Abstract
Objectives
The aim of this study was to develop and validate a radiomics signature for predicting survival and chemotherapeutic benefits of patients with lower-grade gliomas (LGG).
Methods
Radiomics features were extracted from precontrast axial fluid-attenuated inversion recovery (FLAIR) and contrast-enhanced axial T-1 weighted (CE-T1-w) sequence. Lasso Cox regression model was used for feature selection and radiomics signature building. The radiomics signature was developed in a primary cohort that consisted of 149 LGG patients and was then validated on an entirely new validation cohort that contained 66 LGG patients. A radiomics nomogram for the prediction of OS was established by adding the radiomics to clinicopathologic nomogram which developed with clinical data.
Results
A radiomics signature derived from joint CE-T1-w and FLAIR images showed better prognostic performance (C-index, 0.798) than signatures derived from CE-T1-w (C-index, 0.744) or FLAIR (C-index, 0.736) sequences alone. Multivariable Cox regression revealed that the radiomics signature was an independent prognostic factor. One radiomics nomogram integrated the radiomics signature from joint CE-T1-w and FLAIR sequences with the clinicopathologic nomogram outperformed the clinicopathologic nomogram based on clinicopathologic data alone in predicting OS of LGG (C-index, 0.821 vs. 0.692; p < 0.001). Further analysis revealed that patients with higher radiomics signature were prone to benefit from chemotherapy.
Conclusions
The radiomics signature was independent with clinicopathologic data and was a noninvasive pretreatment predictor for LGG patients’ survival. Moreover, it could predict which patients with LGG benefit from chemotherapy.
Key Points
• A radiomics signature derived from joint CE-T1-w and FLAIR sequences showed better prognostic performance than signatures derived from either single imaging modality.
• The radiomics signature is an independent prognostic factor and outperformed clinicopathologic features in predicting overall survival of LGG patients.
• The radiomics signature could help preoperatively identify LGG patients who may benefit from chemotherapy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Gliomas are the most common primary malignant brain tumors in adults. The lower-grade gliomas (LGG) comprised of diffuse low-grade and intermediate-grade gliomas (the World Health Organization (WHO), grade II and III gliomas [1,2,3] have relatively favorable but highly variable prognosis [1]. While most of the patients survive a long time, there is still a large subset of patients who have a very short lifespan [4, 5]. This results in the need for identifying high-risk LGG patients with poor survival outcomes, for whom more intensified treatment was needed. Moreover, existing studies have shown that one group of patients does not benefit from the current guidelines recommend chemotherapy of glioma, represented mainly by temozolomide [6, 7]. Therefore, accurate identification of potential LGG patients who may benefit from temozolomide (TMZ) chemotherapy will contribute to more personalized treatment. Although molecular biomarkers such as IDH1 mutation and 1p/19q codeletion appear to have the ability to stratify survival of LGG more accurately [1, 8,9,10,11,12] and confirming MGMT promoter methylation status could help to predict the efficacy of chemotherapy [13, 14], the approach to obtain this information is always invasive, expensive, and not available until the end of some treatment course. Therefore, developing noninvasive and preoperative biomarkers will be of great importance for prognostic counseling and treatment planning of patients in LGG.
Radiomics is an emerging methodology that extending radiological images into comprehensive and quantitative features, which may provide insights into personalized medicine [15,16,17,18]. Most recently, a radiomics approach has been successfully utilized in predicting the prognosis in response to treatment for a variety of cancers [19,20,21,22]. Aerts et al demonstrated that 440 radiomics features extracted from computed tomography data of 1019 patients with lung or head-and-neck cancer had prognostic value [23].
As for LGG, few studies have investigated the association between radiomics features and survival [24, 25]. Zhou and et al showed MR imaging data can be used to predict tumor progression [24]. Liu and et al showed progression-free survival can be predicted noninvasively in patients with LGGs by a group of radiomics features [25]. However, these studies only used a few kinds of image modalities or extracted a small number of radiomics features, which caused limited mining of imaging data. In terms of TMZ chemotherapy response of LGG, although there are few studies that have evaluated the potential of MR radiomics features to predict MGMT methylation [26], a prognostic biomarker, no study has made effort to predict how much benefit LGG patients would derive from TMZ chemotherapy using radiomics features.
