspring. We replicated substrain differences in heart, liver, and spleen, and brain weight, and confirmed a low incidence of abnormally small corpus callosum in both BALB/c substrains. We also made several observations that were inconsistent with the existing literature, including that BALB/cJ mice showed more anxiety-like behavior, less depression-like behavior, and less aggression than the BALB/cByJ substrain. The third finding was an especially stark departure from the long-standing literature reporting high levels of aggression in BALB/cJ male mice (Ciaranello et al. 1974; Kessler et al. 1977; Velez et al. 2010). We note that certain parameters of the tests may be different among the studies and are sometimes unknown (e.g., Ciaranello et al. did not specify whether aggression testing occurred during light or dark cycle). Furthermore, several of the behavioral differences we observed were not as robust as expected, and the F2 and N2 crosses did not show the expected degree or pattern of variation. Given these findings, we did not carry out our original plan to perform linkage mapping for these traits. We have provided detailed tables summarizing prior findings on substrain differences for the phenotypes in question. Our data do not identify the reasons for the observed inconsistencies. Possible explanations include 1) incomplete fixation of the causal genetic factors such that the strains are not fully inbred at relevant loci; 2) non-genetic sources of variation, whether epigenetic or environmental, perhaps including litter effects; or 3) fundamental problems with the assays that preclude replication (e.g., Crabbe et al. 1999). Our findings do not support the use of these substrains for linkage mapping for the phenotypes we tested, and may suggest caution for the use of other pairs of sister strains for similar purposes.
We are not the first group to observe inconsistent behavioral differences between BALB/cJ and BALB/cByJ mice. We have prepared supplemental tables that highlight all prior phenotypic studies of these two substrains. Notably, less than half of cross-study findings are consistent even when discrepant sex differences are not included as conflicts (Supplementary Tables 1–4). One study reported inconsistencies for depression-like behavior between two experiments reported in the same paper (Jiao et al. 2011). The most surprising finding of the current study, that BALB/cJ males were less aggressive than BALB/cByJ, is not unprecedented; an earlier paper showed no difference in aggressive behavior toward a NIH Swiss mouse in the RI paradigm between the BALB/cJ and BALB/cByJ males (Hilakivi and Lister 1989). Another recent experiment using the social approach test found that BALB/cJ attacked less than C57BL/6 J males, and that the BALB/cJ mice had similar levels of social behavior as compared to B6 mice (Li et al. 2012). Finally, others have seen that certain BALB/cJ mice are highly sociable, while others are not (Sankoorikal et al. 2006; Fairless et al. 2008). In terms of morphological differences, the anatomy of the corpus callosum, which has been suggested to underlie social behavior, is also highly variable (see Supplementary Table 5). In 1974, BALB/cJ mice were reported to have an absence of the corpus callosum (Wahlsten 1974). In 1977, BALB/c mice were reported to show an 11 % incidence of corpus callosum defects. In 1989, they were reported to have a 20 % incidence of corpus callosum defects (Wahlsten 1989a, b). More recently, all BALB/c brains were not found to have any corpus callosum deficits (Bohlen et al. 2012). It is not clear whether behavioral and morphological differences are correlated with each other, nor is it clear what might underlie these inconsistencies.
There are several potential explanations for the striking discordance among comparisons of the BALB/cJ and BALB/cByJ substrains. One possible explanation for the marked inconsistencies is that there are polymorphisms that have not reached fixation in the BALB/c substrains. A related possibility is that these substrains exhibit genetic instability, whether due to copy number variants (Williams et al. 2009), transposable elements (Palmer and Dulawa 2010), somatic mosaicism, or a high rate or of sporadic germline mutations. It is also possible that stable genetic polymorphisms interact with environmental or purely stochastic factors to produce incomplete penetrance. While these phenomena are not amenable to linkage analysis, they are potentially worth of study using other approaches (Tarantino et al. 2011). In addition to genetic causes, epigenetic factors could underlie shifting strain differences such as those observed in the present study. Our studies were not designed to examine trans-generation heritability of these behavioral differences, but such studies, perhaps in combination with examination of epigenetic markers, might provide further insights. Finally, it is known that some behavioral assays are stable, while others show high variability due to a variety of known and unknown experimental factors (Crabbe et al. 1999; Chesler et al. 2002; Fonio et al. 2012). Thus, the non-replication we observed may be phenotype-specific and we cannot rule out the possibility that these strains could be used successfully to map more stable phenotypes such as alcohol preference (Blizard et al. 2004). While the observed inconsistency was undesirable for the linkage study we had planned, these two substrains offer an exciting opportunity to explore factors that likely confound both model organism and human genetic studies.
