Abstract
A crossed population between Iberian × Landrace pigs consisting of 321 F2, 87 F3, and 85 backcross individuals has been analyzed to refine the number and positions of quantitative trait loci (QTL) affecting shape, growth, fatness, and meat quality traits in SSC4. A multitrait multi-QTL approach has been used. Our results suggest that carcass length and shoulder weight are affected by two loci. The first one, close to the AFABP gene, has a very strong pleiotropic effect on fatness, whereas the second one, in the interval between S0073 and S0214, also affects live weight, although to a lesser extent. This latter QTL would correspond to the FAT1 locus described initially in pigs. It seems that SSC4’s loci play an important role in redistributing total weight, and the Landrace allele increases shoulder weight and carcass length much more than ham or total weight. Furthermore, there is also strong evidence of additional loci influencing pH and color in more distant, telomeric positions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A large number of quantitative trait loci (QTL) mapping programs have been developed for the pig over the last decade, most of them consisting of whole-genome scans in crosses between divergent breeds. Many of these experiments have succeeded in identifying multiple QTL related to growth, fatness, meat quality, or reproductive traits (see Bidanel and Rothschild 2002 for a review). The first QTL reported in pigs was located in Chromosome 4 (Andersson et al. 1994) and, despite the high variability of QTL studies, the porcine Chr 4 QTL affecting fatness and/or growth stands as one of the most repeatable results. It has been confirmed in most if not all successive studies (Knott et al. 1998; Walling et al. 1998; Wang et al. 1998; Paszek et al. 1999; Pérez–Enciso et al. 2000; Walling et al. 2000; Bidanel et al. 2001; De Koning et al. 2001; Milan et al. 2002 ). The main effect reported initially was on fatness; this locus was named FAT1 after Marklund et al. (1999), but other studies found that it affected mostly growth De Koning et al. 1999; Rattink et al., 2000). To add complexity, some studies do not give clear single peaks on the chromosome scan for growth QTL (Walling et al. 1998). Thus, it is not clear yet how many loci affecting body composition are segregating on this region of SSC4.
The main limitation to discerning between these alternative hypotheses has been the poor precision of QTL mapping, with confidence intervals of 20–30 cM, complicating the identification of the causal mutation underlying the QTL effect. To refine the QTL position, additional generations can be used to increase the number of recombinants available (Darvasi 1998). It has been shown that multitrait analyses of correlated traits can improve the identification of QTL whose effects are too small to be detected in single-trait analyses Gilbert and Le Roy 2003; Turri et al. 2004). Moreover, multitrait methods allow testing of whether more than one linked QTL is segregating or whether a QTL affects more than one trait (Knott and Haley 2000; Varona et al. 2004). When a pleiotropic QTL is present, using information from different traits simultaneously increases the precision of its location estimate Knott and Haley 2000. Despite its potential advantages, however, multitrait methods have not become popular mainly because of the lack of friendly software and because of computing constraints.
Previously, we developed an F2 cross between Iberian × Landrace breeds in order to detect QTL related to growth, carcass, and meat-quality traits (Pérez–Enciso et al. 2000; Ovilo et al. 2002; Varona et al. 2002. In this article we focus on chromosome 4, where we have typed six additional markers and developed an F3 and backcross. The main goal was to determine whether one or more QTL are segregating on this chromosome and estimate their most likely position and effects. We focus on the complex relationship between growth, fatness, and shape traits that are affected by loci in this chromosome, although meat-quality traits are also analyzed. A new and powerful multitrait approach, implemented in Qxpak (Pérez–Enciso and Misztal 2004), has been used.