In this study, we will extract comprehensive radiomics features from the FLAIR and CE-T1-w sequence and construct a radiomics signature based on these features. We will test the hypothesis that the radiomics signature is an independent prognostic factor of LGG patients and investigate whether it could be used to identify patients who might benefit from temozolomide treatment. Our ultimate aim is to develop and validate a radiomics signature for predicting survival and chemotherapeutic benefits of patients with lower-grade gliomas.
Materials and methods
Patients
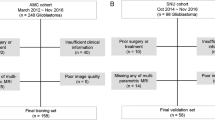
Ethical approval for this retrospective analysis was obtained and the need to obtain informed consent from the patients was waived. The primary cohort of this study comprised an evaluation of the medical records database of Linyi People’s Hospital from February 2011 to November 2016 to identify patients with histologically confirmed LGG who underwent surgical resection. A total of 149 patients were consecutively included in the primary cohort according to the following inclusion and exclusion criteria. The inclusion criteria were (a) newly diagnosed patients without any surgical history, (b) both two MRI sequences (FLAIR and CE-T1-w) were available before any treatment, (c) pathologically confirmed typical histological-based WHO classification was available, (d) MRI scans with diagnostic image quality, (e) the follow-up time not less than 24 months, (f) TMZ was used as the therapeutic drug if chemotherapy was conducted. The exclusion criteria were: (a) not undergoing surgery at Linyi People’s Hospital or Qilu Hospital of Shandong University, (b) insufficient MRI quality to obtain measurements. From February 2016 to April 2017 at Qilu Hospital of Shandong University, an independent validation cohort consisted of 66 patients meeting the above criteria was enrolled. The patient recruitment pathway, as well as the inclusion and exclusion criteria, was presented in Fig. S1. Overall survival (OS) of all patients in two cohorts was calculated from the date of surgery until death or last follow-up visit.
MRI acquisition and image preprocessing
Patients in two cohorts underwent pretreatment 3.0-T MRI scans using the MR system in the same version (Magnetom Verio, Siemens Healthineers) but with different parameters. Detailed imaging acquiring parameters (slice thickness, voxel size, and slice gap, etc.) used for each sequence in each cohort were shown in Supplementary Material S1.
To reduce the discrepancy of imaging parameters that were employed in different hospitals, a postprocessing pipeline including bias correction, skull stripe, registration, and intensity normalization was applied on all MRI scans. First, within-patient registration was performed, in which the FLAIR and CE-T1-w sequences of the same patient were aligned into the same geometric space using the General Registrations (BRAINS) Toolbox in 3D Slicer. In the second step, bias correction was applied on FLAIR and CE-T1-w images to compensate for intensity non-uniformities using N4 algorithm implemented in “extrantsr” package. In the third step, to remove the effect of grey values’ skull on intensity normalization in the next step, brain tissue was extracted from the rest of the image using “fslr” package which wraps FSL commands to use in R. Finally, accounting for scanner differences, intensities of images were normalized using “WhiteStripe” package in R which conducts white stripe normalization procedure. An overview of the image postprocessing pipeline is shown in Fig. 1a.
Radiomics analysis pipeline for solid tumor. a Preprocessing pipeline including within-patient registration, bias correction, brain extraction, and intensity normalization. b (I) Tumor contours delineated on FLAIR sequence was saved in a label volume and then placed on both the post-processed FLAIR and CE-T1-w sequence. (II) Features quantifying first-order, shape/size, and texture were extracted from within the defined tumor contours on the original and wavelet filtered MRI images. (III) Feature selection and radiomics signature building were performed using LASSO logistic model in terms of prediction for short- versus long-term survival. The prediction performance was assessed using ROC analysis and waterfall plot
Segmentation and feature extraction
After within-patient image registration, tumor regions of interest (ROI) were semi-automatically segmented slice by slice using 3D Slicer (www.slicer.org) on FLAIR sequence, because the FLAIR sequence is well accepted in identifying tumor borders of lower-grade gliomas. The ROI of each patient was saved in a label volume and then placed on the CE-T1-w sequence (Fig. 1b). The ROI segmentation was performed by two radiologists (Xuejun Zheng and Rui Jing) with more than 10 years of experience in brain MRI study interpretation. Both two radiologists were blinded to the patient cohort, clinicopathological data, and survival. The interobserver agreement of radiomics feature extraction was assessed using the intraclass correlation coefficients (ICC) [27], considering an ICC greater than 0.75 to represent good agreement.
