Abstract
The Tmhs gene codes for a tetraspan transmembrane protein that is expressed in hair cell stereocilia. We previously showed that a spontaneous missense mutation of Tmhs underlies deafness and vestibular dysfunction in the hurry-scurry (hscy) mouse. Subsequently, mutations in the human TMHS gene were shown to be responsible for DFNB67, an autosomal recessive nonsyndromic deafness locus. Here we describe a genetically engineered null mutation of the mouse Tmhs gene (Tmhs tm1Kjn) and show that its phenotype is identical to that of the hscy missense mutation, confirming the deleterious nature of the hscy cysteine-to-phenylalanine substitution. In the targeted null allele, the Tmhs promoter drives expression of a lacZ reporter gene. Visualization of β-galactosidase activity in Tmhs tm1Kjn heterozygous mice indicates that Tmhs is highly expressed in the cochlear and vestibular hair cells of the inner ear. Expression is first detectable at E15.5, peaks around P0, decreases slightly at P6, and is absent by P15, a duration that supports the involvement of Tmhs in stereocilia development. Tmhs reporter gene expression also was detected in several cranial and cervical sensory ganglia, but not in the vestibular or spiral ganglia. We also describe a new nontargeted mutation of the Tmhs gene, hscy-2J, that causes abnormal splicing from a cryptic splice site within exon 2 and is predicted to produce a functionally null protein lacking 51 amino acids of the wild-type sequence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The similarity in structure and function of the mouse inner ear to that of humans and the ability to examine all developmental stages make the mouse an ideal model for studying the hearing process and its associated disorders. Hair cells are the specialized mechanoreceptors found in the sensory organs of the inner ear. Modified microvilli called stereocilia extend from the apical surfaces of hair cells and are clustered into highly organized bundles. The precise formation and patterning of these hair bundles is critical to their function in stimulus reception and mechanoelectrical transduction. Mouse mutations have helped identify several genes that are essential for the development and maintenance of hair cell stereocilia, including Myo7a (Self et al. 1998) and Ush1c (Johnson et al. 2003), which are required for proper hair bundle organization; Myo6, which is thought to anchor stereocilia to the cuticular plate (Self et al. 1999); Myo15a (Belyantseva et al. 2003) and Whrn (Mburu et al. 2003), which are involved in the elongation and maintenance of stereociliary length; Espn (Zheng et al. 2000), which is an actin-bundling protein; Rdx (Kitajiri et al. 2004) and Clic5 (Gagnon et al. 2006), which are associated with the actin cytoskeleton of stereocilia; and Cdh23 (Siemens et al. 2004; Michel et al. 2005), Pcdh15 (Ahmed et al. 2006), and Gpr98 (McGee et al. 2006), which appear to be constituents of stereociliary links. The protein products of some of these genes have been shown to interact and may form networks that underlie the cohesion of the hair bundle (Adato et al. 2005; van Wijk et al. 2006).
Tmhs, which encodes a small four-pass membrane protein, is a recently characterized gene that also appears to play an important role in hair bundle morphogenesis and integrity. We reported previously that a missense mutation in Tmhs causes deafness and vestibular defects in hurry-scurry (hscy) mutant mice and that the TMHS protein localizes to the stereociliary bundles of hair cells in the organ of Corti of the cochlea, the cristae ampullaris of the semicircular canals, and the maculae of the saccule and utricle (Longo-Guess et al. 2005). More recently, mutations in the human TMHS gene have been shown to underlie the autosomal nonsyndromic deafness disorder DFNB67 (Kalay et al. 2006; Shabbir et al. 2006). The mutation in the hscy mutant mouse, a G-to-T transversion in exon 2 of Tmhs, causes a substitution of phenylalanine for cysteine at amino acid position 161. This cysteine is highly conserved among species, suggesting that there is a strong functional constraint against its alteration. The conserved cysteine may be involved in forming a disulfide bond that is required for proper conformation, and its disruption could lead to protein degradation or mislocalization.
The important role predicted for the mutated cysteine residue, coupled with the lack of detectable protein expression in the mutant mouse, led us to believe that the hscy mutation was functionally null. To test this hypothesis, we genetically engineered mice with a null mutation of the Tmhs gene to allow for comparisons with the phenotype observed in hscy mutant mice. We also incorporated a lacZ reporter gene into the knockout construct to enable the examination of Tmhs gene expression from late embryonic stages throughout the first four postnatal weeks of life. Consistent with TMHS protein localization results (Longo-Guess et al. 2005; Shabbir et al. 2006), we show that the Tmhs reporter gene is highly expressed in hair cells of the inner ear and coincides with the time period of hair cell stereocilia development. The similarities of gene expression patterns and protein localization suggest that TMHS may associate with CDH23 in hair bundle morphogenesis or maintenance. Tmhs reporter expression also was detected in several cranial and cervical ganglia, although TMHS-specific antibodies have failed to detect protein in these structures (Longo-Guess et al. 2005).
We also report here on the molecular characterization of a recently discovered Tmhs mutant allele, designated Tmhs hscy-2J, that occurred in a colony of ENU-mutagenized mice in the Neuroscience Mutagenesis Facility at The Jackson Laboratory. We show that the hscy-2J mutation is a 6-bp deletion that inactivates the normal donor splice site of exon 2 and causes alternative use of a noncanonical cryptic splice site, which is predicted to produce a mutant protein lacking 51 amino acids of the wild-type sequence.
Materials and methods
All mice used in this study were generated and maintained at The Jackson Laboratory (Bar Harbor, ME). All animal procedures were approved by the Institutional Animal Care and Use Committee.