References
Blizard DA, Vandenbergh DJ, Jefferson AL et al (2004) Effects of periadolescent ethanol exposure on alcohol preference in two BALB substrains. Alcohol 34:177–185. doi:10.1016/j.alcohol.2004.08.007
Bohlen MO, Bailoo JD, Jordan RL, Wahlsten D (2012) Hippocampal commissure defects in crosses of four inbred mouse strains with absent corpus callosum. Genes Brain Behav 11:757–766. doi:10.1111/j.1601-183X.2012.00802.x
Bryant CD, Zhang NN, Sokoloff G et al (2008) Behavioral differences among C57BL/6 substrains: implications for transgenic and knockout studies. J Neurogenet 22:315–331. doi:10.1080/01677060802357388
Bryant CD, Graham ME, Distler MG et al (2009) A role for casein kinase 1 epsilon in the locomotor stimulant response to methamphetamine. Psychopharmacology 203:703–711. doi:10.1007/s00213-008-1417-z
Bull KR, Rimmer AJ, Siggs OM et al (2013) Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS Genet. doi:10.1371/journal.pgen.1003219
Chesler EJ, Wilson SG, Lariviere WR et al (2002) Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci Biobehav Rev 26:907–923. doi:10.1016/S0149-7634(02)00103-3
Ciaranello RD, Lipsky A, Axelrod J (1974) Association between fighting behavior and catecholamine biosynthetic enzyme activity in two inbred mouse sublines. Proc Natl Acad Sci U S A 71:3006–3008. doi:10.1073/pnas.71.8.3006
Cook MN, Bolivar VJ, McFadyen MP, Flaherty L (2002) Behavioral differences among 129 substrains: implications for knockout and transgenic mice. Behav Neurosci 116:600–611. doi:10.1037/0735-7044.116.5.756
Couppis MH, Kennedy CH, Stanwood GD (2008) Differences in aggressive behavior and in the mesocorticolimbic DA system between A/J and BALB/cJ mice. Synapse 62:715–724. doi:10.1002/syn.20545
Crabbe JC, Wahlsten D, Dudek BC (1999) Genetics of mouse behavior: interactions with laboratory environment. Science 284:1670–1672. doi:10.1126/science.284.5420.1670
Crowley JJ, Blendy JA, Lucki I (2005) Strain-dependent antidepressant-like effects of citalopram in the mouse tail suspension test. Psychopharmacology 183:257–264. doi:10.1007/s00213-005-0166-5
Crowley JJ, Brodkin ES, Blendy JA et al (2006) Pharmacogenomic evaluation of the antidepressant citalopram in the mouse tail suspension test. Neuropsychopharmacology 31:2433–2442. doi:10.1038/sj.npp.1301065
Dow HC, Kreibich AS, Kaercher KA et al (2011) Genetic dissection of intermale aggressive behavior in BALB/cJ and A/J mice. Genes Brain Behav 10:57–68. doi:10.1111/j.1601-183X.2010.00640.x
Dulawa SC, Holick KA, Gundersen B, Hen R (2004) Effects of chronic fluoxetine in animal models of anxiety and depression. Neuropsychopharmacology 29:1321–1330. doi:10.1038/sj.npp.1300433
Englander MT, Dulawa SC, Bhansali P, Schmauss C (2005) How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci 25:648–651. doi:10.1523/JNEUROSCI.3895-04.2005
Eppig JT, Blake J a, Bult CJ et al (2012) The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res 40:D881–D886. doi:10.1093/nar/gkr974
Fairless AH, Dow HC, Toledo MM et al (2008) Low sociability is associated with reduced size of the corpus callosum in the BALB/cJ inbred mouse strain. Brain Res 1230:211–217. doi:10.1016/j.brainres.2008.07.025
Fonio E, Golani I, Benjamini Y (2012) Measuring behavior of animal models: faults and remedies. Nat Methods 9:1167–1170. doi:10.1038/nmeth.2252
Hilakivi LA, Lister RG (1989) Comparison between BALB/cJ and BALB/cByJ mice in tests of social behavior and resident-intruder aggression. Aggress Behav 15:273–280
Holick KA, Lee DC, Hen R, Dulawa SC (2008) Behavioral effects of chronic fluoxetine in BALB/cJ mice do not require adult hippocampal neurogenesis or the serotonin 1A receptor. Neuropsychopharmacology 33:406–417. doi:10.1038/sj.npp.1301399
JAX Notes (1990) A brief history of the two substrains of BALB/c, BALB/cJ, and BALB/cByJ. Available from Animal Resources. 443. http://jaxmice.jax.org/jaxnotes/archive/443a.html
Jiao J, Nitzke A, Doukas D et al (2011) Antidepressant response to chronic citalopram treatment in eight inbred mouse strains. Psychopharmacology 213:509–520
Keane TM, Goodstadt L, Danecek P et al (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. doi:10.1038/nature10413
Kessler S, Elliott GR, Orenberg EK, Barchas JD (1977) A genetic analysis of aggressive behavior in two strains of mice. Behav Genet 7:313–321
Koboldt DC, Steinberg KM, Larson DE et al (2013) The next-generation sequencing revolution and its impact on genomics. Cell 155:27–38. doi:10.1016/j.cell.2013.09.006
