Abstract
The microbial diversity of faecal communities co-existing with mega fauna is not well understood even though these faecal communities are critical for health and development. Additionally, the transfer of microbial taxa among host animals is little studied. Here, we used 16S sequences obtained from clone libraries to characterise the faecal microbiota of Weddell seals breeding in McMurdo Sound and at White Island, Antarctica. Faecal bacterial communities were dominated by four phyla; Actinobacteria (20 %), Bacteroidetes (13 %), Firmicutes (23 %), and Proteobacteria (13 %). We also used automated ribosomal intergenic spacer analysis to examine the dispersal of bacteria between populations of Weddell seals breeding at White Island and in McMurdo Sound. The Weddell seals at White Island are isolated by the Ross Ice Shelf from the larger population of Weddell seals breeding in McMurdo Sound. We found that the faecal bacteria communities of the seals at White Island had lower diversity and that the community composition was significantly different compared with the seals in the McMurdo Sound area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial ecology drives the earth’s ecology (Curtis et al. 2002), yet the diversity and biogeography of bacterial communities are often poorly understood. The predominant paradigm for microbial diversity has been that “everything is everywhere” and that the environment selects (Cho and Tiedje 2000). However, studies of bacteria in symbiotic relationships with eukaryotes and bacteria in soils are revealing that endemism exists within the microbial world, bacterial diversification is ongoing (Fulthorpe et al. 1998; Cho and Tiedje 2000; Funk et al. 2000), and that bacterial communities are isolated by distance. One major challenge associated with identifying bacterial diversification is untangling the effects of isolation from the potentially confounding effects of environmental selection (Whitaker 2006).
The enteric bacteria of wild animals with their more constant internal environment offer one potential solution for disentangling environmental selection from isolation (for example, Ley et al. 2005, 2006, 2008; Li et al. 2008; Banks et al. 2009; Turnbaugh et al. 2009; Zhang et al. 2010) and Weddell seals in McMurdo Sound may be an ideal example for studying the distribution of faecal bacteria. The Weddell seal populations in McMurdo Sound have been monitored for several decades with extensive tagging of adults and pups each year providing extensive demographic data (Hastings and Testa 1998). Weddell seal pups are born on the sea ice from mid-October to November in approximately 11 colonies located along perennial cracks in ice resulting from tidal action and glacial movements (Hastings and Testa 1998). Seal pups in McMurdo Sound wean after approximately 6 weeks and disperse to the ice covering the surface of McMurdo Sound (Stirling 1969). One particularly notable colony of Weddell seals breeds on the Ross Ice Shelf (floating glacier) around White Island. The approximately 24 seals in this colony have been isolated from the open sea and other seals for at least 50 years as a consequence of the expansion of the Ross Ice Shelf. However, a small number of openings in the ice shelf are maintained by glacial and tidal movements around White Island allowing the seals access to the sea directly under the ice (Stirling 1972). Because of the thickness and extent of the glacial ice away from the island, there is no possibility of seals swimming to the open sea approximately 18 km away. In essence, these seals are “trapped” near their breathing holes (Heine 1960). Despite this restricted existence, the seals at White Island are significantly larger than their counterparts in McMurdo Sound. However, only four to nine pups are born each year and pup mortality is high (Stirling 1972; Gelatt 2001).
Isolated populations are of considerable interest to parasitologists in that the small size of host populations, and the patchy distribution of parasites, usually means that the hosts have fewer commensal taxa than their source host populations (Paterson et al. 1999, 2003). Commensal populations in small, isolated host populations are also thought to have a greater risk of extirpation than taxa in larger populations because of stochastic effects (Paterson et al. 2003). The combination of a limited distribution of parasites and higher risk of extinction results in isolated host populations having a lower diversity of parasites than larger interconnected host populations (Rozsa 1993; Clayton et al. 2003). Conversely, larger populations are thought to have better dispersal power (MacArthur and Wilson 1967) and to parasitise more host species; for example, Banks and Paterson (2005) found that multi-host ectoparasites (chewing lice) were more abundant than host-specific ectoparasites. Dispersal limitation has also been shown for bacterial species. For example, analysis of Salmonella enterica strains isolated from marine and terrestrial Galápagos iguanas found that dispersal events were limited and this limitation gave rise to distinct S. enterica serovar assemblages at each site (Lankau et al. 2012). Likewise, Linnenbrink et al. (2013) showed that geographical distance among mice was the most significant factor contributing to mouse gut microbial diversity.
