Abstract
Eisenia nordenskioldi subsp. nordenskioldi is an earthworm species inhabiting a large part of Asia, from approximately the 40° of north latitude to the shores of the Arctic Ocean. This taxon is known to have very high genetic diversity and contains several cryptic genetic lineages. In this study, we investigated phylogeography of E. n. nordenskioldi populations from the northernmost part of its range, from tundra and north taiga, using mitochondrial cox1 sequences. The studied populations were mostly represented by the 9th lineage of this subspecies. This lineage was estimated to have diverged about a million years ago, and many of its populations probably survived several glacial cycles in situ. Judging by phylogeographic patterns, the ancestral range of the 9th lineage was most probably located in southern Yakutia. From this area, it dispersed to the rest of the current distribution, where specific haplotype groups were detected in certain geographic regions: the Urals, the Kolyma river basin and the shore of the East Siberian Sea, the Taui bay, the Anadyr river valley, and Kamchatka. It is noteworthy that the Verkhoyansk ridge did not act as a barrier to earthworm dispersal, as populations from the Lena, the Yana, and the Indigirka basins were genetically similar but distinct from the populations of the Kolyma river basin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptic diversity in some earthworm species has long been demonstrated using chromosome and molecular genetic analysis (e.g., Omodeo et al. 1952; Graphodatsky et al. 1982; Vsevolodova-Perel 1997; Heethoff et al. 2004; Harper et al. 2006; Lentzsch and Golldack 2006). King et al. (2008) hypothesized that high diversity is the rule in this group: they found several most common cosmopolitan earthworm species to contain two to five highly diverged mitochondrial lineages. This finding was corroborated by plenty of subsequent studies of cosmopolitan earthworm taxa, e.g., Aporrectodea caliginosa species complex (see Pérez-Losada et al. 2009; Fernández et al. 2011, 2012, 2013; Porco et al. 2013), A. icterica (see Torres-Leguizamon et al. 2014), Allolobophora chlorotica (see Dupont et al. 2011; Porco et al. 2013), Lumbricus rubellus (see Donnelly et al. 2013, 2014; Kille et al. 2013; Porco et al. 2013). All these results, however, remain disputable, since DNA sequencing of type specimens is required to be sure about the taxonomic identity of the studied species, as argued by Blakemore et al. (2010). At least in some cases, it was shown that no gene flow occurs in sympatry between these genetic lineages.
Although there are fewer studies on endemic earthworm species, they are also known to possess high genetic diversity, e.g., species of the genus Hormogaster (see Novo et al. 2009, 2010, 2011, 2015), Pontoscolex corethrurus (see Cunha et al. 2014), several Postandrilus species (see Pérez-Losada et al. 2011), as well as some other studies. Some authors suggest that high genetic diversity may be a promising tool for paleogeographic studies (Fernández et al. 2013; Novo et al. 2015).
Although all aforementioned studies were performed on earthworms inhabiting low and moderate latitudes, we hypothesized that species from northern regions might also have deep phylogenetic patterns. During the last glacial maximum (LGM), as well as some previous glaciations, northwestern Europe and northern parts of North America were covered by solid glaciers. The eastern part of Europe and northern Asia was not glaciated because of low precipitation (Hopkins 1976; Glushkova 1984) and contained only local mountain glaciers, i.e., on the Polar and North Ural mountains, the Putorana Plateau, northeastern Taymyr, the Verkhoyansk mountain range, the Suntar-Khayata, and mountains of the Kamchatka Peninsula (Velichko 2002). Although the climate was harsh, certain species were sufficiently adapted to it and could survive its fluctuations in situ.
The only native earthworm in Northern Asia (in addition to several synantropic cosmopolites) is Eisenia nordenskioldi subsp. nordenskioldi (Eisen 1879). This subspecies has the uttermost cold tolerance (Berman and Meshcheryakova 2013; Meshcheryakova and Berman 2014) and is found on the shores of the Arctic ocean and on some Arctic islands, sometimes north of the 70th parallel (Perel 1979; Vsevolodova-Perel and Leyrikh 2014). Its southern distribution spans up to about the 45th parallel; it is also found to the west of the Urals (Blakemore 2016). Within E. n. nordenskioldi, several races with different ploidy, body size, and ecological characteristics are known (Perel 1979; Viktorov 1997; Vsevolodova-Perel and Bulatova 2008). Recent studies demonstrated that this subspecies consists of several highly diverged genetic lineages, which are represented by high-order bootstrap-supported branches on phylogenetic trees (Blakemore 2013; Shekhovtsov et al. 2013, 2014, 2016). These genetic lineages were characterized by high percentage of nucleotide substitutions both within and among them, and were estimated to have diverged several million years ago (Shekhovtsov et al. 2013, 2017).