Material and methods
Animal material
A comprehensive study of the phenotypic differences between the founder breeds, Iberian Guadyerbas and Landrace, has been published Serra et al. 1998. Full details of the F2 experiment are given in Varona et al. (2002) and references therein. In addition to the 321 F2 animals genotyped in Varona et al. (2002), we included 85 backcross (BC) produced after mating four F2 boars × 22 Landrace gilts and 87 F3-pigs, offspring of three F2 boars × 16 F2 gilts. BC and F3 animals were slaughtered in three and two batches, respectively. Boars contributing to the F3 and BC generations were selected based on marker heterozygosity for Chr 4 and 6 [SSC6 also harbored a large-effect QTL for fatness (Ovilo et al. 2000)]. Three of the boars were sires in both the F3 and BC.
Traits analyzed
The measures of the phenotypic traits are described elsewhere Ovilo et al. 2000; Pérez–Enciso et al. 2000. The traits analyzed here were back fat thickness at the shoulder level (BF1), subcutaneous fat thickness between the third and fourth last ribs (BF34), live weight at slaughter (LW), carcass length (CL), shoulder weight (SW), ham weight (HW), pH measured 24-h postmortem in Semimembranosus muscle (pH24), muscle color Minolta measurements (L* for lightness and a* for redness) and % of pigment content (% Haem) measured in the Longissimus thoracis muscle. BF34, color, and % Haem content measurements were available only in the F2 individuals.
Genotyping
Genomic DNA from parental, F1 and F2 individuals was extracted using a saline precipitation protocol (Miller et al. 1988), and DNA from F3 and BC was obtained from blood using a phenol/chloroform extraction (Ausubel et al. 1987). Animals were genotyped for 14 markers— 13 microsatellites and 1 single nucleotide polymorphism (SNP) (2,4-dienoyl CoA reductase 1 gene (DECR1)) (Clop et al., 2002), Sw2404 (0.0), S0301 (37.2), S0001 (57.0), Sw317 (67.5), Sw35 (69.0), AFABP (69.8), Sw839 (74.2), DECR (79.3), S0073 (91.6), S0214 (95.9), Sw524 (114.0), Sw445 (128.0), Sw58 (130.5), and S0097 (145.9). Numbers in parentheses are the positions (in cM) obtained with the option “build” of the CRI-MAP program version 2.4 (Green et al. 1990). Six of these microsatellites (Sw317, Sw35, AFABP, S00073, Sw524, and Sw58) are new compared with the previous report Pérez–Enciso et al. 2000. Polymerase chain reactions (PCR) were carried out in an automatic PCR ABI PRISM 877 integrated thermal cycler (Applied Biosystem, Fostercity, CA). Amplified PCR products were analyzed by capillary electrophoresis with fluorescent detection in an ABI PRISM 3100 Genetic analyzer (Applied Biosystems). Finally, genotypes were analyzed with Genescan Analysis software v3.7 (Applied Biosystems) and stored in the Gemma database Iannuccelli et al. 1996.
QTL analyses
For this QTL study we have used mixed-model techniques, which offer a much wider modeling flexibility than traditional regression-based techniques. In particular, it allows us to use different models for each trait in multitrait analyses, adding an infinitesimal genetic residue, missing records in an arbitrary manner, or allowing for genetic variability within purebreds. In addition, the required identity by descent probabilities is computed using all animals in the pedigree (F0, F1, F2 F3, and BC) simultaneously. We have used recently developed software, Qxpak, which implements these features (Pérez–Enciso and Misztal 2004). Qxpak allows for fitting the QTL effect as either “fixed,” which assumes that alternative alleles are fixed in each breed, or “mixed,” which allows the QTL to be segregating within lines. We have not found any evidence of variability within lines, so only the results with the fixed option are reported here. The general univariate model for all traits is
where y i is the ith individual record, β is the covariate coefficient, c is carcass weight (except for LW itself, that is corrected by age), a is the QTL additive effect, d is the dominant effect, u is the infinitesimal genetic effect, and e is the residual. The coefficients P (g i ≡ VW) are the probabilities, obtained via a Monte Carlo Markov chain algorithm, of the ith individual having alleles of breed origin V and W at the position of interest (Pérez–Enciso and Misztal 2004). The Haldane mapping function is assumed to obtain these probabilities. The dominant QTL effect d is included only when it is significant (nominal p value < 5%). The infinitesimal genetic effect is treated as random, with covariance A σ2u, A being the numerator relationship matrix. A single residual variance is assumed for all generations (F2, F3 and BC). The traits carcass length, ham weight and shoulder weight are also corrected by age. We call these “shape” traits because they measure how body weight is distributed across the different body parts. Rough, approximate confidence intervals are obtained using the LOD-drop method.