The enormous population sizes of microbes, in comparison with multicellular organisms, are thought to result in a ubiquitous distribution for bacteria (Fenchel 1993) which in turn reduces the probability of distinct communities developing (Finlay and Clarke 1999; Linnenbrink et al. 2013). Here, we used genetic analyses (clone libraries and ARISA) to test whether faecal bacterial communities follow the same patterns of distribution as those followed by multicellular parasites. Specifically, we tested the hypothesis that White Island seals would have faecal communities that were different to those in the wider McMurdo Sound. We predicted that if the distribution of bacteria is restricted by host isolation, the faecal bacterial communities of seals at White Island would differ from the communities of seals in McMurdo Sound, and that the White Island seals would have fewer bacterial taxa than their conspecifics in McMurdo Sound.
Methods
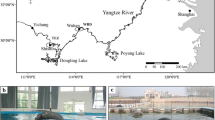
Adult Weddell seals were restrained using the head bag technique of Stirling (1966b); pups were restrained by simply holding them. Faecal samples were collected in November 2006 from seals at Marble Point (77.42°S 163.80°E), Strand Moraines (77.76°S 164.53°E), Tent Island (77.69°S 166.37°E) and White Island (78.02°S 167.40°E) (Fig. 1) by inserting sterile rayon swabs (LP Italiana Spa, Milan, Italy) directly into the anus. Swabs were frozen immediately in the field in liquid nitrogen and then stored at −80 °C on return to the laboratory. Genomic DNA was extracted from the swabs using the Power Soil kit (Mo Bio, West Carlsbad, California, USA) following the manufacturer’s instructions. DNA was re-suspended in 50 μL of elution buffer. Negative DNA extractions using swabs dipped in HPLC grade water were conducted with each batch of extractions.
Clone library construction and analysis
We amplified a portion of the 16S rRNA gene using the primers ITSReub (Cardinale et al. 2004) and EubB (Lane 1991) from two adult females and two pups at White Island, one pup at Marble Point, one pup at Strand Moraines and one adult female at Tent Island (a total of seven seals). Each reaction consisted of 2.5 µL of 10× buffer with MgCl2 (15 mmol L−1) and dNTP (8 mmol L−1), 0.625 µL of each primer (10 mmol L−1), 1.5 units of Taq (Roche), 15.75 µL of water. PCR conditions were 94 °C for 10 min, 25 cycles of 94 °C for 40 s, 55 °C for 40 s and 72 °C for 3 min, with a final step at 72 °C for 7 min. Negative controls using HPLC water as a template were run with each batch of samples.