A large set of quantitative radiomics features were extracted using the PyRadiomics [28], an open-source python package for the extraction of radiomics features from medical images. The images and ROI contours were resampled to a spatial resolution of 1 × 1 × 6 mm using the “sitkBSpline” interpolation and gray level was discretized with a bin width of 25. In total, 1576 radiomics features were extracted per patient (788 radiomics features were extracted from FLAIR and CE-T1-w sequence, respectively). More detailed information about the radiomics features can be found in Supplementary Material S2.
Data analysis
Construction of the radiomics signature
The LASSO Cox regression algorithm was used to select the most useful prognostic features in the primary cohort. The features with nonzero coefficients were selected when the optimal model was determined by 5-fold cross-validation based on minimum criteria. Weighted by their respective LASSO coefficients, these selected features were then combined into a radiomics signature. The predictive efficacy of the radiomics signature was quantified by Harrell’s concordance index (C-index) in both the primary and validation cohorts.
Validation of radiomics signature
To assess the potential association of radiomics signature with OS, we divided the patients into high risk and low risk based on the median value of individual radiomics signatures [29]. Patients whose radiomics signature larger than the threshold were placed in high-risk groups. Survival curves of two groups were evaluated using Kaplan–Meier methods and the difference in the survival curves was tested using the log-rank test in the primary and validation cohorts. Multivariate Cox regression analysis was further performed to identify whether the radiomics signature is an independent prognostic factor.
Assessment of the incremental value of radiomics signature
To demonstrate the incremental value of the radiomics signature to the clinical-pathologic risk factors for assessment of OS in LGG patients, both a radiomics nomogram and a clinical nomogram were developed in the primary cohort. The clinical nomogram was constructed based on a multivariable Cox regression which incorporated the independent clinicopathologic variables. The radiomics nomogram integrated the radiomics signature and all the independent clinicopathological risk factors in the clinical nomogram.
The incremental value of the radiomics signature to the clinicopathological risk factors was assessed by comparing the performance of the radiomics nomogram in respect of calibration and discrimination with that of the clinical nomogram. The calibration performance of two nomograms was compared using calibration curves which assess the agreement between the observed survival and the predicted survival. The discrimination performance was measured using C-index.
Investigation of the prediction ability of radiomics signature for the chemotherapeutic benefits
Moreover, to explore the ability of radiomics signature to predict chemotherapeutic benefits, we investigated whether patients in high- and low-risk groups defined above could benefit from postoperative TMZ chemotherapy. We first tested the interaction effect between risk group and TMZ treatment in a multivariable Cox regression adjusting other independent clinicopathologic variables. A subset analysis was then conducted to test the difference of survival between patients who received TMZ chemotherapy or not in each risk subgroup. The corresponding Kaplan–Meier survival curves, which compared the TMZ chemotherapy group with the no chemotherapy group by risk group, were evaluated using Kaplan–Meier methods and the difference in the survival curves was tested using log-rank test in the low- and high- risk subgroups separately. The interaction effect test and subset analysis were performed in the primary cohort and validated in the validation cohort.
All statistical analyses were performed with R software, version 3.5.2 (http://www.R-project.org). All statistical tests were two-sided with a statistical significance of p < 0.05.
Results
Clinical characteristics of the patients
The clinical characteristics of the primary cohort and validation cohort are summarized in Table 1. No significant differences were found between the training and validation cohorts in terms of age, gender, grade, radiotherapy treatment, chemotherapy treatment, seizure history, and survivors (p > 0.05). More detailed characteristics between grade II and grade III in two cohorts were presented in Supplementary Table S1.
Feature selection and radiomics signature building
The interobserver ICCs ranged from 0.796 to 0.928, indicating favorable interobserver reproducibility of feature extraction. To build the radiomics signatures derived from CE-T1-w sequence and derived from the FLAIR sequence, thirteen features from CE-T1-w sequence, and seventeen features from FLAIR sequence were selected with nonzero coefficients in the primary cohort, respectively. To build the radiomics signature derived from joint CE-T1-w and FLAIR sequence, twelve features from CE-T1-w sequence and seventeen features from FLAIR sequence with nonzero coefficients were selected in the primary cohort. Weighted by their respective LASSO coefficients, these selected features were then combined into a radiomics signature derived from the respective sequence. The detailed information of the optimum LASSO logistic regression model and the radiomics signature calculation formula are presented in Supplementary Materials S3.