Generation of B6.129-Tmhs tm1Kjn mice
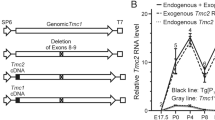
We used polymerase chain reaction (PCR) to amplify homology arms from coisogenic genomic DNA of R1 embryonic stem (ES) cells (Fig. 1). The homology arms corresponded to the regions immediately flanking the 5′ end of exon 1 and the 3′ end of exon 2 of Tmhs. The 3.6-kb 5′ arm was subcloned into the XmaI site of plasmid p231 between the vector backbone and the lacZ reporter gene, and the 2.5-kb 3′ arm was cloned into the SalI site between the gene that causes neomycin resistance (Neo) and the thymidine kinase (TK) cassette. The resulting construct was linearized with AclI and electroporated into R1 ES cells. Homologous recombination was identified by Southern blots of DNA digested with EcoRI and hybridized with a probe corresponding to the 3′ end of the construct, which produces a 5.1-kb band in the correctly targeted cells. Presence of the construct was confirmed by Southern blot using a probe corresponding to the 5′ end of the construct, which produces a 7.8-kb band when digested with PstI. Correctly targeted ES clones were injected into C57BL/6J (B6) blastocyst-stage mouse embryos (3.5 dpc), followed by surgical transfer into a host female. The embryos were allowed to develop to term and chimeras arising from an admixture of host embryo and introduced cells were identified and bred to B6 mice for germline transmission. Mice carrying the neomycin resistance cassette were identified by PCR using the Neo Touchdown assay developed by the Induced Mutant Resource at The Jackson Laboratory (http://www.jaxmice.jax.org/pub-cgi/protocols/protocols.sh?objtype=protocol&protocol_id=701). Mice carrying the targeted mutation were repeatedly backcrossed with B6 mice until the N10 generation, then inbred by sister-brother matings to develop the B6.129-Tmhs tm1Kjn strain.
Targeted disruption of the Tmhs gene and replacement with a lacZ reporter gene. A Schematic diagram illustrating the targeting construct, the wild-type Tmhs allele, and the recombinant allele with exons 1 and 2 replaced by a lacZ/Neo cassette. The 15.5-kb targeting construct included the plasmid vector, the 5′ and 3′ homology arms, a lacZ reporter gene and neomycin-resistance (Neo) cassette, and a thymidine kinase (TK) negative selection cassette. B The 5′ and 3′ homology arms of the targeting construct were amplified by high-fidelity PCR from genomic DNA of R1 ES cells. C After electroporation of the linearized targeting construct into R1 ES cells and antibiotic selection, positive clones were expanded and screened for recombinant alleles by Southern blot analysis, first with a 3′ probe hybridized to EcoRI-digested DNA followed by verification with a 5′ probe hybridized to PstI-digested DNA. The properly targeted ES cells were then microinjected into B6 blastocyst-stage embryos (3.5 dpc). R1 ES cells are derived from the hybrid strain 129X1/SvJ × 129S1/Sv-p + Tyr + Kitl Sl-J/J
X-gal staining and immunofluorescence
X-gal detection of the lacZ reporter gene product was performed as described (Oberdick et al. 1994) with a few modifications. Ears from mice older than one week of age were dissected from the skull and decalcified in 7% EDTA in phosphate-buffered saline (PBS) before staining. Mice younger than one week were decapitated. Bisected heads and thoracic sections of the vertebral column (containing spinal cord) were fixed overnight in 2% paraformaldehyde (PFA) in 0.1 M modified PIPES buffer. The specimens then were placed in 30% sucrose in PBS for cryoprotection and cryo-embedded in Tissue-Tek OCT Compound (Sakura Finetek, Torrance, CA). Sections (10 μm) were cut using a Leica cryostat model CM 1900 and mounted onto Superfrost plus slides (Fisher Scientific, Suwanee, GA). Slides were stained in X-gal buffer overnight at 37°C, washed three times in PBS, and then counterstained with neutral fast red for 1 min. Slides were coverslipped and photographed with an Olympus BX40 light microscope and Olympus DP70 camera (Olympus Optical Co., Tokyo, Japan). Whole-mount preparations of dissected ears and sensory epithelia from X-gal-stained half heads of mice were placed in glycerol and viewed with a Nikon E800 microscope (Farinas et al. 2001; Fritzsch et al. 2005).
Newborn mouse ears were prepared for immunofluorescence by bisecting the heads and fixing them in 4% PFA in PBS overnight at 4°C. The bisected heads were embedded in paraffin for sagittal sectioning and 6-μm sections were cut through the ears. Tissue sections were stained with anti-TMHS antibody as previously described (Longo-Guess et al. 2005).
Phalloidin staining of hair cell actin filaments
Organ of Corti surface preparations and phalloidin-staining procedures were adapted from previously described methods (Beyer et al. 2000). Heads were bissected and fixed overnight in 4% PFA. Inner ears were dissected away from the head and bone and stria vascularis were removed from the cochlea to expose the organ of Corti. Inner-ear tissue was permeabilized in 0.3% Triton-X-100 for 5 min followed by a 30-min incubation in rhodamine phalloidin (Molecular Probes, Eugene, OR) diluted 1:100 in PBS. Samples were rinsed several times in PBS. The cochlear turns were separated from the modiolus and mounted on glass slides with Vectashield mounting media (Vector Laboratories, Burlingame, CA).