PCR products were gel-purified using gel extraction kits (Qiagen) and cloned using TOPO TA (Invitrogen, Carlsbad) cloning kits. Plasmids were cultured on media containing kanamycin to identify those containing an insert, and then, the plasmid was extracted using Quickclean II plasmid miniprep kits (Catalogue number L00420, Genscript Piscataway, USA). Plasmid inserts were sequenced using the primer M13F supplied with the cloning kit on an Applied Biosystems 3130xl genetic analyzer. Sequences were checked for chimeras using DECIPHER (Wright et al. 2012). Operational taxonomic units were assigned using MOTHUR (Schloss et al. 2009) using a minimum of 97 % or 99 % sequence similarity to classify clones to a phylotype. Clones were assigned to taxonomic groups using the sequence match function in the Ribosomal Database Project (RDP) (Cole et al. 2007; Wang et al. 2007) and BLASTN searches in GenBank (http://www.ncbi.nlm.nih.gov/). Sequences from the clones were assembled in BioEdit 7.0.5.3 (Hall 1999) and aligned in ClustalX 2.1 (Thompson et al. 1997). MOTHUR was also used to calculate Shannon indices (H′) (Magurran 1988) to assess community diversity, and the Chao-1 richness estimate (Chao 1987) to estimate the richness of the faecal bacterial community. Shannon diversity indices (H) combine estimates of richness (OTUs in this study) and evenness (relative abundance of each OTU), and trend towards zero as communities become dominated by a single OTU.
Automated ribosomal intergenic spacer analysis (ARISA)
We also used automated ribosomal intergenic spacer analysis (ARISA) (Fisher and Triplett 1999) to characterise faecal bacterial communities from 16 seals (two adult females and two pups from White Island, Marble Point, Strand Moraines and Tent Island). Polymerase chain reactions were conducted on the DNA extracted from the swabs using the primers ITSF and hex-labelled ITSReub (Cardinale et al. 2004). Each reaction consisted of 5 μL of 10× buffer with MgCl2 (15 mmol L−1) and dNTP (8 mmol L−1), 0.625 μL of each primer (10 μmol L−1), 1.5 units of Taq (Roche), 10 ng of genomic DNA and water to make a final reaction volume of 25 μL. Reaction temperatures and times were those used by Cardinale et al. (2004). Negative controls using HPLC water as a template were run with each batch of samples. PCR products were purified with Quick Clean purification kits (GenScript Corporation). Amplicon lengths were resolved on a Megabace 500 series capillary sequencer (Amersham Pharmacia, Sunnyvale).
Fragment length was assigned to bins of three nucleotides, as replicate analyses of artificial communities suggested that this was the level of precision of ARISA (Wood et al. 2008), and peaks that were less than three standard deviations above the baseline ‘noise’ in the output from the sequencer were removed from the analysis using the software T-RFLP Stats (Abdo et al. 2006). Peaks were transformed to presence absence data using the Primer6 software (Plymouth Routines in Multivariate Ecological Research, Primer-E Ltd, Plymouth, Clarke and Warwick 2001). Bray Curtis similarity indices were calculated from the transformed data using Primer6. Descriptive statistics and t tests were conducted using Systat 9.01 (SPSS 1998). Comparisons of the White Island faecal bacterial communities with the McMurdo Sound communities were carried out using the ANOSIM (analysis of similarity) test in Primer 6, Version 6.1.13(Clarke and Warwick 2001), with a null hypothesis that the communities were not different.
Results
Community composition
We obtained sequences for 274 clones from the seven seal faecal samples. MOTHUR assigned the sequences to 75 OTUs based on at least 99 % sequence similarity (Genbank accession numbers KM100368–KM100441) and 48 OTUs at 97 % sequence similarity. The average similarity between the 75 OTUs and the sequences in Genbank was 97 % indicating that many of the seal OTUs (Online Resource 1) have not been previously sequenced. On average each seal had 13.1 clone OTUs (range 8–19) and each clone was found in an average of 1.16 seals (range 1–3 seals) Of the 75 OTUs, 57 % were unique to a single host.
Where the bacteria could be classified to phylum by RDP, Firmicutes and Actinobacteria made up the largest proportion of the clone libraries, with Bacteroidetes and Proteobacteria making up smaller proportions of the communities. These four phyla made up 68 % of the clone libraries from the seal faecal samples (Fig. 2).