Our recent study of several E. n. nordenskioldi populations from Northeastern Eurasia suggested that this region harbors a distinct genetic lineage of this subspecies, which we further refer to as lineage 9 (Shekhovtsov et al. 2015). It is only distantly related to other lineages of the species (Shekhovtsov et al. 2015). Different populations of this lineage from high latitudes diverged hundreds of thousand years ago, which implies that they survived several glaciation cycles in situ.
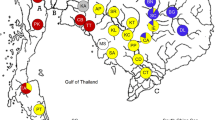
Therefore, we suggested that the phylogeography of E. n. nordenskioldi in northern Asia may shed light on its dispersal history. The aim of this study was to investigate genetic variation of E. n. nordenskioldi populations from the tundra and taiga zones of Asia, and to attempt to reconstruct the history of this subspecies. We collected individuals from 37 populations (Fig. 1; Table 1) and sequenced a fragment of the mitochondrial cox1 gene, which is a high-resolution sequence that is most commonly used in earthworm phylogeography. We focus on the 9th lineage as it was the most numerous in this region and had the highest genetic diversity.
Eisenia nordenskioldi nordenskioldi locations sampled in this study. Circles indicate lineage 9; boxes, lineages 1 and 3. Location numbers refer to Table 1. Dashed lines represent major mountain ranges. The red line indicates the current tundra/taiga boundary
Most studies suggest that major rivers serve as phylogeographic borders in northern Asia both for animals and plants, e.g., the Lena, the Kolyma, and the Indigirka (Hope 2011; Eidesen et al. 2013). For earthworms, however, rivers are not natural barriers but dispersal agents. In our sample, we had several population groups confined to river basins or coastal regions (shores of the East Siberian Sea and the Taui bay) (Fig. 1; Table 1). As a null hypothesis, we suggested that populations within a river basin would be genetically more closely related to each other than to those from other basins. Moreover, we expected these differences to be higher when river basins are separated by geographic barriers (mountain ranges). Several mountain ranges are present in the studied region: the Lena basin is separated from the Yana and the Indigirka by the Verkhoyansk ridge and the Suntar-Khayata. The Anadyr river basin (location 32) is separated from the rest of the territory by the ridges of Chukotka, and the western shore of Kamchatka (location 33), by the Eastern and Middle ridges and the Koryak Upland.
We must note that mountain ridges can only be considered relative barriers, as during the Ice ages sea level receded and large areas of shelf were probably habitable for earthworms. However, we hypothesized that mountain ranges should significantly limit gene flow. Moreover, we intended to use genetic data to delimit “ancient” populations that survived several glaciation cycles and “young” ones.
Materials and methods
Earthworm individuals were collected in 2012–2015 (Table 1; Fig. 1). The studied individuals are stored in the collection of the Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia. Total DNA was extracted from several caudal segments using silica columns (BioSilica, Novosibirsk). Amplification of a fragment of the cox1 gene (658 bp without primers) was performed using primers HCO2198 (5′-TAAAC-TTCAG-GGTGA-CCAAAAAATC-A-30; Folmer et al. 1994) and LCO1490 m (5′-TACTC-AACAA-ATCACAAAGA-TATTG-G-3′; Folmer et al. 1994).
A portion of the studied sample was published earlier in Shekhovtsov et al. (2015); those sequences were not initially submitted to Genbank, and were 7 bp shorter as they were amplified using other primers. For this study, we resequenced that sample. Sequences were deposited in GenBank under the accessions KX601279-KX601656. We also used a single sequence (JX531501) from our previous study (Shekhovtsov et al. 2013).
Molecular diversity indices, pairwise nucleotide and amino acid distances and Fst values were calculated using ARLEQUIN v.3.1 (Excoffier et al. 2005). We also used ARLEQUIN to perform mismatch distribution analysis; the obtained tau parameter of demographic expansion was used to estimate divergence time of the studied sample. Precise generation time for E. n. nordenskioldi is unknown. However, it is certain that it takes longer than a year for an earthworm to reach maturity and leave offspring in cold northern soils, so we adopted the minimal estimate of two years.
Bayesian analysis was performed using MRBAYES v.3.2.0 (Ronquist and Huelsenbeck 2003). The GTR + I+G model was suggested by MRMODELTEST (Nylander 2004). The estimate of cox1 mutation rate of 3.5% per million years was taken from Chang and Chen (2005). Two simultaneous independent analyses were run from different random starting trees using four chains of ‘metropolis coupled Monte Carlo simulations’ for 20,000,000 generations, sampling a tree every 1000 generations; 25% of generations were discarded as burn-in. Molecular clock analysis using fossil calibrations was based on the protocol of Marchán et al. (2017). All sequences were translated to amino acids; identical sequences were discarded from the sample; outgroup sequences were taken from GenBank.