Auditory-evoked brainstem response (ABR)
Hearing assessment was performed as previously described (Zheng et al. 1999). Mice were anesthetized with tribromoethanol, and then placed on a heating pad in a sound-attenuating chamber. Needle electrodes were placed just under the skin, with the active electrode placed between the ears just above the vertex of the skull, the ground electrode between the eyes, and the reference electrode underneath the left ear. High-frequency transducers were placed just inside the ear canal and computer-generated sound stimuli were presented at defined intervals. Thresholds were determined for a broadband click and for 8-, 16-, and 32-kHz pure-tone stimuli by increasing the sound pressure level (SPL) in 10-dB increments followed by 5-dB increases and decreases to determine the lowest level at which a distinct ABR wave pattern could be recognized. Stimulus presentation and data acquisition were performed using the Smart EP evoked potential system (Intelligent Hearing Systems, Miami, FL).
Scanning electron microscopy (SEM)
SEM was performed essentially as described (Furness and Hackney 1986). Inner ears were dissected out of the skull, fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 3-4 h at 4°C, and then washed three to four times in 0.1 M phosphate buffer. Hair cells of the organ of Corti were exposed by carefully dissecting away the overlying bone and membrane. The tissues then were processed in osmium tetroxide-thiocarbohydrazide (OTOTO), dehydrated with ethanol, and critical-point dried with hexamethyldisilazane (Electron Microscopy Sciences, Hatfield, PA). Samples were mounted onto aluminum stubs and sputter-coated to produce a 15-nm gold coat. Samples were examined at 20 kV with a Hitachi 3000N VP Scanning Electron Microscope with an EDAX X-ray Microanalysis unit and PCI Quartz Image Management software (Vancouver, BC).
ENU mutagenesis
The detailed procedure used for ENU-induced mutagenesis and production of mutant mice by the Neuroscience Mutagenesis Facility at The Jackson Laboratory is described on their website: http://www.nmf.jax.org/protocols.html. Briefly, a B6 male mouse injected with ENU that had recovered his fertility (designated G0) was mated to a female wild-type mouse. The male offspring from this mating (G1 founders), which each represent one mutagenized gamete, were mated back to B6 females. The resulting G2 generation female offspring were then backcrossed to the male G1 founder to recover recessive phenotypes in the G3 progeny.
DNA sequencing, genotyping, and transcript analysis of the hscy-2J mutation
To screen for the hscy-2J mutation, genomic DNA was isolated from tail tips, and PCR primers flanking each exon of the Tmhs gene (Table 1) were used to amplify products for DNA sequence analysis. PCR products were purified with the Exo-SAPIt reagent (USB Corp, Cleveland, OH) according to the manufacturer’s protocol and then sequenced using an Applied Biosystems (Foster City, CA) 3700 DNA sequencer with an optimized Big Dye Terminator method.
To genotype mice for the hscy-2J mutation, primers were designed to amplify the Tmhs genomic region surrounding the hscy-2J 6-bp deletion (Table 1). PCR amplification with these primers produced a 104-bp product from wild-type DNA and a 98-bp product from hscy-2J DNA, which were distinguished by electrophoretic separation in a 6% polyacrylamide gel.
To examine the effects of the hscy-2J mutation on transcription, total RNA was isolated from mouse brain tissue using Trizol reagent (Invitrogen, Carlsbad, CA). Mouse cDNA for PCR analysis was synthesized from total RNA by using SuperScript II reverse transcriptase according to the manufacturer’s instructions (Invitrogen). A forward primer corresponding to exon 1 of Tmhs and a reverse primer corresponding to exon 4 (Table 1) were used to amplify products from cDNAs, which then were purified and sequenced as described above.
Results
Targeted knockout of the Tmhs gene
We used homologous recombination in ES cells to replace exons 1 and 2 of the Tmhs gene with a lacZ reporter and neomycin cassette (Fig. 1). We screened approximately 400 ES cell clones and found two that had undergone the appropriate targeting event. One of these clones was injected into B6 blastocysts. Pups with a predominantly agouti coat color were selected as possible high-ES-cell-content chimeras and bred to B6 mice. Germline transmission of the targeted allele was confirmed by PCR using primers specific to genomic sequence in the 5′ arm of the construct and a reverse primer designed from the lacZ reporter gene sequence. Carrier mice were crossed back to B6 strain mice for ten generations and then inbred to produce strain B6.129-Tmhs tm1Kjn.
An allelism test was performed between the genetically engineered mutation and the spontaneous hscy mutation of the Tmhs gene. A mating between a Tmhs +/hscy female and a Tmhs +/tm1Kjn male produced 13 offspring, 3 of which exhibited the same circling and head-tossing phenotype as the original Tmhs hscy/hscy mutant mouse, thus confirming allelism. We then used a specific antibody to test whether the TMHS protein is absent as predicted in homozygous Tmhs tm1Kjnmutant mice (Tmhs -/-). As described previously, TMHS localizes to the hair bundles of both the outer and inner hair cells in the cochlea of wild-type mice (Longo-Guess et al. 2005). We examined TMHS expression in the cochleae of a P0 Tmhs -/- mouse and a heterozygous littermate control. In the control mouse, TMHS localized to hair cell stereocilia as expected (Fig. 2A); however, TMHS expression was not detected anywhere in the inner ear of the Tmhs -/- knockout mouse (Fig. 2B). The lack of immunofluorescence in cochleae of mutant mice is due to the absence of TMHS protein and not due to missing or dysmorphic hair bundles as evidenced by organ of Corti surface preparations stained with phalloidin, which clearly show the presence of normal-appearing hair bundles in mutant mice at P0 (Fig. 2 C,D).