In addition to single-trait single-QTL analyses, we have carried out a series of multitrait and multi-QTL analyses, outlined in Table 1. The statistical model is the same as in Eq. (1), but here the variance of the infinitesimal genetic effects is, for a two-trait model,
with the subindices 1 and 2 referring to traits 1 and 2, respectively, and ⊗ denotes the Kronecker product. Multitrait models allow us to fit the same or different QTL effects for any of the traits. Models 1, 2, 3, and 8 in Table 1 are single-QTL models where several sets of traits were analyzed jointly assuming a single pleiotropic QTL, e.g., in model 1 we analyzed BF1 and BF34, both corrected by carcass weight and assuming that the QTL effect was strictly additive. Model 4 is also a single-QTL model and helped us to determine whether there is one or more QTL affecting the shape traits; it was compared with models 5, 6, and 7. These last three models involve multivariate multi-QTL analyses. In model 5, SW is affected by two QTL, the first one also influencing fatness (BF1) and the second one influencing growth (LW). In contrast, in models 6 and 7 no trait is affected by more than one QTL. Either the same QTL affects BF1 and SW (model 6) or SW and LW (model 7). The likelihood of models 5, 6, and 7 was compared against model 4 and against a model without any QTL. The second QTL (QTL2) could be dominant, whereas the first one is strictly additive. In order to spare computing time, QTL1 was fitted only between positions 60 and 80 cM, and QTL2 was fitted between 60 and 100 cM for models 1–7 (carcass traits) and between 90 and 145 cM for models 8 and 9 (quality traits). These ranges should cover all potential positions for the QTL.
Likelihood ratio tests were computed to assess the significance of the QTL effects. The p values so obtained were nominal; permutation techniques cannot be applied here because we included an infinitesimal genetic value and randomization of the data would break the family structure. But tentative approximations can be used: in a previous study (Pérez–Enciso et al. 2000) we showed that chi-squared values with 2 degrees of freedom corresponding to 5% and 1% chromosome-wise significance p values were 10.5 and 14.2; these figures in turn roughly corresponded to nominal p values of 0.005 and 0.001. Although we cannot claim that this is a completely satisfactory solution, we can reasonably argue that nominal p values < 0.001 are significant for the single QTL test. Most p values reported here are well beyond that bound.
Results and discussion
A glance at (Fig. 1), which shows the univariate analyses, clearly shows that this is a “rich-QTL” chromosome. Closer inspection reveals at least three promising regions around positions 70, 85–95, and 110–120 cM. Fatness and growth QTL are located in the first two regions and color-related QTL are positioned in the last region. The shape traits (carcass length and shoulder weight) tend to show two maxima about positions 70 and 90 cM, the bimodal profile being much more pronounced when corrected by age. It is likely that the pH QTL is located in another more telomeric position. The main results are presented in Table 2. Note that p values are highly significant for the traits shown, except ham weight. Gene action is additive for all traits, except for live weight, with the heterozygote closer to the homozygous Iberian genotype.
Back fat thickness traits
The most significant QTL found are those affecting back fat thickness (BF1 and BF34); they map at positions 73 and 85 cM, respectively (Table 2). As expected, the Iberian allele increases fatness. Since the two positions are in different marker intervals, the first important question is whether one or two fatness QTL are segregating and which is the most likely QTL position(s). BF34 p value profile is multimodal and more significant than BF1, whereas the BF1 p value profile is unimodal (Fig. 1). The significance in BF34 is higher than in BF1 because BF34 is a more precise measurement; it is done in the lab with a cross section of the carcass, whereas BF1 is measured in the slaughterhouse. The BF1 QTL region agrees with the positions previously described in this experiment that used a smaller number of markers and animals (Pérez–Enciso et al. 2000; Varona et al. 2002). It seems that both traits have the same genetic basis, as the estimated genetic correlation between BF1 and BF34 is 0.81 and the environmental correlation is smaller, about 0.45, probably because the measurement errors are quite different.