Fossil calibrations were set as lognormal distributions so that 95% distribution density fell within the range from the minimum estimate to minimum estimate ×2. Relaxed molecular clock (ucld.mean) was set as a uniform distribution: initial value, 0.035; upper, 0.1; lower, 0.001. MCMC simulations were performed as described above.
Results
Total genetic diversity
We obtained a total of 379 cox1 sequences of E. n. nordenskioldi individuals from 34 locations. Within this sample, 331 belonged to lineage 9 of this subspecies; 41, to lineage 1; and 7, to lineage 3 (Table 1; Fig. 1). One cox1 haplotype (JX531501) was taken from Shekhovtsov et al. (2013). All sequences were 658 bp in length (without primers) and contained no indels. All three detected genetic lineages of E. n. nordenskioldi form strongly diverged bootstrap-supported clades on the cox1 tree (Fig. 2).
Bayesian phylogenetic tree constructed using the cox1 gene. Three Eisenia nordenskioldi nordenskioldi lineages found in our sample (of the total known nine) are shown. While all genetic lineages are by definition monophyletic, relationships between them have not been resolved yet. The 9th lineage represents the majority of our sample and is shown in detail. The most important clades are shown in color (branch names and colors refer to Fig. 1; if not specified, a branch belongs to the Yakutia group). Although detailed relationships within the 9th lineage could not be reconstructed, one can see that certain haplotype groups from the south of Yakutia are located at the base of the tree, and the rest are scattered throughout the tree, while haplotypes from other geographical regions form compact groups and are monophyletic. The tree is rooted with Eisenia uralensis, a congeneric endemic from the Urals. Numbers near branches indicate Bayesian posterior probabilities
Genetic variation was high both within and among lineages. Within lineage 9 alone, we found 186 haplotypes and 187 nucleotide substitutions, 167 of which were parsimony-informative; no mutations leading to stop-codons or indels were detected. Haplotypes within this sample differed by up to 7.6%, with the average distance of 4%. Given this high variation level within a single lineage, it is reasonable to suspect if these haplotypes represent nonfunctional nuclear pseudogenes of mitochondrial origin (NUMTs). However, we can refute this hypothesis as the number of amino acid substitutions was low: 11 per 8 polymorphic positions, four of which led to changes in amino acid class, and none resulted in gene truncation, much less than one would expect under loss of function. Of the eight polymorphic positions, half were singletons and the other half, parsimony-informative. Most individuals differed by only one or two amino acid substitutions. Thus, we can conclude that despite very high nucleotide substitution rate the amino acid sequence of the cox1 gene is under strong purifying selection, and thus the haplotypes obtained by us are functional mitochondrial sequences.
Lineage 1 was detected in seven locations (Table 1) and was represented by three haplotype groups. Individuals from locations 34 and 35 had cox1 sequences very closely related to haplotypes found in various populations from the south of West Siberia (Shekhovtsov et al. 2013). The haplotype from location 34 was identical to one of the individuals from the Tomsk oblast and that from location 35 differed by only five nucleotide substitutions from another one from the same region.
The most numerous group of lineage 1 haplotypes contained individuals from locations 10, 11, 36, and all specimens except one from location 8 had sequences closely related or identical to those of the populations from the Zhuya river (to the east of the Baikal). Another group that included one specimen from location 8, a single one from location 9, and all from location 37 was not closely related to the rest of the lineage 3 haplotypes, differing from them by about 7% of nucleotide substitutions.
E. n. nordenskioldi lineage 3 was found only in location 37, along with lineage 1 (Table 1). There were only three haplotypes and two polymorphic sites. It was most closely related to haplotypes of this lineage from the southern Far East (Primorye krai, Amur oblast and Jewish autonomous oblast; our sequences, unpublished), differing from them by about 6% or 40 substitutions.
The 9th lineage
The southernmost location from Yakutia (location 17) was invariably recovered at the base of 9th lineage (Fig. 2). Other basal branches were formed by haplotypes from locations 11, 12, 14, and 15, also from southern Yakutia. The rest of the tree lacked phylogenetic resolution; however, several groups formed by haplotypes from certain geographic regions were detected. About a quarter of the total sample of the 9th lineage formed a bootstrap-supported clade (shown as the Y1 subgroup in Figs. 1 and 2) that included specimens from many locations from the Anabar, Lena, Yana, and Indigirka basins. Haplotypes from the Taui group (locations 29–31) were rather closely related and formed a separate branch on the tree. The same holds true for specimens from West Siberia (locations 1–3). Another bootstrap-supported branch included populations from the Kolyma basin and the shore of the East Siberian Sea, except for haplotypes from location 28 that were closely related to some individuals from location 20 from the lower reaches of Indigirka (further referred to as the Pevek-Indigirka group). Haplotypes from the only studied population from the Anadyr river basin were not closely related to any other haplotypes and formed three braches on the tree. Two specimens from Kamchatka (location 33) also formed a separate branch.