Absence of protein expression and hair bundle degeneration in knockout mice (Tmhs -/-). TMHS-specific immunofluorescence was detected in stereocilia (indicated by downward-pointing arrowheads) of inner hair cells (IHC) and outer hair cells (OHC) in +/+ control mice (A), as previously reported (Longo-Guess et al. 2005), but was absent in cochleae of Tmhs -/- mice (B). Organ of Corti surface preparations were stained with phalloidin to visualize hair bundles in control (C) and mutant (D) newborn mice. Normal-appearing stereociliary bundles are clearly present on inner (bottom row) and outer hair cells (three upper rows) of P0 mutant mice. Hair bundle morphology in adult mice was examined by SEM of organ of Corti surface preparations. By P28, stereocilia degeneration is apparent in OHC bundles of Tmhs -/- mice (F) as compared with the normal bundle morphology seen in age-matched +/+ control mice (E). All scale bars, 10 μm
Tmhs -/- mutant mice begin to exhibit a slight behavioral abnormality at P11 and by three weeks of age begin to circle rapidly and toss their heads. Hearing in mutant and control mice at 34 days of age was evaluated by auditory brainstem response (ABR) to broadband clicks and to 8-, 16-, and 32-kHz pure tones. Auditory stimuli up to 100 dB SPL failed to elicit ABRs in the two Tmhs -/- mutant mice tested, indicating that they are profoundly deaf. All five heterozygous controls exhibited normal ABR thresholds according to previously established criteria (Zheng et al. 1999). The behavioral and hearing phenotype of the Tmhs -/- knockout mice was thus very similar to that of Tmhs hscy mutant mice (Longo-Guess et al. 2005).
To investigate hair bundle morphology in cochleae of adult mice, we examined organ of Corti surface preparations by scanning electron microscopy (SEM). We looked at two Tmhs -/- mice and one B6 control mouse at approximately one month of age. In the control cochlea, the outer hair cells were neatly organized into three rows with rigidly patterned V-shaped bundles, and the inner hair cells were properly oriented in a single neat row (Fig. 2E). In the Tmhs -/- cochlea, the outer hair cell bundles were disorganized and many stereocilia had degenerated (Fig. 2F); the stereocilia of the inner hair cells exhibited a splayed appearance. There were patches in the basal region of the cochlea where outer hair cell bundles were missing completely. The hair cell pathology that we observed in Tmhs -/- mice is very similar to our previous observations of Tmhs hscy/hscy mice (Longo-Guess et al. 2005).
LacZ reporter gene expression in the B6.129-Tmhs tm1Kjn mice
The lacZ reporter was included in the Tmhs targeting construct to further investigate normal Tmhs gene expression in heterozygous mice. Sagittal sections from embryos at ages E15.5, E16.5, and E17.5, and postnatal mice at ages P0, P3, and P6 were evaluated for Tmhs expression. Whole-mount inner-ear preparations from P0, P2, P15, and P30 mice were also examined. Tmhs expression was detected in all hair cells of the cochlea, saccule, utricle, and semicircular canals, but only in mice younger than P15 (Figs. 3 and 4). Cochlear whole-mount preparations of 2-day-old mice showed expression in hair cells along the entire length of the cochlea except at the extreme apex where hair cells are sparse and less developed (Fig. 3F,G).
Tmhs reporter gene expression in the inner ear and sensory ganglia. The lacZ reporter gene for Tmhs expression was visualized by histochemical reaction of its product, β-gal. A,B Two views of whole-mount preparations of inner ears from P0 mice show that Tmhs expression is limited to the neurosensory epithelia: the cristae of the posterior (PC), anterior (AC), and horizontal (HC) semicircular canals, the maculae of the utricle (U) and saccule (S), and the organ of Corti of the cochlea (Co). C-H Surface preparations of the dissected organ of Corti of P2 mice show expression in the three rows of outer hair cells (OHC) and single row of inner hair cells (IHC) along the entire length of the cochlear duct except for the most extreme apical region. I-K Surface preparations of the dissected vestibular sensory regions show expression in the cristae of the posterior (PC), anterior (AC), and horizontal (HC) semicircular canals and in the maculae of the utricle and saccule. Tmhs expression also was detected in cranial and cervical ganglia (L-N), including the geniculate ganglion (GnG, part of the VII cranial nerve), the glossopharyngeal ganglion (IX cranial, see Fig. 4A), the vagal ganglion (X cranial), both proximal and distal parts, the trigeminal ganglion (V cranial), and the superior cervical sympathetic ganglion (SCG), but was not detected in the vestibular ganglion (VG) nor in the spiral ganglion (see Fig. 4A). All scale bars, 100 μm
Tmhs reporter gene expression in inner-ear cross sections. Midmodiolar cross section through the cochlea of a P0 mouse (A) shows β-gal expression in the hair cells of the basal and middle turns (arrowheads) but not in the apex. Expression also is seen in cells of the glossopharyngeal (IX cranial) ganglion, but not in spiral ganglion (SG) cells. Cochlear expression in inner hair cells (IHC) and outer hair cells (OHC) is strongest at P0 (B), diminishes slightly by P6 (C), and is no longer detectable at P15 (not shown). The Tmhs reporter gene is also strongly expressed in vestibular hair cells of the utricle (not shown), saccule (D), and crista ampularis (E). Expression can be detected as early as E15.5 in vestibular hair cells (E), but not until E17 in cochlear hair cells (not shown). Scale bars: A, 200 μm; B-E, 50 μm
We first noted Tmhs expression in the crista ampullaris at E15.5 (Fig. 4E), but did not see expression in the organ of Corti until E16.5 (not shown). Expression in the cochleae of mice at E16.5 and E17.5 was seen as punctate dots in the inner and outer hair cells. At P0-P2 the lacZ staining was more diffuse throughout the cell body of the hair cells, and the staining was strongest at this age. Staining was slightly fainter at P6, and by P15 we could no longer detect any Tmhs expression.