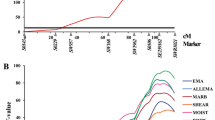
To resolve the question of the most likely position, we ran a series of multitrait analyses (Fig. 2) restricted to the most interesting region, 50–120 cM. Previously, we had found that an additional QTL was not significant in a two-QTL model for BF34 (results not presented). The joint analysis of BF34 and BF1 shows no clear indication of the most likely QTL position but suggests that it is approximately 73 cM, i.e., coincident with BF1 estimate. Next, we added the shape traits (shoulder weight and carcass length). Then the joint analysis clearly pinpointed position 73 cM, very close to the AFABP gene. It is interesting to note that adding carcass length did not significantly improve the results once BF1, BF34, and SW were included in the model. These results suggest that there is only one QTL affecting fatness and is located on position 73 cM of our map (close to AFABP and Sw839) rather than in position 85, i.e., close to S0073.
Log10 p-value profiles in the multitrait, single-QTL analyses. Note that in order to save computing time, the whole chromosome was not scanned. Description of the models is in Table 1; the null model implies no QTL in any trait.
Growth traits
As noted above, the effect on growth of SSC4s loci has been debated in the literature. In an early study of only 200 F2 animals (Pérez-Enciso et al. 2000), we did not find an effect on growth, but here, with more data, the nominal p value obtained for LW was 5.7 × 10−4 (Table 2), much higher (i.e., less significant) than the p values observed for fatness but still significant. The differences in effects reported between different QTL studies may be caused in part by lack of power, although the stage of growth recorded also has influence (Wang et al. 1998; Walling et al. 2000; Bidanel et al. 2001; Knott et al. 2002). The most likely position for growth obtained in this study, between markers S0073 and S0214, is consistent with the position described previously (Varona et al. 2002). The profile of ham weight QTL runs parallel to that of live weight but the genetic correlation between both traits is small (0.30), suggesting that overall their genetic architectures are rather different.
Shape traits
Although the most significant effect found in this work is fatness, “shape” traits (carcass length and shoulder weight) are also highly significant and exhibit almost completely parallel profiles, indicating that they share the same genetic basis regarding this chromosome. The estimated genetic correlation between both traits is rather high and positive, 0.77. Both profiles are clearly bimodal; the first maximum coincides with the fatness QTL position and the second coincides with the growth QTL. The bimodal profile is much more evident when the traits are corrected by age; in this case, the likelihoods at the two maxima are almost identical. Thus, it seems that the SSC4 loci have a strong effect on redistributing total weight: The Landrace allele favors an increase in shoulder weight and carcass length, whereas the effect on total weight is much more moderate and almost negligible on ham weights. In short, the SSC4 QTL change(s) the shape of the pig.
From the univariate profiles (Fig. 1), we conjectured that there may exist at least two QTL, one affecting growth and the other affecting fatness, and both affecting the shape of the animal (carcass length and shoulder weight). In order to test this hypothesis, we ran a series of multitrait multi-QTL models as described in Table 1; the main results are in Table 3. The fatness QTL (QTL1) is additive and the growth QTL (QTL2) shows dominance. SW and BF1 are corrected by carcass weight and LW is corrected by age at slaughter. For the sake of comparison, the corresponding single-QTL model is also shown (model 4). Two p values are shown: p value0 corresponds to the likelihood ratio against a non-QTL model and p value1 corresponds to the likelihood ratio against model 4. Models 6 and 7 do not result in very coherent results since the two QTL are located in the same region, including that for LW. As a result, a test of model 6 or 7 against 4 does not allow us to reject the null hypothesis (model 4). In contrast, model 5 results agree with univariate analyses (Fig. 1), in particular, the two QTL are in the expected regions. Note that the effects of the two QTL affecting SW would be similar and that when only one QTL is fitted, this absorbs the whole effect. The p value1 borders significance (it would be significant at the nominal level). Note that the dominant effect is not significant for SW (0.19 ± 0.11). If we removed that effect from the model, the degrees of freedom and the p value would decrease. Unfortunately, Qxpak does not allow the same QTL to be fit with different genic actions for each trait.