We calculated pairwise Fst values among the geographic regions given in Table 2. One can see that most of them correspond to high levels of differentiation (over 0.25). Moderate Fst values were detected between the Lena basin and other rivers within the Yakutia group. In addition, the populations from the Anadyr river basin and Kamchatka also had the lowest Fst distance from the Lena river basin. These results could partly be explained by the differences in sample size. However, the Kolyma river basin sample was close in size to the Lena river basin sample but had high Fst distances in comparison to other groups (Table 2).
Mismatch distribution analysis suggested that the 9th lineage experienced recent demographic expansion (Fig. 3a), and the tau parameter value was high (26.53 (95% CI 20.31–33.18)). Using the tau parameter, we can estimate divergence time for lineage 9 given assumptions about mutation rates and generation times. Under such assumptions, divergence of all haplotypes of the 9th lineages occurred 1,075,000 years ago (95% CI 823,000–1,345,000 years). The analysis performed using the 3.5% divergence rate yielded a similarly old age of this lineage: 1,128,000 years (95% CI 788,000–1,486,000 years). The outgroup fossil calibration method (Marchán et al. 2017) gave much higher estimates 4,039,000 years (95% CI 1,933,000–6,624,000 years). We might guess that these results may be affected by the magnitude of differences between calibration points and the age of estimated groups (hundreds of millions of years vs. hundreds of thousands). Anyway, all estimates suggest that the divergence time of the 9th lineage is much older than LGM events.
In addition, we performed mismatch distribution analysis for locations that include over 15 individuals. Some of those are shown in Fig. 3b–d. Some locations exhibited the pattern characteristic for recent population expansion (Fig. 3b). Others experienced recent bottleneck events (Fig. 3b), while some contained a mixture of several not very closely related founder haplotype groups (Fig. 3d).
Discussion
High genetic diversity within lineage 9
Molecular genetic studies (Heethoff et al. 2004; King et al. 2008; and countless subsequent papers) suggest that very high intraspecific genetic diversity is the rule for earthworms. While certain results (Blakemore 2010) need to be checked by detailed taxonomic investigations, including studies of taxonomic types restoration of valid synonyms, intraspecific genetic diversity in earthworms is still high in most cases (see Blakemore 2013). Within a species, there are usually several genetic lineages that can be defined as reciprocally monophyletic clusters differing by multiple nucleotide substitutions (approximately 10–20% for mtDNA) with significantly lower diversity within lineages. This situation also holds true for E. nordenskioldi (Shekhovtsov et al. 2013, 2015, 2016, 2017), which is unsurprising considering its huge distribution. According to molecular clock estimates, genetic lineages of this species diverged in Pliocene (Shekhovtsov et al. 2013).
In this study, we focused on genetic diversity of one E. n. nordenskioldi lineage, the 9th lineage that is found in Northeastern Eurasia. We demonstrated that one more layer of diversity exists within this lineage, with several of its branches sometimes found in sympatry. The observed pattern of nucleotide substitutions suggests that all this diversity is represented by functional mitochondrial sequences rather than nuclear NUMTs (see Results).
Two factors contribute to this high diversity. The first is the huge distribution of the 9th lineage. Another reason is that this distribution is patchy. Most soils in Northeast Eurasia are unsuitable for E. nordenskioldi because of acidic pH (3.5–4) caused by low carbonate content in parent rock and low temperature that promotes the accumulation of acid peat. Only certain habitats (aspen grooves on southern slopes, edges of steppe patches, mountain tundra meadows, river valleys) have suitable pH values (over 4.2). Most of these patches, however, do not exceed 10 ha and are separated from other patches by long stretches of unsuitable soil. Thus, most local populations of the 9th lineage are small and experience pronounced impact of genetic drift and founder effect.
Although calibration of a molecular clock for earthworms is problematic, we can still use the obtained values as a rough estimate. Our results suggest that the divergence of all sampled haplotypes of the 9th lineage of E. n. nordenskioldi took place about one million years ago. This fact and high genetic structure in all parts of its distribution suggests that many local populations of lineage 9 could survive several glacial cycles.
We can suggest that southern Yakutia is the center of genetic diversity of the 9th lineage and most probably its ancestral area. First, populations from southern Yakutia occupy the basal position on the trees (Fig. 2), with the southernmost location (location. 17) being the most basal. Second, genetic diversity of other haplotypes from the Lena basin was much higher compared to those from other regions (we should note that this could be partly explained by higher number of sampled individuals and populations). Southern Yakutia obviously had more stable climatic conditions than areas further to the north, so we could hypothesize that local populations were less affected by climatic perturbations. However, more detailed sampling of this area is needed to make strong conclusions.