We also detected lacZ expression in cranial and cervical sensory ganglia, including both the proximal and the distal vagal ganglion, the superior sympathetic ganglion, the geniculate ganglion, the trigeminal ganglion (Fig. 3L-N), and the glossopharyngeal ganglion (Fig. 4A). We did not detect β-gal activity in the vestibular ganglion (Fig. 3L) or in the spiral ganglion (Fig. 4A) nor in brain tissue at any age. We also did not detect any Tmhs reporter gene expression in the spinal cord or in the dorsal root ganglia in cross sections from the thoracic region of a P0 mouse.
Characterization of the hscy-2J mutation
The hscy-2J mutant allele originated in a colony of mice from an ENU mutagenesis screen undertaken by the Neuroscience Mutagenesis Facility at The Jackson Laboratory (http://www.nmf.jax.org/). The recessive mutation, which is on a B6 genetic background, was originally designated nmf430 (for neuroscience mutagenesis facility mutant #430). Mutant mice homozygous for the Tmhs hscy-2J allele can be identified at approximately three weeks of age by their rapid circling behavior and their slight head shaking. The two Tmhs hscy-2J mutants evaluated for hearing by ABR at 32 days of age showed no response to auditory stimuli up to 100 dB SPL, indicating that they are profoundly deaf. The two age-matched heterozygous controls exhibited normal ABR thresholds.
Linkage analysis of 41 F2 progeny from an intercross of (B6-nmf430 × BALB/cByJ) F1 hybrids localized the nmf430 mutation to Chr 17, between D17Mit133 at 24.6 Mb and D17Mit175 at 31.6 Mb (NCBI m36 assembly). This interval contains the Tmhs gene, which is located at 28.3 Mb. Negative complementation test results from matings between mice with the nmf430 and Tmhs hscy mutations provided further evidence that nmf430 is a remutation of the Tmhs gene. A mating between a heterozygous +/nmf430 female and a homozygous Tmhs hscy/hscy male produced two litters with a total of seven mutants and five normal-appearing mice. The nm430 mutation was therefore redesignated Tmhs hscy-2J .
We designed PCR primers flanking each of the four exons of the Tmhs gene (Table 1) and used them to amplify genomic DNA from two different hscy-2J mutants. We compared the DNA sequences of these products to the corresponding sequences of the Tmhs gene from the mouse genome database (Ensembl gene ID ENSMUSG00000062252; NCBI m36 assembly). A 6-bp deletion was discovered in the intron immediately flanking the 3′ end of exon 2 in mutant DNA, which disrupts the normal splice donor recognition site (Fig. 5A,B). No other sequence abnormalities of the Tmhs gene were detected in mutant DNA. RT-PCR performed using primers spanning exons 1-4 produced a product in the mutant DNA that was 153 bp smaller than that of the control (Fig. 5D). Sequence analysis of this smaller mutant product confirmed that disruption of the wild-type splice recognition site causes alternative splicing from a cryptic donor site in the middle of exon 2, thereby omitting 153 nucleotides encoded by the 5′ end of this exon (Fig. 5B,C). The mutant transcript is spliced in frame with exon 3, leaving the last three amino acids and the stop codon intact (Fig. 5C). The 153 bases missing in the mutant transcript code for 51 amino acids that form the fourth transmembrane domain and part of the second extracellular loop of the TMHS protein. To test whether the predicted loss of 51 amino acids affects protein localization, we examined TMHS expression in cochlear cross sections from P0 hscy-2J mutants and heterozygous littermate controls by immunofluorescence. TMHS localized to hair bundles as expected in control mice (Fig. 6A) but was not detected in cochleae of hscy-2J mutant mice (Fig. 6B), providing further evidence that the hscy-2J mutation is functionally null.