The Shannon diversity index for all seven clone libraries combined at 99 % sequence similarity was 3.69 (95 % confidence interval 3.53–3.84). Shannon diversity indices for individual libraries ranged from 1.19 to 2.74. Chao-1 estimated that there were 175 OTUs (at 99 % similarity, 95 % confidence interval 118–308) for the seven clone libraries combined. Chao-1 estimates of diversity for clone libraries obtained from individual seals ranged from 17.3 to 70 OTUs. At 97 % OTU similarity, Chao-1 estimated a species richness of 98 (95 % confidence interval 65–189).
Mother pup comparisons
The faecal bacterial communities of seal mothers, as measured by ARISA, did not differ significantly from those of the pups (ANOSIM, Global R = −0.160, P two tailed = 99.5). Weddell seal mothers had slightly more ARISA OTUs than the pups (Mean number of ARISA OTUs, mothers = 23.1, pups = 19.9). However, the difference was not statistically significant (Student’s t test, t = 1.14, P two tailed = 0.27).
Diversity of White Island seal microbiota communities compared with McMurdo Sound seal communities
On average each seal had 21.5 ARISA OTUs. Most ARISA OTUs were identified from a single faecal sample although one OTU was found from 14 of the 16 faecal samples (Fig. 3). White Island seals had significantly fewer ARISA OTUs than McMurdo Sound seals (White Island mean OTUs = 17.75, McMurdo Sound mean OTUs = 22.75, two sample t test, t = 2.414, df = 13.6, P two tailed = 0.03, Fig. 4).
The faecal bacterial communities of White Island Weddell seals were also significantly different from the bacterial communities of seals in McMurdo Sound (ANOSIM Global R = 0.431, P two tailed < 0.01, Fig. 5).
Discussion
Community diversity
Weddell seal faecal microbiota appears to have slightly higher levels of diversity to that found for other seal species. We detected 48 OTUs (at 97 % 16S sequence similarity) in Weddell seal faecal bacterial communities, slightly more than detected from 16S clone libraries generated for hooded seals, Cystophora cristata, (28 OTUs), harbour seals, Phoca vitulina, (12–39 OTUs), and grey seals, Halichoerus grypus, (20–27 OTUs) based on 97 % similarity for 16S sequences (Kristiansen 2007; Glad et al. 2010). Chao-1 calculations from the clone libraries estimated the true species richness as 98 OTUs at 97 % sequence similarity. Our estimate of OTU richness is higher than the richness estimated for hooded seals (43 OTUs), harbour seals (15 OTUs), and grey seals (22 OTUs) estimated by Glad et al. (2010) and for grey seals (39 OTUs) estimated by Kristiansen (2007) using Chao 1 but comparable to Kristiansen’s estimate for harbour seals (97 OTUs).
Shannon diversity indices (a measure of species abundance and evenness of a community) from Weddell seals were also similar to the indices reported for other seal species. Shannon diversity indices ranged from 1.91 to 2.74 for individual Weddell seal faecal microbial communities characterised by cloning. Overall the Shannon index for all seven communities combined was 3.69 which is slightly higher than that calculated by Kristiansen (2007) for harbour seals (H = 3.25) and markedly higher than that calculated for grey seals (H = 2.91). Glad et al. (2010) calculated a Shannon diversity index of 2.64 for hooded seals, 1.76 for harbour seals and 2.59 for grey seals off the coast of northern Norway.
Community composition
The dominance of the faecal flora by the four bacterial phyla, Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria is similar to the results that Smith et al. (2013) obtained from Australian fur seals, Arctocephalus pusillus doriferus, collected north of the Ross Sea. Clone libraries generated from the faeces of wild southern elephant seals and leopard seals from western Antarctica also showed that the communities were dominated by Firmicutes, Bacteroidetes, and Proteobacteria but Actinobacteria were a minor component of the communities of these species (Nelson 2012). Glad et al. (2010) found that colon bacterial communities from hooded, harbour, and grey seals were dominated by Bacteroidetes and Firmicutes; however, Proteobacteria and Fusobacteria were present only in hooded seal bacterial communities.