Populations from other regions formed several branches on phylogenetic trees. Unsurprisingly, three populations of the 9th lineage (locations 1–3) that were found over 2000 km from other locations formed a separate branch on the tree (the Ural branch in Figs. 1 and 2). In the easternmost part of the distribution, distinct groups were isolated from the rest of the sample by mountain ridges: specimens from the coast of the Taui bay (locations 29–31) stand separate on the tree, as well as those from Kamchatka (location 33). Earthworms from the single population from the Anadyr river basin were quite diverse and formed as many as three separate branches that were not closely related to haplotypes from other regions. In all these cases, geographic isolation could explain genetic differences.
In contrast, for Yakutia and western Chukotka the observed genetic patterns do not correspond to geographic features. Populations from the Anabar, Olenek, Lena, Yana, and Indigirka river basins were genetically close to each other, although not forming a monophyletic clade. Certain haplotypes from, e.g., the basins of the Anabar (locations 4 and 5) and Indigirka (location 19) differed only by a few substitutions, suggesting very close genetic relationship. There are no geographic barriers between the Lena valley and the rivers of the North Siberian Lowland (Olenek and Anabar), so genetic similarity between E. n. nordenskioldi populations is not unexpected. However, the Yana and the Indigirka are separated from the Lena by the Verkhoyansk and the Suntar-Khayata ridges that are formidable barriers to earthworm dispersal. Contacts between these river systems could have been possible during glaciations along the Laptev Sea coast when ocean level was lower; however, we would still expect drastically reduced gene flow.
At the same time, populations from the Indigirka and Kolyma basins were not closely related. The latter (locations 22–23) formed a clade with populations from the coast of the Northeastern Sea (locations 24–27). It is somewhat unanticipated as there are no geographic barriers between the Kolyma and the Indigirka, and these rivers are geographically closer to each other than the Indigirka and the Yana.
We should note that in this region one more group of haplotypes was detected (Fig. 1) that included several specimens from location 20 in upper reaches of Indigirka and location 28 from the Chaun bay that are located over 500 km apart. This group was not closely related to any other haplotypes from that region. On the whole, such phylogeographic patterns that are not totally dependent on geography may reflect complex history of E. n. nordenskioldi and certain dispersal or vicariance events that occurred throughout several glaciation cycles.
It is noteworthy that genetic differences appear to be independent of community type: from Fig. 1, it is obvious that there are no genetic differences between populations from tundra and taiga. It indicates that, while soil physicochemical properties are important for E. n. nordenskioldi, they can adapt to different community types.
Lineages 1 and 3
We found three distinct genetic lineages of E. n. nordenskioldi in our sample. While lineage 9 was widespread throughout the whole region studied by us, lineages 1 and 3 had smaller distributions. In the north of West Siberia, lineage 1 was found alongside lineage 9 (Fig. 1). The fact that local haplotypes are closely related or identical to those found in the south of West Siberia clearly indicates that the northern populations of lineage 1 are recent invaders there. The most probable dispersal route is the Ob river, as rivers are pathways of earthworm migrations.
Lineage 1 was also found in six locations in the Lena basin, in one instance alongside lineage 3 (Fig. 1; Table 1). It is noteworthy that lineage 1 is evidently widespread in the south, in upper reaches of Lena and Vilyui, but to the north it was detected only in location 8 (Fig. 1; Table 1). While populations of lineage 1 from the north of West Siberia originated in the south of that region, populations from Yakutia have Far Eastern affinities; denser sampling is needed to clarify this issue.
History of Eisenia n. nordenskioldi in northern biomes
During Pleistocene glaciations, northern parts of Europe and North America were repeatedly covered by ice sheets, which led to complete erasing of all local biota (Hewitt 2000). While some animals could retreat southwards, this was obviously not the case for earthworms. At the same time, only local mountain glaciers existed in northern Asia due to low precipitation. Climatic oscillations still had a high impact on flora and fauna, resulting in transformation of communities from tundra and boreal forest to tundra steppe.
The distribution of lineage 9 once encompassed the whole north of Northern Asia. However, glaciations caused its contraction in West Siberia. When glaciers retreated, earthworm populations began moving northwards from refugia. This is clearly reflected in genetic patterns of locations 1–3: haplotypes of the Ural group are closely related, which is in stark contrast to patterns observed in the eastern part of the distribution. Along with lineage 9, this region was also colonized by lineage 1 of E. n. nordenskioldi (see above).
Deep phylogeographic differences observed among the populations in the eastern part of the distribution suggest that climatic oscillations and the associated community changes were not catastrophic for earthworms. In the northeast of Northern Asia, the 9th lineage seems to be the only genetic lineage of E. n. nordenskioldi. Only in the upper and middle reaches of the Lena and its tributaries we observed lineages 1 and 3 of E. n. nordenskioldi. We also failed to find Dendrobaena octaedra, which is widespread in the north of Europe.