Thus, it seems that the largest-effect QTL on SSC4 affects primarily fatness, and it is located in interval 70 cM, whereas a second QTL that affects growth lies in position 95 cM. Both QTL affect the shape of the animal. Alternatively, it can be argued that the QTL in position 95 cM primarily affects the shape and that the effect on growth is a secondary, pleiotropic effect. The choice of these two alternative hypotheses will have an effect on the kind of positional candidate genes chosen for future studies. The appearance of loci that affect fatness and shape, in particular, carcass length, in genome locations that are quite close is extremely interesting when seen through the perspective of the 19th century breeders. In the case of the Danish Landrace breed, a harmonious animal with a long body was especially valued. Some Landrace individuals were reported to have 16–17 rib pairs instead of the usual 14–15 pairs. In contrast it seems that no special attention was paid in the early days to the size of the shoulders or hams. In his review, Jonsson (1975) noted a decrease of −0.86 cm in back fat thickness and an increase of 2.2 cm in body length between 1958 and 1972. Nothing is said, though, about shoulder weights. The pig shoulder develops earlier than the ham and given that selection for growth and leanness has resulted in a more juvenile animal, it can be expected that genes with an effect on fatness may have an indirect effect on development. Moller et al. (2004) recently published a detailed comparative mapping of the FAT1 regions of pig, human, and mouse. Interestingly, the FAT1 locus corresponds to the second QTL described in this work, which affects mostly growth and is homologous to HSA1. In contrast, the greatest effect of the fatness QTL described here lies in a region homologous to HSA8. This result affects the choice of positional candidate genes for further studies
Quality traits
So far we have focused on carcass traits, but more QTL seem to be segregating on this particularly complex chromosome. A significant QTL for muscle color (L* and a*) is located at positions 108 and 114 cM, respectively. Another OTL for pigment content (% Haem) also maps at position 114 cM (Table 2, Fig. 1). These results are coincident with the positions previously described (Ovilo et al. 2002), confirming the presence of these QTL. This region coincides with the position of the putative QTL for color reported by Wang et al. (1998). The Iberian allele increases pigment content and meat redness but decreases lightness, in agreement with the breed differences (Serra et al. 1998). In addition, a QTL for pH 24h maps to position 134. In these cases, an increment of pH occurs when Iberian alleles are present, again in agreement with breed phenotypes (Serra et al. 1998). The pH QTL was not found in our previous analysis of this experiment, with less individuals and markers, on this chromosome, although one was detected on Chromosome 3 by Ovilo et al. (2002). Multitrait analyses were carried out, paralleling those for carcass traits (Table 1). Results for model 8 are shown in Fig. 2, with a maximum located at position 110 cM. Next, we compared a two-QTL (model 9) with a one-QTL model (model 8, Table 1); the difference in likelihood was 1.98 (i.e., p > 0.05). Thus, we do not have enough evidence to reject the one-QTL model.