Prognostic validation of the radiomics signature
In the primary cohort, the radiomics signature derived from the CE-T1-w sequence yielded a C-index of 0.744 (95% CI: 0.673–0.814). The radiomics signature from the FLAIR sequence yielded a C-index of 0.736 (95% CI: 0.663–0.808). The radiomics signature from joint CE-T1-w and FLAIR sequences yielded the highest C-index, which was 0.798 (95% CI: 0.735–0.861). The consensus results were found in the validation cohort; the radiomics signature derived from CE-T1-w sequence, FLAIR sequence, and joint two sequences yielded the C-index of 0.604 (95% CI: 0.480–0.728), 0.635 (95% CI: 0.511–0.759), and 0.678 (95% CI: 0.554–0.802), respectively. Therefore, the radiomics signature derived from joint CE-T1-w and FLAIR sequence was adopted in the next analysis.
The median of the radiomics signature in the primary cohort was used to stratify patients into high-risk group (radiomics signature ≥ - 3.002) and low-risk group (radiomics signature < - 3.002). Then, the same threshold values were applied to the validation cohort. Patients in the low-risk group had a significantly favorable OS. The radiomics signature was associated with the OS in the primary cohort (p < 0.001; HR = 8.376, 95% CI: 3.756–18.678) (Fig. 2a), and this finding was confirmed in the validation cohort (p = 0.030; HR = 3.820; 95% CI: 1.135–12.854) (Fig. 2b).
Construction of clinical nomogram and radiomics nomogram
We first constructed a multivariable Cox regression model which included all the clinicopathologic variables with a p value lower than 0.05 in the univariable Cox regression model. The multivariable Cox regression analysis identified age, histologic grade, and IDH1 mutation as independent risk factors (Table S2). We first developed a clinicopathologic nomogram based on this multivariable Cox regression model (Fig. 3a). Next, we included the radiomics signature in the multivariate Cox regression, which indicated that the radiomics signature remained an independent prognostic factor even after adjusted clinicopathological variables (Table S3). No significant difference was found in radiomics signature between subgroups of each clinicopathological variable (Table S4). A radiomics nomogram was then developed by combining the radiomics signature and the significant clinicopathologic variables (Fig. 3c). The calibration curves for the probability of survival at 1, 2, or 3 years after surgery showed good agreement between the estimation with the clinicopathologic nomogram and actual observation (Fig. 3b). The calibration curve of the radiomics nomogram demonstrated better performance (Fig. 3d).
Use of the constructed the clinical-pathologic nomogram and radiomics nomogram to estimate the OS for LGG, along with the assessment of the model calibration. a Clinical-pathologic nomogram. To use the nomogram, locate an individual patient’s value on each variable axis and draw a line upward to the points axis to determine how many points were received for each variable value. Then, sum the numbers and locate it on the total points axis. A line is drawn downward to the survival axes to determine the likelihood of 1-, 2-, or 3-year survival. b The calibration curve of clinical-pathologic nomogram for predicting patient survival at 1-, 2-, 3-year survival which presented with the red, blue, purple line, respectively. Nomogram predicted probability of OS is plotted on the x-axis while the actual OS is plotted on the y-axis. A closer lining with the diagonal dotted line represents a better estimation. c Radiomics nomogram which was constructed with integrating the radiomics signature with the clinical-pathological nomogram. d The calibration curve of the radiomics nomogram
The incremental value of the radiomics signature when added to the clinical data
In the primary cohort, the radiomics nomogram that combining the radiomics signature overperformed the clinicopathologic nomogram based clinicopathological risk factors alone (C-index: 0.821; 95% CI: 0.768–0.874 vs. 0.692; 95% CI: 0.623–0.762). When tested in the validation cohort, the radiomics nomogram also yielded a better prediction accuracy (C-index: 0.763; 95% CI: 0.639–0.887) than the clinicopathologic nomogram (C-index: 0.675; 95% CI: 0.553–0.797).