As stated above, ancestral distribution of the 9th lineage was most probably located in southern Yakutia. Colonization of West Siberia and various regions of Northeastern Siberia occurred independently, which is indicated by the lack of close genetic relations among these groups, especially between the Kolyma and the Taui bay and populations from Anadyr and Kamchatka, which we would expect to be closely related if dispersal proceeded stepwise as could be anticipated based on geographic distribution (Fig. 1). Current differences between the Kolyma and Indigirka groups, as well as the presence of an additional group in that region are most probably the result of ancient allopatry.
Ecological characteristics of the 9th lineage of E. n. nordenskioldi imply low population size and, consequently, frequent local extinction and recolonization events. Mismatch distribution analysis of individual populations (Fig. 3b–d) suggests that different populations are indeed on various stages of these processes. Some, e.g., location 25 (Fig. 3b) and location 31 (not shown), have a pattern characteristic for a recent expansion of an initially small and genetically uniform populations. Others, e.g., location 6 (Fig. 3c) and location 22 (not shown), have strong deviations from the model mismatch distribution, indicating bottleneck events. Several populations have several peaks on mismatch distribution profiles [e.g., locations 32 (Fig. 3d) and 14 (not shown)] implying several diverged founder groups.
Conclusions
Earthworms seem rather delicate organisms, vulnerable to cold and dry. However, genetic evidence proved that the representatives of the 9th lineage of E. n. nordenskioldi managed to survive several climatic oscillations and the associated turnover of communities in the northern latitudes of Asia. Although local populations appear to be on different states of evolution, this lineage as a whole appears to be very viable in this extreme and constantly changing environment. Genetic diversity of this lineage is very high and can be used in paleogeographic reconstructions in particular northern regions. It is also reasonable to suggest that other representatives of soil fauna may also possess similarly deep phylogeographic patterns and can thus be promising object for molecular genetic studies.
References
Berman DI, Meshcheryakova EN (2013) Ranges and cold hardiness of two earthworm subspecies (Eisenia nordenskioldi, Lumbricidae, Oligochaeta). Biol Bull 40:719–727
Blakemore RJ (2013) Earthworms newly from Mongolia (Oligochaeta, Lumbricidae, Eisenia). ZooKeys 285:1–21
Blakemore RJ (2016) Cosmopolitan earthworms VI: an eco-taxonomic guide to the peregrine species of the world. VermEcology, Yokohama
Blakemore RJ, Kupriyanova EK, Grygier MJ (2010) Neotypification of Drawida hattamimizu Hatai, 1930 (Annelida, Oligochaeta, Megadrili, Moniligastridae) as a model linking mtDNA (COI) sequences to an earthworm type, with a response to the ‘Can of Worms’ theory of cryptic species. ZooKeys 41:1–29
Chang C-H, Chen J-H (2005) Taxonomic status and intraspecific phylogeography of two sibling species of Metaphire (Oligochaeta: Megascolecidae) in Taiwan. Pedobiologia 49(6):591–600
Cunha L, Montiel R, Novo M, Orozco-terWengel P, Rodrigues A, Morgan AJ, Kille P (2014) Living on a volcano’s edge: genetic isolation of an extremophile terrestrial metazoan. Heredity 112(2):132–142
Donnelly RK, Harper GL, Morgan J, Orozco-Terwengel P, Pinto-Juma GA, Bruford MW (2013) Nuclear DNA recapitulates the cryptic mitochondrial lineages of Lumbricus rubellus and suggests the existence of cryptic species in an ecotoxological soil sentinel. Biol J Linn Soc Lond 110(4):780–795
Donnelly RK, Harper GL, Morgan J, Pinto-Juma GA, Bruford MW (2014) Mitochondrial DNA and morphological variation in the sentinel earthworm species Lumbricus rubellus. Eur J Soil Biol 64:23–29
Dupont L, Lazrek F, Porco D, King RA, Rougerie R, Symondson WOC, Livet A, Richard B, Decaëns T, Butt KR, Mathieu J (2011) New insight into the genetic structure of the Allolobophora chlorotica aggregate in Europe using microsatellite and mitochondrial data. Pedobiologia 54(4):217–224