Molecular characterization of the hscy-2J mutation. A Sequence chromatograms of genomic DNA from a B6 +/+ control and a hscy-2J/hscy-2J mutant mouse. The hscy-2J 6-bp deletion is indicated by dashed lines. The control and mutant 9-bp donor splice site recognition sequences for exon 2 of Tmhs are enclosed in boxes. The hscy-2J deletion inactivates the normal donor splice site. B Genomic structure of Tmhs illustrating the effect of the hscy-2J mutation on transcription. The Tmhs gene comprises four exons, here represented as rectangles with black regions indicating protein-coding sequence and connecting lines representing introns. The hscy-2J mutation causes abnormal splicing from a cryptic, noncanonical donor splice site within exon 2. C DNA sequence analysis of mutant transcripts. The hscy-2J deletion causes the alternative use of a cryptic donor splice site (boxed region) within exon 2. Thus, mutant transcripts do not contain the 153 nt shown in red, which are encoded by the skipped 3′ region of exon 2. Although mutant transcripts are spliced in-frame with exon 3 (splice sites are indicated by arrows), their translated protein product lacks the 51 amino acids shown in blue. D RT-PCR analysis of Tmhs transcripts. Primers corresponding to exon 1 and exon 4 (Table 1) were used to amplify cDNA from brain tissue of two +/+ and two hscy-2J/hscy-2J mice. DNA sequencing verified that the +/+ product (358 bp, lanes 1 and 2) is properly spliced and that the smaller size of the hscy-2J/hscy-2J product (205 bp, lanes 3 and 4) is caused by the abnormal splicing detailed in panels B and C. A negative control without RT is shown in lane 5. E PCR genotyping assay. A simple PCR assay can be used to genotype mice for the 6-bp hscy-2J deletion, with primers that amplify a 104-bp product from wild-type DNA and a 98-bp product from the hscy-2J DNA (Table 1). The PCR products can be separated on a 6% polyacrylamide gel to distinguish +/hscy-2J (lanes 1 and 3), hscy-2J/hscy-2J (lanes 2 and 4), and +/+ (lane 5) genotypes
Absence of TMHS protein expression in cochleae of hscy-2J mutant mice. TMHS-specific immunofluorescence was detected in stereocilia (indicated by downward-pointing arrowheads) of inner hair cells (IHC) and outer hair cells (OHC) in +/+ control mice at P0 (A), but was absent in age-matched hscy-2J mutant mice (B). Scale bars, 10 μm
Discussion
In the mouse inner ear, hair bundles first appear in the vestibular organs at E13.5 (Mbiene and Sans 1986; Denman-Johnson and Forge 1999) and in the cochlea at E15 (Anniko 1983). We first detected Tmhs expression at E15.5 in the sensory regions of the vestibular organs and at E16.5 in the organ of Corti, just after the initial emergence of stereocilia but prior to their period of elongation and staircase patterning. We have determined the peak expression period of the Tmhs gene to be between P0 and P3; it is slightly reduced at P6 and can no longer be detected at P15. This period of expression coincides with the period of stereocilia maturation (Nishida et al. 1998; Mburu et al. 2006). Hair bundles in Tmhs mutant mice begin to degenerate by P8 (Longo-Guess et al. 2005), which is about the same time that Tmhs expression ceases. Tmhs is expressed in a gradient along the organ of Corti from the base to the apex. This expression pattern mirrors the pattern of development of the stereocilia, which also occurs in a basal-to-apical fashion (Zine and Romand 1996), but not the pattern of hair cell mitosis, which progresses from the apex to the base (Matei et al. 2005). The temporal and spatial pattern of Tmhs reporter gene expression in hair cells of the inner ear (Figs. 3 and 4), the previously determined subcellular localization of the TMHS protein (Longo-Guess et al. 2005), and the hair bundle pathology observed in Tmhs mutant mice (Fig. 2D) strongly support a role for TMHS in hair bundle morphogenesis.
Expression patterns of other genes associated with the hair bundle have been determined by protein immunodetection or mRNA analysis, but none have been evaluated by reporter gene expression as described here for Tmhs. The expression pattern of Cdh23 appears most similar to that of Tmhs. Cdh23 expression is first detected at E16.5 in inner and outer hair cells along the length of the cochlea except for the apical turn; it then progresses toward the apical turn by late embryonic stages (Boeda et al. 2002; Lagziel et al. 2005). Cdh23 expression in the cochlea is highest at P1-P7, is reduced by P14, and reaches a low but stable level at P21 that persists throughout adulthood (Rzadzinska et al. 2005). CDH23 has been shown to interact with harmonin b (the product of Ush1c), MYO7A, and PCDH15 in the developing hair bundle to form a stable structural unit (Boeda et al. 2002; Adato et al. 2005), and it is possible that TMHS may also be involved in the same hair bundle complex. TMHS is a member of the four transmembrane spanning (tetraspan) family of proteins. Some members of this family have been shown to associate with cadherins and are thought to play a role in cellular adhesion and epidermal morphology (Simske et al. 2003; Kearsey et al. 2004). Recently, the mouse tetraspan protein TMEM47 (transmembrane protein 47) was shown to colocalize with the tips of actin filaments in differentiating kidney podocytes (Bruggeman et al. 2007). Similar to TMHS, the overall expression of TMEM47 peaked at P4 and disappeared upon differentiation. TMHS, possibly in association with CDH23, may thus be involved in hair bundle morphogenesis in a manner analogous to that of the TMEM47-cadherin interaction associated with podocyte differentiation.
In this study of lacZ-stained B6.129-Tmhs tm1Kjnmice, we detected strong Tmhs expression in the hair cells of the inner ear, yet the Unigene expressed sequence tag (EST) profile for mouse Tmhs (Mm.284760) does not include any EST sequences from inner-ear libraries. This seeming discrepancy can be explained by the restricted expression of Tmhs in the inner ear, which is limited to hair cells of mice older than E15 and younger than P15. Hair cells make up a small portion of the total inner-ear tissue mass, and more than 75% of the Unigene inner-ear ESTs are from libraries derived from adults or otocysts, stages when Tmhs is not expressed. Northern blot results have shown Tmhs expression in adult brain tissue (Longo-Guess et al. 2005), and the Unigene EST expression profile (Mm.284760) includes brain, spinal cord, endocrine glands, and eye. The lacZ reporter gene results reported here give no indication of Tmhs expression in these tissues but do show positive expression in several cranial and cervical sensory ganglia. It is possible that some peripheral ganglion cells that express Tmhs were unintentionally included in the brain or spinal cord preparations used as sources for the EST libraries and Northern blots, which would then give a false indication of Tmhs expression in those tissues.