The prediction ability of radiomics signature for the chemotherapeutic benefits
Furthermore, we investigated whether patients in the high- and low-risk groups could benefit from postoperative chemotherapy. A significant interaction effect between radiomics signature and chemotherapy on OS in patients with LGG was found after adjusted age, histologic grade, and IDH1 mutation, which indicated that the benefit from TMZ chemotherapy was superior among patients with high-radiomics signature (primary cohort: HR 0.126 (0.060–0.267), p < 0.001, p = 0.029 for interaction; and validation cohort: HR 0.031 (0.007–0.144), p < 0.001, p = 0.042 for interaction; Table 2) than among those with low-radiomics signature. The corresponding Kaplan–Meier survival curves, which comprehensively compared TMZ chemotherapy with no chemotherapy treatment by risk group, are shown in Fig. 4. The log-rank test results from the subset analysis within each radiomics signature-based risk group revealed that chemotherapy significantly increased OS in the high-radiomics signature group (primary cohort: p < 0.001; and validation cohort: p < 0.001), but had no significant effect in the low-radiomics signature group (primary cohort: p = 0.994; and validation cohort: p = 0.583). Consequently, these results suggested that the radiomics signature could successfully identify patients with LGG who tend to benefit from chemotherapy.
Discussion
In this study, we extracted more comprehensive features than previous studies from FLAIR and CE-T1-w sequence, which enable more deep mining of radiologic images. A radiomics signature based on those features was developed as a novel prognostic factor for LGG patients. We then integrated it with traditional clinicopathological prognostic factors to construct a radiomics nomogram as a novel approach for estimating the overall survival of LGG patients. This nomogram was trained on a primary cohort and sufficiently validated on an entirely new, consistent validation cohort. Furthermore, we demonstrated that the radiomics signature is an effective tool for identifying patients who might benefit from temozolomide treatment.
Radiomics applies advanced computational methods to extract high-throughput features from routinely acquired imaging data, which noninvasively evaluate tumor heterogeneity and provides important insights into tumor microenvironment [17, 30]. To our knowledge, there have been few radiomics-based models for LGG prognosis, with only three studies in this area [24, 25, 31]. However, previous studies are limited by few image modalities, few features, or lacking independent consistent validation cohort. In the current study, we demonstrated the radiomics signature from joint CE-T1-w and FLAIR sequences achieved better prognostic performance than the radiomics signature from either CE-T1-w or FLAIR sequences alone. Compared with the other two similar studies, our signature using radiomics feature of multi-sequence MRI showed higher prognostic efficiency. And more importantly, we validated our radiomics signature in an entire newly independent and consistent cohort, which ensured the robustness of our results.
We then incorporated the radiomics signature with clinicopathologic risk factors into a radiomics nomogram which provided better estimation of overall survival for LGG patients than either the radiomics signature or clinicopathologic nomogram-based clinicopathological risk factors alone. A nomogram is a graphical representation of a statistical model that could provide individualized survival probability estimation. Several previous studies have made effort to develop a prognostic nomogram for LGG patients. The most recent one was developed by Gittleman et al, in which tumor grade, age at diagnosis, Karnofsky Performance Status (KPS), and molecular subtype were included [2]. The molecular subtype in the Gittleman nomogram was defined based on IDH mutation and 1p/19q codeletion status, which is in accordance to the suggestion of the TCGA Research Network and the WHO [1, 32]. Indeed, our nomogram will be more relevant if it includes these molecular biomarkers. However, 1p/19q codeletion was not available in our dataset because it still has not been widely tested for LGG patients in most hospitals in China. In addition, KPS was excluded due to its wide variability of measurement by different observers. We also excluded variables related to radiotherapy or chemotherapy, as their use in clinical practice was less consistent among patients. Hence, our radiomics nomogram included almost all the variables in the Gittleman nomogram exception to postoperative 1p/19 codeletion and KPS, and added radiomics signature and seizure which has traditionally been identified as a prognostic factor. We demonstrated that it was an effective tool for providing individualized survival probabilities for newly diagnosis LGG patients rather than a group estimate based on specific patient-level characteristics and validated its robustness in an independent cohort.