The average distance between each clone obtained from the Weddell seals and its closest match in GenBank was 97 % suggesting that many of the OTUs had not been sequenced before. However, this average similarity was higher than that seen for OTUs identified from clone libraries obtained with the same primers from faecal samples obtained from Adelie penguins in the same region (Banks et al. 2009) possibly because more faecal communities have been characterised for mammals. Operational taxonomic units more than 97 % similar are generally considered to be similar to metazoan species (Hagström et al. 2000) although OTUs that are 99 % similar for 16S may show considerable phenotypic differences (Ward 1998).
Specific matches obtained from GenBank for the 16S bacterial sequences included clone 1603Q1 which was a 100 % match to Enterococcus faecalis that Goyache et al. (2003) found in 35 % of cloacal samples obtained from Magellanic penguins, Spheniscus magellanicus from Peninsula Valdés, Argentina. However, Banks et al. (2009) did not find Enterococcus faecalis in cloacal samples collected from Adelie penguins in McMurdo Sound suggesting that this bacterium may be less common in McMurdo Sound. One of the clones was 99 % similar to Arcanobacterium phocae, a bacterium that Johnson et al. (2003) isolated from California sea lions (Zalophus californianus), Pacific harbour seals (Phoca vitulina richardii), northern elephant seals (Mirounga angustirostris), southern sea otters (Enhydra lutris nereis), and common dolphin (Delphinus delphis). Johnson et al. (2003) suggested that A. phocae is a significant pathogen of stranded marine mammals. Another of the clones (1671T) was identical to Streptococcus phocae a bacterium that Skaar et al. (1994) found was associated with opportunistic secondary bacterial infections of harbour and grey seals that were infected by phocine distemper virus, a morbillivirus. Given that we did not observe any obvious signs of illness in the seals that we sampled, it is likely that these two bacterial species are a common inhabitant of the seal gut flora and that numbers can increase when animals are ill or stressed. One seal (#1603) produced a clone with 99 % similarity to Edwardsiella tarda a bacterium that has been isolated from a range of species around the world including Antarctica. Leotta et al. (2009) identified the bacterium in Antarctic sea birds and penguins, and Weddell seals. Although the bacterium has been associated with diseased fish (Xiao et al. 2008), E. tarda is considered a common bacterium of Antarctic wildlife that probably does not cause any clinical signs of disease (Leotta et al. 2009).
Several of the bacterial clones’ closest matches have been identified from the gastrointestinal tracts of other seal species. Escherichia coli is a known inhabitant of the vertebrate gut (Bergey and Holt 1994). Fusobacterium varium have been isolated from southern elephant seals Mirounga leonine and leopard seals Hydrurga leptonyx (Nelson 2012), Moxarella phenylpyrivuca was renamed Pyschrobacter phenylpyruvicus by Bowman et al. (1996) and other Psychrobacter bacteria (e.g., P. lutiphocae) have been isolated from seals (Yassin and Busse 2009). Atopobacter phocae is a facultatively anaerobic Gram-negative bacterium originally isolated from the small intestine of a common seal (Lawson et al. 2000). Streptococcus marimammalium was isolated from grey and common seals (Lawson et al. 2005). The links between some of our clone and Genbank sequences were not as clear. For example, Lysobacter koreensis (Clone 1613T1) was originally isolated from a ginseng field (Lee et al. 2006). Furthermore, several of the closest matches in Genbank had less than 97 % similarity suggesting that many of the bacterial species inhabiting seal faeces have yet to be characterised.
White Island microbial isolation
Two Weddell seals were first recorded at White Island in 1958, shortly after the establishment of McMurdo Station in 1956 and Scott Base in 1957 (Heine 1960). Gelatt (2001) estimated from genetic, demographic and glaciological data that a few seals had colonised the tide cracks around the northern tip of White Island following a brief break in the ice shelf between 1947 and 1956. Numbers of seals at White Island have ranged from nine in 1964 (Stirling 1972) up to 26 adults in 1994 (Testa and Scotton 1999) and on average 4.5 pups were born each year from 1990 to 1999 (Gelatt 2001). Movement of tagged seals between White Island and the open water in Erebus Bay has not been recorded despite 80 % of seals in Erebus Bay carrying tags and the tagging of all pups born in Erebus Bay since 1982 (Hadley et al. 2007).