Eidesen PB, Ehrich D, Bakkestuen V, Alsos IG, Gilg O, Taberlet P, Brochmann C (2013) Genetic roadmap of the Arctic: plant dispersal highways, traffic barriers and capitals of diversity. New Phytol 200(3):898–910
Eisen G (1879) On the Oligochaeta collected during the Swedish expeditions to the arctic regions in the years 1870, 1875 and 1876. Kongl Sv Vetensk Akad Handl 15(7):1–49
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fernández R, Almodóvar A, Novo M, Gutiérrez M, Díaz Cosín DJ (2011) A vagrant clone in a peregrine species: phylogeography, high clonal diversity and geographical distribution in the earthworm Aporrectodea trapezoides (Dugès, 1828). Soil Biol Biochem 43(10):2085–2093
Fernández R, Almodóvar A, Novo M, Simancas B, Díaz Cosín DJ (2012) Adding complexity to the complex: new insights into the phylogeny, diversification and origin of parthenogenesis in the Aporrectodea caliginosa species complex (Oligochaeta, Lumbricidae). Mol Phylogenet Evol 64(2):368–379
Fernández R, Almodóvar A, Novo M, Gutiérrez M, Díaz Cosín DJ (2013) Earthworms, good indicators for palaeogeographical studies? Testing the genetic structure and demographic history in the peregrine earthworm Aporrectodea trapezoides (Dugès, 1828) in southern Europe. Soil Biol Biochem 58:127–135
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Glushkova OY (1984) Morphology and paleogeography of Late Pleistocene glaciations in the Northeast of the USSR. In: Bespaly VG (ed) Pleistocene glaciations of East Asia. SVKNII FESC AS USSR, Magadan, pp 28–42 (In Russian)
Graphodatsky AS, Perel TS, Radzhabli SI (1982) Chromosome sets of two forms of Eisenia nordenskioldi (Eisen) (Lumbricidae, Oligochaeta). Doklady AN USSR 262:1514–1516 (In Russian)
Harper GL, Cesarini S, Casey SP, Morgan AJ, Kille P, Bruford MW (2006) Microsatellite markers for the earthworm Lumbricus rubellus. Mol Ecol Notes 6(2):325–327
Heethoff M, Etzold K, Scheu S (2004) Mitochondrial COII sequences indicate that the parthenogenetic earthworm Octolasion tyrtaeum (Savigny 1826) constitutes of two lineages differing in body size and genotype. Pedobiologia 48(1):9–13
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913
Hope AG (2011) Mammalian diversification across the Holarctic: spatiotemporal evolution in response to environmental change. University of New Mexico, Albuquerque
Hopkins DM (1976) Sea level in Beringia in the last 250 000 years. In: Kontrimavichus VL (ed) Beringia in the Caenozoic. FESC AS USSR, Vladivostok, pp 9–27 (In Russian)
Kille P, Andre J, Anderson C, Ang HN, Bruford MW, Bundy JG, Donnely R, Hodson ME, Juma G, Lahive E, Morgan J, Stürzenbaum SR, Spurgeon DJ (2013) DNA sequence variation and methylation in an arsenic tolerant earthworm population. Soil Biol Biochem 57:524–532
King RA, Tibble AL, Symondson WOC (2008) Opening a can of worms: unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol Ecol 17(21):4684–4698
Lentzsch P, Golldack J (2006) Genetic diversity of Aporrectodea caliginosa from agricultural sites in Northeast Brandenburg Germany. Pedobiologia 50(4):369–376
Marchán DF, Fernández R, de Sosa I, Díaz Cosín DJ, Novo M (2017) Pinpointing cryptic borders: fine-scale phylogeography and genetic landscape analysis of the Hormogaster elisae complex (Oligochaeta, Hormogastridae). Mol Phylogenet Evol 112:185–193
Meshcheryakova EN, Berman DI (2014) The cold hardiness and geographic distribution of earthworms (Oligochaeta, Lumbricidae, Moniligastridae). Zool Zh 93:53–64 (In Russian)
Novo M, Almodóvar A, Díaz Cosín DJ (2009) High genetic divergence of hormogastrid earthworms (Annelida, Oligochaeta) in the central Iberian Peninsula: evolutionary and demographic implications. Zool Scripta 38(5):537–552
Novo M, Almodóvar A, Fernández R, Trigo D, Díaz Cosín DJ (2010) Cryptic speciation of hormogastrid earthworms revealed by mitochondrial and nuclear data. Mol Phylogenet Evol 56(1):507–512
Novo M, Almodóvar A, Fernández R, Giribet G, Díaz Cosín DJ (2011) Understanding the biogeography of a group of earthworms in the Mediterranean basin—The phylogenetic puzzle of Hormogastridae (Clitellata: oligochaeta). Mol Phylogenet Evol 61(1):125–135
Novo M, Fernández R, Fernández Marchán D, Trigo D, Díaz Cosín DJ, Giribet G (2015) Unearthing the historical biogeography of Mediterranean earthworms (Annelida: hormogastridae). J Biogeogr 42(4):751–762