All cranial ganglia express the bHLH gene Neurod1 and require the pou domain factor POU4F1 for differentiation (Huang et al. 2001; Kim et al. 2001), whereas hair cells seem to express Neurod1 late in development and require the pou domain factor POU4F3 for development (Fritzsch et al. 2006). Sympathetic ganglia, while of neural crest origin, require the bHLH gene Ascl1 for differentiation (Anderson et al. 1997). Thus, there appears to be no common denominator for the observed expression of the Tmhs gene in cranial and sympathetic ganglia and in hair cells. The expression of the Tmhs gene in cranial and cervical ganglion cells may have little or no functional significance because TMHS protein could not be detected in these cells (Longo-Guess et al. 2005), suggesting a rapid degradation or low concentration. Tmhs mutant mice exhibit circling and head-tossing behaviors typical of inner-ear dysfunction, but do not show any of the abnormalities that have been associated with cranial nerve deficiencies (Cordes 2001; Mar et al. 2005). Light microscopic examinations of the cranial ganglia in Tmhs-deficient mutant mice did not reveal any gross morphologic abnormalities. The apparent inconsequence of TMHS deficiency in the sensory ganglion cells might also be explained by functional redundancies with similar proteins.
The hscy-2J mutation, which was discovered in a colony of ENU-mutagenized mice, is a 6-bp deletion that disrupts the splice donor recognition site of exon 2 and causes alternative use of a cryptic, noncanonical donor splice site within exon 2. Although 98.7% of known mammalian splice sequences contain canonical GT-AG junctions at the 5′ and 3′ ends of introns, 0.6% have noncanonical GC-AG splice site pairs (Burset et al. 2001), so the noncanonical cryptic splice site (GG/GCAAGT) activated in hscy-2J is not unprecedented. A splicing defect similar to what we have observed for the mouse hscy-2J mutation of Tmhs has been described for a mutant allele of the human CDC73 gene (Bradley et al. 2005). In both cases a donor splice site mutation activates a cryptic noncanonical donor splice site and results in abnormally spliced transcripts that encode mutant proteins with a loss of amino acids. The 51 amino acids omitted from the mutant protein of hscy-2J mice form part of the second extracellular loop, the entire fourth transmembrane domain, and part of the C-terminal region of the wild-type TMHS protein. The predicted loss of such a large portion of the protein and the similarity of the hscy-2J mutant phenotype with that of the Tmhs knockout mouse indicate that hscy-2J is also functionally null. TMHS-specific antibody staining of cochlear cross sections from hscy-2J mutant and control mice at P0 confirmed the absence of TMHS protein in mutant mice (Fig. 6).
ENU is known to induce point mutations. The great majority of ENU-induced mouse mutations are single nucleotide substitutions; fewer than 1% are deletions, and most, if not all, of these are deletions of a single base pair (Barbaric et al. 2007). The 6-bp deletion reported here for hscy-2J represents either an extremely rare ENU-induced mutational event or a mutation that occurred spontaneously in an ENU-mutagenized colony of mice. Because the mutagenesis and subsequent breeding history involved only the B6 strain, it is impossible to distinguish between these two possibilities.
In summary, we report the generation and characterization of two new mutant alleles of the Tmhs gene: a targeted knockout allele (Tmhs tm1Kjn) and a nontargeted mutation discovered in a colony of ENU-mutagenized mice (Tmhs hscy-2J). Mutant mice for both of these alleles exhibit a rapid circling and head-tossing behavior and are profoundly deaf at the earliest age tested (4 weeks). SEM analysis at this age shows that hair bundles have already severely degenerated. The two new mutant alleles reinforce the importance of the Tmhs gene in inner-ear function and emphasize its likely role in hair bundle morphogenesis. The targeted null allele with the lacZ reporter is particularly useful because it provides a sensitive means to study the temporal and spatial expression patterns of the Tmhs gene and could be useful as a hair cell or ganglion cell marker for future studies.
References
Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam KN, et al. (2005) Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet 14:347–356
Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, et al. (2006) The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci 26:7022–7034
Anderson DJ, Groves A, Lo L, Ma Q, Rao M, et al. (1997) Cell lineage determination and the control of neuronal identity in the neural crest. Cold Spring Harb Symp Quant Biol 62:493–504
Anniko M (1983) Postnatal maturation of cochlear sensory hairs in the mouse. Anat Embryol (Berl) 166:355–368
Barbaric I, Wells S, Russ A, Dear TN (2007) Spectrum of ENU-induced mutations in phenotype-driven and gene-driven screens in the mouse. Environ Mol Mutagen 48:124–142
Belyantseva IA, Boger ET, Friedman TB (2003) Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci U S A 100:13958–13963
Beyer LA, Odeh H, Probst FJ, Lambert EH, Dolan DF, et al. (2000) Hair cells in the inner ear of the pirouette and shaker 2 mutant mice. J Neurocytol 29:227–240
Boeda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, et al. (2002) Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J 21:6689–6699
Bradley KJ, Cavaco BM, Bowl MR, Harding B, Young A, et al. (2005) Utilisation of a cryptic non-canonical donor splice site of the gene encoding PARAFIBROMIN is associated with familial isolated primary hyperparathyroidism. J Med Genet 42:e51
Bruggeman LA, Martinka S, Simske JS (2007) Expression of TM4SF10, a Claudin/EMP/PMP22 family cell junction protein, during mouse kidney development and podocyte differentiation. Dev Dyn 236:596–605
Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29:255–259
Cordes SP (2001) Molecular genetics of cranial nerve development in mouse. Nat Rev Neurosci 2:611–623
Denman-Johnson K, Forge A (1999) Establishment of hair bundle polarity and orientation in the developing vestibular system of the mouse. J Neurocytol 28:821–835