White Island seals had fewer OTUs than McMurdo Sound seals. This could be because the seals establishing the colony at White island had only a subset of the faecal flora that co-exists with the seals in McMurdo Sound. Alternatively, restricted colonisation of the White Island seals combined with extirpation of bacterial taxa would also result in reduced bacterial diversity. In either case, our results suggest that bacteria are not dispersing freely between seals in McMurdo Sound and White Island.
The faecal bacterial communities of seals at White Island were also significantly different from the seals in McMurdo Sound. Stirling (1966a) speculated that the White Island seals may have migrated from open water using a series of cracks in the ice shelf adjacent to the Dailey Islands, Brown Island, and Black Island as “stepping stones”. This stepping stone hypothesis was supported by our multidimensional scaling analysis of the faecal communities which found that the White Island communities most closely resembled the communities of seals breeding at the Strand Moraines and two of the Marble Point communities which are the open-water colonies closest to the start of the “stepping stones”.
It could also be that the White Island bacterial communities differ markedly from McMurdo Sound communities because of dietary differences or differences in energy intake. Adult and pups at White Island are significantly larger with a girth to length ratio 16 % larger than seals in McMurdo Sound (Stirling 1972). Genetically obese mice have been shown to have different gut microbial communities compared with lean mice (Ley et al. 2005; Turnbaugh et al. 2006; Vijay-Kumar et al. 2010). Obese humans have a bacterial community that differs from those of lean humans, and obese people who dieted and lost weight developed a microbial community that was more similar to the lean humans (Ley et al. 2006). A comparison of the diets of White Island and McMurdo Sound Weddell seals may resolve this alternative hypothesis.
The Weddell seals at White Island present a unique opportunity to understand founder effects in symbionts when host populations are established from small populations. Studying the prey DNA present in the faeces of these seals may provide information on dietary differences between the White Island population and the seals in the McMurdo Sound colonies. This study will further enhance our understanding of the roles that environmental and biogeographical factors play in determining the distribution and prevalence of gut micro flora.
References
Abdo Z, Schüette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P (2006) Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol 8:929–938
Banks JC, Paterson AM (2005) Multi-host parasite species in cophylogenetic studies. Int J Parasitol 35:741–746
Banks JC, Hogg ID, Cary SC (2009) The phylogeography of Adelie penguin faecal bacteria. Environ Microbiol 11:577–588
Bergey DH, Holt DH (1994) Bergey’s manual of determinative bacteriology, vol 2, 9th edn. Williams and Wilkins, Philadelphia
Bowman JP, Cavanagh J, Austin JJ, Sanderson K (1996) Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46:841–848. doi:10.1099/00207713-46-4-841
Cardinale M et al (2004) Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol 70:6147–6156
Chao A (1987) Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783–791
Cho J, Tiedje JM (2000) Biogeography and the degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66:5448–5456
Clarke KR, Warwick RM (2001) Change in marine communities: an approach to statistical analysis and interpretation, 2nd edn. Primer-E limited, Plymouth
Clayton DH, Al-Tamimi S, Johnson KP (2003) The ecological basis of coevolutionary history. In: Page RDM (ed) Tangled trees. Phylogeny, cospeciation, and coevolution. University of Chicago Press, Chicago, pp 310–341