Conclusion
This work reports a novel multitrait multi-QTL analysis with mixed-model techniques. It also reports the analyses with new individuals and more markers. We have shown the usefulness of multitrait methods, allowing us to increase the power of the detection of QTL compared with single-trait analyses, and we have highlighted the importance of modeling, e.g., compare the two profiles of SW when correcting by weight or by age (Fig. 1). A potential drawback of multitrait maximum likelihood techniques is that the number of possible models to be tested is very large, making biological interpretation of the results difficult. Here we have presented only a subset of all possible tests that can be performed. We have argued that there must exist at least two loci with an effect on the shape of the animal, with the effect on growth being most likely an indirect, pleiotropic effect. There must exist several mutations on SSC4 with a dramatic effect on the phenotype, and some of these mutations probably were selected for since the 19th century because of their effect on the shape of the animal. The fact that the fatness QTL are in the same region must have accelerated the change in this chromosome region once that leanness became an important target in modern breeding. Because artificial selection has changed not only the performance but also the shape of the animal, analyzing these changes shed light on the nature of the economically important QTL. Unfortunately, very little attention has been paid to the genetic architecture of shape in domestic animals, which contrasts to studies in other species like mice (e.g., Klingenberg et al. 2001) or plants (Frary et al. 2004). In the future, specialized allometric techniques could be used (e.g., Wu et al. 2002).
References
Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, et al. (1994) Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263, 1771–1774
Ausubel F, Brent R, Kingston R, Moore D, Seidman J, et al. (1987) Current protocols in molecular biology. (New York: Greene Publishing Associates and Wiley-Interscience)
Bidanel JP, Milan D, Iannuccelli N, Amigues Y, Boscher MY, et al. (2001) Detection of quantitative trait loci for growth and fatness in pigs. Genet Sel Evol 33, 289–309
Bidanel JP, Rothschild M (2002) Current status of quantitative trait locus mapping in pigs. Pig News and Information 23, 39–54
Clop A, Cercos A, Tomas A, Pérez–Enciso M, Varona L, et al. (2002) Assignment of the 2,4-dienoyl-CoA reductase (DECR) gene to porcine chromosome 4. Anim Genet 33, 164–165
Darvasi A (1998) Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet 18, 19–24
De Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ, et al. (1999) Detection of quantitative trait loci for back fat thickness and intramuscular fat content in pigs (Sus scrofa). Genetics 152, 1679–1690
De Koning DJ, Rattink AP, Harlizius B, Groenen MAM, Brascamp EW, et al. (2001) Detection and characterization of quantitative trait loci for growth and reproduction traits in pigs. Livest Prod Sci 72, 185–198
Frary A, Fritz L, Tanksley S (2004) A comparative study of the genetic bases of natural variation in tomato leaf, sepal, and petal morphology. Theor Appl Genet 109, 523–533
Gilbert H, Le Roy P (2003) Comparison of three multitrait methods for QTL detection. Genet Sel Evol 35; 281–304
Green P, Falls K, Crooks S (1990) Documentation for CRIMAP. Unpublished mimeo. Available at: http ://biobase.dk/Embnetut/Crimap/
Iannuccelli N, Woloszyn N, Arhainx J, Gellin J, Milan D (1996) GEMMA: a database to manage and automate microsatellite genotyping. In Proceeding of the International Society of Animal Genetics Conference, Tours, France. (Oxford, UK: Blackwell) p 88
Jonsson P (1975) Methods of pig improvement through breeding in the European countries: A review. Livest Prod Sci 2, 1–28
Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM (2001) Genetic architecture of mandible shape in mice: Effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157, 785–802
Knott SA, Haley CS (2000) Multitrait least squares for quantitative trait loci detection. Genetics 156, 899–911