Nylander JAA (2004) MrModeltest v2. Evol. Biol. Centre, Uppsala Univ
Omodeo P (1952) Cariologia dei Lumbricidae. Caryologia 4(2):173–275
Perel TS (1979) Range and regularities in the distribution of earthworms of the USSR fauna. Nauka, Moscow (In Russian)
Pérez-Losada M, Ricoy M, Marshall JC, Domínguez J (2009) Phylogenetic assessment of the earthworm Aporrectodea caliginosa species complex (Oligochaeta: lumbricidae) based on mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 52(2):293–302
Pérez-Losada M, Breinholt JW, Porto PG, Aira M, Domínguez J (2011) An earthworm riddle: systematics and phylogeography of the Spanish lumbricid Postandrilus. PLoS ONE 6(11):e28153
Porco D, Decaens T, Deharveng L, James SW, Skarzynski D, Erséus C, Butt KR, Richard B, Hebert PDN (2013) Biological Invasions in soil: DNA barcoding as a monitoring tool in a multiple taxa survey targeting European earthworms and springtails in North America. Biol Invasions 15(4):899–910
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Shekhovtsov SV, Golovanova EV, Peltek SE (2013) Cryptic diversity within the Nordenskiold’s earthworm, Eisenia nordenskioldi subsp. nordenskioldi (Lumbricidae, Annelida). Eur J Soil Biol 58:13–18
Shekhovtsov SV, Golovanova EV, Peltek SE (2014) Invasive lumbricid earthworms of Kamchatka (Oligochaeta). Zool Stud 53(1):52
Shekhovtsov SV, Berman DI, Peltek SE (2015) Phylogeography of the earthworm Eisenia nordenskioldi nordenskioldi (Lumbricidae, Oligochaeta) in Northeastern Eurasia. Dokl Biol Sci 461(1):1–4
Shekhovtsov SV, Golovanova EV, Peltek SE (2016) Mitochondrial DNA variation in Eisenia n. nordenskioldi (Lumbricidae) in Europe and Southern Urals. Mitochondrial DNA Part A 27(6):4643–4645
Shekhovtsov SV, Berman DI, Golovanova EV, Peltek SE (2017) Genetic diversity of the earthworm Eisenia nordenskioldi (Lumbricidae. Vavilov J Genet Breeding, Annelida). doi:10.18699/VJ17.24-o (In Russian)
Torres-Leguizamon M, Mathieu J, Decaëns T, Dupont L (2014) Genetic structure of earthworm populations at a regional scale: inferences from mitochondrial and microsatellite molecular markers in Aporrectodea icterica (Savigny 1826). PLoS ONE 9(7):e101597
Velichko AA (2002) Dynamics of landscape components and internal seas of North Eurasia in the last 130 000 years. GEOS, Moscow (In Russian)
Viktorov AG (1997) Diversity of polyploid races in the family Lumbricidae. Soil Biol Biochem 29(3):217–221
Vsevolodova-Perel TS (1997) The earthworms of the fauna of Russia. Nauka, Moscow (In Russian)
Vsevolodova-Perel TS, Bulatova NS (2008) Polyploid races of earthworms (Lumbricidae, Oligochaeta) in the East European plain and Siberia. Biol Bull 35(4):385–388
Vsevolodova-Perel TS, Leirikh AN (2014) Distribution and ecology of the earthworm Eisenia nordenskioldi pallida (Oligochaeta, Lumbricidae) dominant in Southern Siberia and the Russian Far East. Zool Zh 93:45–52 (In Russian)
Acknowledgements
We thank R.J. Blakemore, B.J. Adams, and an anonymous reviewer for helpful comments. We are grateful to N.E. Bazarova for technical assistance and to A.V. Alfimov, A.B. Babenko, A.V. Barkalov, P.V. Bubyakin, A.P. Burnasheva, N.E. Dokuchaev, N.N. Dvornik, A.Kh. Gabysheva, E.V. Golovanova, E.V. Kirillin, N.S. Kirillin, O.A. Khruleva, A.V. Kondratyev, S.V. Konyaev, A.V. Losorov, O.L. Makarova, E.I. Meshcheryakova, I.M. Okhlopkov, A.A. Popov, K.V. Regel, L.B. Rybalov, R.R. Safronov, E.V. Shemyakin, M.M. Sidorov, A.V. Shestakov, M.M. Sledov, Yu.A. Sleptsov, A.V. Tanasevich, and E.N. Zykov for the earthworm specimens kindly provided to us. This study was supported by the Russian Fund for Basic Research under Grants 14-04-01121_a, 13–04–00156_a, and 16-04-00082_a; by the President of the Russian Federation under Grant MK-6685.2015.4; and by the State Assignment no. 0324-2016-0003.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shekhovtsov, S.V., Berman, D.I., Bulakhova, N.A. et al. Phylogeography of Eisenia nordenskioldi nordenskioldi (Lumbricidae, Oligochaeta) from the north of Asia. Polar Biol 41, 237–247 (2018). https://doi.org/10.1007/s00300-017-2184-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-017-2184-2