Cole JR et al (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucl Acids Res 35:D169–D172
Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. PNAS 99:10494–10499. doi:10.1073/pnas.142680199
Fenchel T (1993) There are more small than large species? Oikos 68:375–378
Finlay BJ, Clarke KJ (1999) Ubiquitous dispersal of microbial species. Nature 400:828
Fisher MM, Triplett EW (1999) Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol 65:4630–4636
Fulthorpe R, Rhodes A, Tiedje J (1998) High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl Environ Microbiol 64:1620–1627
Funk D, Helbling L, Wernegreen J, Moran N (2000) Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc Roy Soc Lond Series B Biol Sci 267:2517–2521
Gelatt TS (2001) Male reproductive success, relatedness, and the mating system of Weddell seals in McMurdo Sound, Antarctica. Ph. D. Thesis, University of Minnesota
Glad T, Kristiansen V, Nielsen K, Brusetti L, Wright A-D, Sundset M (2010) Ecological characterisation of the colonic microbiota in Arctic and sub-Arctic seals. Microb Ecol 60:320–330. doi:10.1007/s00248-010-9690-x
Goyache J et al (2003) Corynebacterium sphenisci sp. nov., isolated from wild penguins. Int J Syst Evol Microbiol 53:1009–1012
Hadley GL, Rotella JJ, Garrott RA (2007) Evaluation of reproductive costs for Weddell seals in Erebus Bay, Antarctica. J Anim Ecol 76:448–458
Hagström A, Pinhassi J, Zweifel UL (2000) Biogeographical diversity among marine bacterioplankton. Aquat Microb Ecol 21:231–244
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hastings KK, Testa JW (1998) Maternal and birth colony effects on survival of Weddell seal offspring from McMurdo Sound Antarctica. J Anim Ecol 67:722–740
Heine A (1960) Seals at White Island. Antarctic 2:271–272
Johnson S, Jang S, Gulland F, Miller M, Casper D, Lawrence J, Herrera J (2003) Characterization and clinical manifestations of Arcanobacterium phocae infections in marine mammals stranded along the central California coast. J Wildl Dis 39:136–144
Kristiansen V (2007) Ampicillin resistance and bacterial diversity in colon content from grey seals (Halichoerus grypus) and harbour seals (Phoca vitulina) at the coast of Northern Norway. Ph. D. Thesis, Universitetet i Tromsø
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lankau EW, Cruz Bedon L, Mackie RI (2012) Salmonella strains isolated from Galápagos iguanas show spatial structuring of serovar and genomic diversity. PLoS ONE 7:e37302. doi:10.1371/journal.pone.0037302
Lawson PA, Foster G, Falsen E, Ohlen M, Collins MD (2000) Atopobacter phocae gen. nov., sp. nov., a novel bacterium isolated from common seals. Int J Syst Evol Microbiol 50:1755–1760
Lawson PA, Foster G, Falsen E, Collins MD (2005) Streptococcus marimammalium sp. nov., isolated from seals. Int J Syst Evol Microbiol 55:271–274. doi:10.1099/ijs.0.63342-0
Lee JW, Im W-T, Kim MK, Yang D-C (2006) Lysobacter koreensis sp. nov., isolated from a ginseng field. Int J Syst Evol Microbiol 56:231–235. doi:10.1099/ijs.0.63955-0
Leotta GA, Piñeyro P, Serena S, Vigo G (2009) Prevalence of Edwardsiella tarda in Antarctic wildlife. Polar Biol 32:809–812. doi:10.1007/s00300-009-0610-9
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI (2005) Obesity alters gut microbial ecology. PNAS 102:11070–11075
Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Human gut microbes associated with obesity. Nature 444:1022–1023
Ley RE et al (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Li M et al (2008) Symbiotic gut microbes modulate human metabolic phenotypes. PNAS 105:2117–2122
Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF (2013) The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol 22:1904–1916. doi:10.1111/mec.12206
MacArthur RH, Wilson EO (1967) The theory of Island biogeography. Princeton University Press, Princeton
Magurran AE (1988) Ecological diversity and its measurement. Croom Helm, London