Knott SA, Marklund L, Haley CS, Andersson K, Davies W, et al. (1998) Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 149, 1069–1080
Knott SA, Nystrom PE, Anderssoneklund L, Stern S, Marklund L, et al. (2002) Approaches to interval mapping of QTL in a multigeneration pedigree: the example of porcine chromosome 4. Anim Genet 33, 26– 32
Marklund L, Nystrom PE, Stern S, Andersson–Eklund L, Andersson L (1999) Confirmed quantitative trait loci for fatness and growth on pig chromosome 4. Heredity 82(Pt 2), 134–141
Milan D, Bidanel JP, Iannuccelli N, Riquet J, Amigues Y, et al. (2002) Detection of quantitative trait loci for carcass composition traits in pigs. Genet Sel Evol 34, 705–728
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16, 1215
Moller M, Berg F, Riquet J, Pomp D, Archibald A, et al. (2004) High-resolution comparative mapping of pig Chromosome 4, emphasizing the FAT1 region. Mamm Genome 15, 717–731
Ovilo C, Pérez–Enciso M, Barragan C, Clop A, Rodriguez C, et al. (2000) A QTL for intramuscular fat and back fat thickness is located on porcine chromosome 6. Mamm Genome 11, 344–346
Ovilo C, Clop A, Noguera JL, Oliver MA, Barragan C, et al. (2002) Quantitative trait locus mapping for meat quality traits in an Iberian × Landrace F2 pig population. J Anim Sci 80, 2801–2808
Paszek AA, Wilkie PJ, Flickinger GH, Rohrer GA, Alexander LJ, et al. (1999) Interval mapping of growth in divergent swine cross. Mamm Genome 10, 117–122
Pérez–Enciso M, Misztal I (2004) Qxpak: a versatile mixed model application for genetical genomics and QTL analyses. Bioinformatics 20, 2792–2798
Pérez–Enciso M, Clop A, Noguera JL, Ovilo C, Coll A, et al. (2000) A QTL on pig chromosome 4 affects fatty acid metabolism: evidence from an Iberian by Landrace intercross. J Anim Sci 78, 2525–2531
Rattink AP, De Koning DJ, Faivre M, Harlizius B, van Arendonk JA, et al. (2000) Fine mapping and imprinting analysis for fatness trait QTLs in pigs. Mamm Genome 11, 656–661
Serra X, Gil F, Pérez–Enciso M, Oliver MA, Vázquez JM, et al. (1998) A comparison of carcass, meat quality and histochemical characteristics of Iberian and Landrace pigs. Livest Prod Sci 56, 215–223
Turri MG, DeFries JC, Henderson ND, Flint J (2004) Multivariate analysis of quantitative trait loci influencing variation in anxiety-related behavior in laboratory mice. Mamm Genome 15, 69–76
Varona L, Ovilo C, Clop A, Noguera JL, Pérez–Enciso M, et al. (2002) QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet Res 80: 145-154
Varona L, Gomez–Raya L, Rauw WM, Clop A, Ovilo C, et al. (2004) Derivation of a Bayes factor to distinguish between linked or pleiotropic quantitative trait loci. Genetics 166, 1025–1035
Walling GA, Archibald AL, Cattermole JA, Downing AC, Finlayson HA, et al. (1998) Mapping of quantitative trait loci on porcine chromosome 4. Anim Genet 29, 415–424
Walling GA, Visscher PM, Andersson L, Rothschild MF, Wang L, et al. (2000) Combined analyses of data from quantitative trait loci mapping studies. Chromosome 4 effects on porcine growth and fatness. Genetics 155, 1369–1378
Wang L, Yu TP, Tuggle CK, Liu HC, Rothschild MF (1998) A directed search for quantitative trait loci on chromosomes 4 and 7 in pigs. J Anim Sci 76, 2560–2567
Wu R, Ma CX, Littell RC, Casella G (2002) A statistical model for the genetic origin of allometric scaling laws in biology. J Theor Biol 219, 121–135
Acknowledgments
This project was funded by Ministerio de Ciencia y Tecnologia (MCYT) (AGF99−0284-CO2) and (CPE03-010) grants. A. Mercadé was funded by a Formació Personal Investigador (FI) fellowship from the Generalitat de Catalunya, and J. Estellé was funded by a Formación Personal Universitario (FPU) grant from the Spanish Ministry of Education and Science (MEC). Part of the analyses was carried out in the Centre de Supercomputació de Catalunya (CESCA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mercadé, A., Estellé, J., Noguera, J.L. et al. On growth, fatness, and form: A further look at porcine Chromosome 4 in an Iberian × Landrace cross. Mamm Genome 16, 374–382 (2005). https://doi.org/10.1007/s00335-004-2447-4
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00335-004-2447-4