Farinas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, et al. (2001) Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci 21:6170–6180
Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, et al. (2005) Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn 233:570–583
Fritzsch B, Pauley S, Beisel KW (2006) Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res 1091:151–171
Furness DN, Hackney CM (1986) High-resolution scanning-electron microscopy of stereocilia using the osmium-thiocarbohydrazide coating technique. Hear Res 21:243–249
Gagnon LH, Longo-Guess CM, Berryman M, Shin JB, Saylor KW, et al. (2006) The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci 26:10188–10198
Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, et al. (2001) Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development 128:2421–2432
Johnson KR, Gagnon LH, Webb LS, Peters LL, Hawes NL, et al. (2003) Mouse models of USH1C and DFNB18: phenotypic and molecular analyses of two new spontaneous mutations of the Ush1c gene. Hum Mol Genet 12:3075–3086
Kalay E, Li Y, Uzumcu A, Uyguner O, Collin RW, et al. (2006) Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum Mutat 27:633–639
Kearsey J, Petit S, De Oliveira C, Schweighoffer F (2004) A novel four transmembrane spanning protein, CLP24. A hypoxically regulated cell junction protein. Eur J Biochem 271:2584–2592
Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, et al. (2001) NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 128:417–426
Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, et al. (2004) Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol 166:559–570
Lagziel A, Ahmed ZM, Schultz JM, Morell RJ, Belyantseva IA, et al. (2005) Spatiotemporal pattern and isoforms of cadherin 23 in wild type and waltzer mice during inner ear hair cell development. Dev Biol 280:295–306
Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, et al. (2005) A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc Natl Acad Sci U S A 102:7894–7899
Mar L, Rivkin E, Kim DY, Yu JY, Cordes SP (2005) A genetic screen for mutations that affect cranial nerve development in the mouse. J Neurosci 25:11787–11795
Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, et al. (2005) Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn 234:633–650
Mbiene JP, Sans A (1986) Differentiation and maturation of the sensory hair bundles in the fetal and postnatal vestibular receptors of the mouse: a scanning electron microscopy study. J Comp Neurol 254:271–278
Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, et al. (2003) Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet 34:421–428
Mburu P, Kikkawa Y, Townsend S, Romero R, Yonekawa H, et al. (2006) Whirlin complexes with p55 at the stereocilia tip during hair cell development. Proc Natl Acad Sci U S A 103:10973–10978
McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, et al. (2006) The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci 26:6543–6553
Michel V, Goodyear RJ, Weil D, Marcotti W, Perfettini I, et al. (2005) Cadherin 23 is a component of the transient lateral links in the developing hair bundles of cochlear sensory cells. Dev Biol 280:281–294
Nishida Y, Rivolta MN, Holley MC (1998) Timed markers for the differentiation of the cuticular plate and stereocilia in hair cells from the mouse inner ear. J Comp Neurol 395:18–28
Oberdick J, Wallace J, Lewin A, Smeyne R (1994) Transgenic expression to monitor dynamic organization of neuronal development: use of the Escherichia coli lacZ gene product, beta-galactosidase. Neuroprotocols 5:54–62
Rzadzinska AK, Derr A, Kachar B, Noben-Trauth K (2005) Sustained cadherin 23 expression in young and adult cochlea of normal and hearing-impaired mice. Hear Res 208:114–121
Self T, Mahony M, Fleming J, Walsh J, Brown SD, et al. (1998) Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 125:557–566
Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, et al. (1999) Role of myosin VI in the differentiation of cochlear hair cells. Dev Biol 214:331–341
Shabbir MI, Ahmed ZM, Khan SY, Riazuddin S, Waryah AM, et al. (2006) Mutations of human TMHS cause recessively inherited nonsyndromic hearing loss. J Med Genet 43:634–640
Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, et al. (2004) Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428:950–955
Simske JS, Koppen M, Sims P, Hodgkin J, Yonkof A, et al. (2003) The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol 5:619–625
van Wijk E, van der Zwaag B, Peters T, Zimmerman U, Te Brinke H, et al. (2006) The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet 15:751–765
Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res 130:94–107
Zheng L, Sekerkova G, Vranich K, Tilney LG, Mugnaini E, et al. (2000) The deaf jerker mouse has a mutation in the gene encoding the espin actin-bundling proteins of hair cell stereocilia and lacks espins. Cell 102:377–385
Zine A, Romand R (1996) Development of the auditory receptors of the rat: a SEM study. Brain Res 721:49–58
Acknowledgments
The authors thank the following personnel of the Jackson Laboratory for their contributions to this article: Patsy Nishina and David Bergstrom for helpful comments on the manuscript, Sandra Gray for mouse colony management, Heping Yu for ABR technical assistance, Peter Finger for SEM assistance, Heather Carlisle for design and assembly of the gene-targeting construct, and personnel of the Cell Biology and Microinjection Services for generating the B6.129-Tmhs tm1Kjnmice. They also thank the personnel of the Neuroscience Mutagenesis Facility and the Fine Mapping Service for the initial phenotyping and mapping of the nmf430 (Tmhs hscy-2J) mutation. This work was supported by National Institutes of Health (NIH) Grants DC004301, RR01183, and DC005590 (BF). The Jackson Laboratory institutional shared services are supported by NIH Grant CA34196.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Longo-Guess, C.M., Gagnon, L.H., Fritzsch, B. et al. Targeted knockout and lacZ reporter expression of the mouse Tmhs deafness gene and characterization of the hscy-2J mutation. Mamm Genome 18, 646–656 (2007). https://doi.org/10.1007/s00335-007-9049-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-007-9049-x