Nelson T (2012) Factors influencing the gut microbiota of Antarctic seals. University of New South Wales, New South Wales
Paterson AM, Palma RL, Gray RD (1999) How frequently do avian lice miss the boat? Implications for coevolutionary studies. Syst Biol 48:214–223
Paterson AM, Palma RL, Gray RD (2003) Drowning on arrival, missing the boat and X-events: how likely are sorting events? In: Page RDM (ed) Tangled Trees. Phylogeny, cospeciation and coevolution. The University of Chicago Press, Chicago, pp 287–309
Rozsa L (1993) Speciation patterns of ectoparasites and “straggling” lice. Int J Parasitol 23:859–864
Schloss PD et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/aem.01541-09
Skaar I, Gaustad P, Tønjum T, Holm B, Stenwig H (1994) Streptococcus phocae sp. nov., a new species isolated from clinical specimens from seals. Int J Syst Bacteriol 44:646–650. doi:10.1099/00207713-44-4-646
Smith SC, Chalker A, Dewar ML, Arnould JPY (2013) Age-related differences revealed in Australian fur seal Arctocephalus pusillus doriferus gut microbiota. FEMS Microbiol Ecol 86:246–255. doi:10.1111/1574-6941.12157
SPSS (1998) Systat 9.01 Statistics. SPSS Inc., San Jose
Stirling I (1966a) The seals at White Island. Antarctic 4:310–313
Stirling I (1966b) A technique for handling live seals. J Mammal 47:543–544
Stirling I (1969) Ecology of the Weddell Seal in McMurdo Sound, Antarctica. Ecology 50:573–586
Stirling I (1972) Regulation of numbers of an apparently isolated population of Weddell Seals (Leptonychotes weddelli). J Mammal 53:107–115
Testa JW, Scotton BD (1999) Dynamics of an isolated population of Weddell seals (Leptonychotes weddellii) at White Island, Antarctica. J Mammal 80:82–90
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1131
Turnbaugh PJ et al (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484
Vijay-Kumar M et al (2010) Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 328:228–231. doi:10.1126/science.1179721
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Ward DM (1998) A natural species concept for prokaryotes. Curr Opin Microbiol 1:271–277
Whitaker RJ (2006) Allopatric origins of microbial species. Philos Trans R Soc B 361:1975–1984. doi:10.1098/rstb.2006.1927
Wood SA, Rueckert A, Cowan DA, Cary SC (2008) Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J 2:308–320
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725. doi:10.1128/aem.06516-11
Xiao J, Wang Q, Liu Q, Wang X, Liu H, Zhang Y (2008) Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquacult Res 40:13–17. doi:10.1111/j.1365-2109.2008.02101.x
Yassin AF, Busse H-J (2009) Psychrobacter lutiphocae sp. nov., isolated from the faeces of a seal. Int J Syst Evol Microbiol 59:2049–2053. doi:10.1099/ijs.0.008706-0
Zhang C et al (2010) Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4:232–241
Acknowledgments
Thanks to Charles Lee, Ian McDonald, Andreas Rueckert, and Susie Wood for advice and comments on the manuscript. Logistic support was provided through Antarctica New Zealand. Financial support was provided by the New Zealand Foundation for Research Science and Technology through a postdoctoral fellowship for JCB (UOWX0504) and research contract UOWX0505, and from the University of Waikato Vice Chancellor’s research fund. This work contributes to the New Zealand Latitudinal Gradient Project (LGP) and the Evolution and Biodiversity in the Antarctic (EBA) programme and its successor Antarctic Thresholds, Resilience and Adaptation (AnT-ERA).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Banks, J.C., Cary, S.C. & Hogg, I.D. Isolated faecal bacterial communities found for Weddell seals, Leptonychotes weddellii, at White Island, McMurdo Sound, Antarctica. Polar Biol 37, 1857–1864 (2014). https://doi.org/10.1007/s00300-014-1567-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-014-1567-x