According to the current guidelines [33], almost all the LGG patients who have undertaken surgery were recommended to undertake following adjuvant chemotherapy represented mainly by temozolomide. However, existing studies showed that one group of patients does not benefit from the current guidelines recommending chemotherapy, represented mainly by temozolomide [6, 7, 34]. A large part of patients gave up chemotherapy due to their poor economic condition and uncertainty about their benefit from chemotherapy. Thus, accurately assessing the response to chemotherapy of patients will be helpful for treatment decisions. Although MGMT promoter methylation has important clinical significance in screening potential patients who may benefit from TMZ chemotherapy [35, 36], confirming MGMT status requires a large tissue sample in an invasive way and is always not available before surgery. In the current study, we demonstrated that the radiomics signature is not only a prognostic tool for LGG patients but also could be used as a tool for assessing the response to temozolomide chemotherapy. After discretizing the radiomics signature into two levels with a threshold of - 3.002, we found a significant interaction effect between radiomics signature and chemotherapy on OS in patients with LGG. The temozolomide treatment provided a better survival benefit to LGG patients classified as high-radiomics signature but did not present a significant effect on the prognosis of those classified as low-radiomics signature. This finding indicated that those patients with high-radiomics signature have relatively unfavorable prognosis but a clearer benefit from temozolomide treatment at the same time. For this subgroup of patients, undertaking chemotherapy is more essential. Therefore, further use of the radiomics signature provides a preoperative and low-cost tool for assessing whether LGG patients will benefit from temozolomide chemotherapy and should be useful to clinicians for counseling patients and their families regarding treatment decisions. Interestingly, our study suggests that the radiomics signature we constructed is both a prognostic tool for LGG patients and a tool for assessing the response to temozolomide chemotherapy, which is similar to the findings in a previous study performed in gastric cancer [21, 37].
Potential reasons for this observation are as follows: first, the radiomics features take in important archetypal imaging characteristics associated with MGMT methylation and thus the response to treatment. Previous studies have reported that certain radiological characteristics on MR images such as tumor necrosis and tumor location are associated with MGMT promoter methylation [38, 39]. Second, radiomics features are able to assess tumor heterogeneity and reflect tumor microstructure and cancer biological processes associated with treatment response. For example, tumor hypoxia, one of the major reasons for chemotherapy treatment failure, will result in lower blood supply and thus may reduce the effectiveness of drug transportation [40, 41]. Hypoxic regions usually have lower enhancement than other regions of solid tumors. Previous studies showing high correlation between genomic expression of hypoxia and the enhancing region on Gd-T1w MRI scans [42]. Although the hypotheses about the relationship between radiomics signature and chemotherapy are preliminary and need to be prospectively evaluated in future radiogenomics studies, our present findings provide an effective preoperative tool to aid in making treatment decisions at low cost.
We recognize that there are some limitations in our study, which mainly come from the experimental design of retrospective analysis. Firstly, the primary cohort contains 149 patients with LGG from a single center. Although our nomogram was validated in a completely independent cohort with significant results, it is undeniable that our study requires more patients from multiple centers to validate the reproducibility and applicability of the results. In addition, our radiomics analysis is based on the most common structural MR images such as CE-T1-w and FLAIR, and it is necessary to further add MRI sequences such as dynamic susceptibility contrast, susceptibility-weighted imaging, and diffusion kurtosis imaging in future studies to further improve the predictive performance. Secondly, our follow-up was not long enough to get the endpoint of all patients, which may limit the power of our models. Future work will keep following up all the current censored patients. Finally, the biological process of radiomics features we extracted deserves further exploration. We hypothesized that the relationship between radiomics signature and temozolomide chemotherapy may be closely related to MGMT promoter methylation. However, due to our retrospective study design, MGMT methylation status was not available in our study cohort and further radiogenomics study is required.
In conclusion, a radiomics signature for predicting overall survival of LGG patients has been built. Our results demonstrated radiomics features extracted from multi-modalities baseline MRI scans were able to identify patients of LGG may have poor survival. Additionally, the radiomics signature we constructed has the potential to screen LGG patients who may benefit most from temozolomide chemotherapy. These findings should be useful in improving treatment strategies and clinical management of patients with LGG.