Kumar V, Kim K, Joseph C et al (2013) C57BL/6 N mutation in Cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342:1508–1512. doi:10.1126/science.1245503
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. doi:10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAM tools. Bioinformatics 25:2078–2079. doi:10.1093/bioinformatics/btp352
Li H, Dow HC, Tan J et al (2012) Sociability and brain development in BALB/cJ and C57BL/6 J mice. Behav Brain Res 228:299–310. doi:10.1016/j.bbr.2011.12.001
Mehta M, Schmauss C (2011) Strain-specific cognitive deficits in adult mice exposed to early life stress. Behav Neurosci 125:29–36. doi:10.1037/a0021952
Miczek KA, O’Donnell JM (1978) Intruder-evoked aggression in isolated and nonisolated mice: effects of psychomotor stimulants and L-dopa. Psychopharmacology 57:47–55. doi:10.1007/BF00426957
Norcross M, Mathur P, Enoch AJ et al (2008) Effects of adolescent fluoxetine treatment on fear-, anxiety- or stress-related behaviors in C57BL/6 J or BALB/cJ mice. Psychopharmacology 200:413–424. doi:10.1007/s00213-008-1297-2
Palmer AA, Dulawa SC (2010) Murine Warriors or Worriers: the saga of comt1, B2 SINE elements, and the future of translational genetics. Front Neurosci 4:177. doi:10.3389/fnins.2010.00177
Potter M (1985) History of the BALB/c family. Curr Top Microbiol Immunol 122:1–5
Poulter MO, Du L, Zhurov V et al (2010) Plasticity of the GABA(A) receptor subunit cassette in response to stressors in reactive versus resilient mice. Neuroscience 165:1039–1051. doi:10.1016/j.neuroscience.2009.11.028
Sankoorikal GMV, Kaercher KA, Boon CJ et al (2006) A mouse model system for genetic analysis of sociability: C57BL/6 J versus BALB/cJ inbred mouse strains. Biol Psychiatry 59:415–423. doi:10.1016/j.biopsych.2005.07.026
Savignac HM, Dinan TG, Cryan JF (2011) Resistance to early-life stress in mice: effects of genetic background and stress duration. Front Behav Neurosci 5:13. doi:10.3389/fnbeh.2011.00013
Takahashi JS, Shimomura K, Kumar V (2008) Searching for genes underlying behavior: lessons from circadian rhythms. Science 322:909–912. doi:10.1126/science.1158822
Tarantino LM, Sullivan PF, Meltzer-Brody S (2011) Using animal models to disentangle the role of genetic, epigenetic, and environmental influences on behavioral outcomes associated with maternal anxiety and depression. Front Psychiatry 2:44. doi:10.3389/fpsyt.2011.00044
Velez Lady, Sokoloff G, Miczek KA et al (2010) Differences in aggressive behavior and DNA copy number variants between BALB/cJ and BALB/cByJ substrains. Behav Genet 40:201–210. doi:10.1007/s10519-009-9325-5
Vijayvargiya N, Fairless AH, Abel T et al (2013) Development of home cage social behaviors in BALB/cJ vs. C57BL/6 J mice. Behav Brain Res 237:338–347. doi:10.1016/j.bbr.2012.08.051
Wahlsten D (1974) Heritable aspects of anomalous myelinated fibre tracts in the forebrain of the laboratory mouse. Brain Res 68:1–18. doi:10.1016/0006-8993(74)90530-7
Wahlsten D (1989a) Genetic and developmental defects of the mouse corpus callosum. Experientia 45:828–838. doi:10.1007/BF01954057
Wahlsten D (1989b) Deficiency of the corpus callosum: incomplete penetrance and substrain differentiation in BALB/c mice. J Neurogenet 5:61–76. doi:10.3109/01677068909167265
Wahlsten D, Colbourne F, Pleus R (2003) A robust, efficient and flexible method for staining myelinated axons in blocks of brain tissue. J Neurosci Methods 123:207–214. doi:10.1016/S0165-0270(02)00359-X
Wang L, Jiao J, Dulawa SC (2011) Infant maternal separation impairs adult cognitive performance in BALB/cJ mice. Psychopharmacology 216:207–218. doi:10.1007/s00213-011-2209-4
Williams R, Lim JE, Harr B et al (2009) A common and unstable copy number variant is associated with differences in Glo1 expression and anxiety-like behavior. PLoS ONE 4:e4649. doi:10.1371/journal.pone.0004649
Yalcin B, Wong K, Agam A et al (2011) Sequence-based characterization of structural variation in the mouse genome. Nature 477:326–329. doi:10.1038/nature10432
Acknowledgments
This study was supported by R01MH079103 to AAP, R25GM066522 to CBC, and a Samsung Scholarship to CJ.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sittig, L.J., Jeong, C., Tixier, E. et al. Phenotypic instability between the near isogenic substrains BALB/cJ and BALB/cByJ. Mamm Genome 25, 564–572 (2014). https://doi.org/10.1007/s00335-014-9531-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-014-9531-1