Abbreviations
- CE-T1-w:
-
Contrast-enhanced axial T-1 weighted sequence
- FLAIR:
-
Fluid-attenuated inversion recovery sequence
- LGG:
-
Lower-grade gliomas
- MGMT:
-
Oxygen 6-methylguanine DNA methyltransferase
- OS:
-
Overall survival
- TMZ:
-
Temozolomide
References
The Cancer Genome Atlas Research Network (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498
Gittleman H, Sloan AE, Barnholtz-Sloan JS (2020) An independently validated survival nomogram for lower-grade glioma. Neuro Oncol 22:665–674
Li Y, Liu X, Xu K et al (2018) MRI features can predict EGFR expression in lower grade gliomas: a voxel-based radiomic analysis. Eur Radiol 28:356–362
Bent MJ, Wefel JS, Schiff D et al (2011) Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol 12:583–593
Bent MJ (2014) Practice changing mature results of RTOG study 9802: another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro Oncol 16:1570–1574
Weller M, Bent MJ, Tonn JC et al (2017) European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 18:e315–e329
Stupp R (2019) Drug development for glioma: are we repeating the same mistakes? Lancet Oncol 20:10–12
Cairncross G, Wang M, Shaw E et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31:337
Abrunhosa-Branquinho AN, Bar-Deroma R, Collette S et al (2018) Radiotherapy quality assurance for the RTOG 0834/EORTC 26053-22054/NCIC CTG CEC.1/CATNON intergroup trial “concurrent and adjuvant temozolomide chemotherapy in newly diagnosed non-1p/19q deleted anaplastic glioma”: individual case review analysis. Radiother Oncol 127:292–298
Baumert BG, Hegi ME, Bent MJ et al (2016) Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol 17:1521–1532
Houillier C, Wang X, Kaloshi G et al (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75:1560–1566
Bent MJ, Brandes AA, Taphoorn MJB et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350
Weller M, Tabatabai G, Kastner B et al (2015) MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR trial. Clin Cancer Res 21:2057–2064
Everhard S, Kaloshi G, Crinière E et al (2006) MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol 60:740–743
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Limkin EJ, Sun R, Dercle L et al (2017) Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 28:1191–1206
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Zhang B, Tian J, Dong D et al (2017) Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res 23:4259–4269
McGarry SD, Hurrell SL, Kaczmarowski AL et al (2016) Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography 2:223–228
Jiang Y, Chen C, Xie J et al (2018) Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine 36:171–182
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non—small cell lung cancer. Radiology 281:947–957
Aerts HJWL, Velazquez ER, Leijenaar RTH et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Zhou H, Vallières M, Bai HX et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19:862–870
Liu X, Li Y, Qian Z et al (2018) A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin 20:1070–1077
Wei J, Yang G, Hao X et al (2019) A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol 29:877–888
Shrout PE, Fleiss JL (1979) Intraclass correlations: Uses in assessing rater reliability. Psychol Bull 86:420–428
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10:7252–7259
Gold S, Bloom J, Hale GR et al (2018) Ability of multiparametric magnetic resonance imaging (MRI) to predict prostate tumor heterogeneity on targeted biopsy. J Clin Oncol 36:113–113
Qian Z, Li Y, Sun Z et al (2018) Radiogenomics of lower-grade gliomas: a radiomic signature as a biological surrogate for survival prediction. Aging (Albany NY) 10:2884–2899
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820
National Comprehensive Cancer Network (2020) NCCN Guidelines Version 1.2020 Central Nervous System Cancers. National Comprehensive Cancer Network, Houston. Available via https://www.nccn.org/professionals/physician_gls/pdf/cns.pdf. Accessed 20 Jun 2020
Liu R, Solheim K, Polley M-Y et al (2009) Quality of life in low-grade glioma patients receiving temozolomide. Neuro Oncol 11:59–68
Hegi ME, Diserens AC, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003
Levin N, Lavon I, Zelikovitsh B et al (2006) Progressive low-grade oligodendrogliomas. Cancer 106:1759–1765
Jiang Y, Yuan Q, Lv W et al (2018) Radiomic signature of 18 F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits. Theranostics 8:5915–5928
Eoli M, Menghi F, Bruzzone MG et al (2007) Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res 13:2606–2613
Drabycz S, Roldán G, de Robles P et al (2010) An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 49:1398–1405
Endrich B, Vaupel P (1998) The role of microcirculation in the treatment of malignant tumours: facts and fiction. In: Molls M, Vaupel P (eds) Blood perfusion and microenvironment of human tumors. Springer, Berlin Heidelberg New York
H€ockel M, Knoop C, Schlenger K et al (1993) Intratumoral pO2predicts survival in advanced cancer of the uterine cervix. Radiother Oncol 26:45–50
Diehn M, Nardini C, Wang DS et al (2008) Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 105:5213–5218
Funding
This study has received funding from the Shandong Province Major Science and Technology Innovation Project (No 2018CXGC1210).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Fuzhong Xue.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 439 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Zheng, X., Zhang, J. et al. An MRI-based radiomics signature as a pretreatment noninvasive predictor of overall survival and chemotherapeutic benefits in lower-grade gliomas. Eur Radiol 31, 1785–1794 (2021). https://doi.org/10.1007/s00330-020-07581-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07581-3