Abstract
Freshwater filamentous green algae with branched thalli are almost unknown from the Antarctic region. They have rarely been recorded from maritime Antarctica and from sub-Antarctic Islands with rich phanerogamic vegetation. In the genus Hazenia, only one unidentified species was reported from several subaerial sites on Signy Island in 1979. However, unique populations of this genus were recently found in the stony littoral zone of two stable shallow lakes in the northern deglaciated region of James Ross Island (NE Antarctic Peninsula). These populations have a specialized ecology; they participate in the microvegetation on the flattened surfaces of stones in the littoral zones of lakes. The dominant green filamentous and richly branched alga from these communities was transferred to monospecific culture and studied in detail. Molecular sequence data (18S ribosomal DNA and the internal transcribed spacer) indicate that it belongs to the genus Hazenia (Bold in Am J Bot 45:737–743, 1958). Based on distinct molecular, morphological, and ecological characters, this alga was described as a new species (Hazenia broadyi spec. nova). A review of the genus Hazenia, based on molecular phylogenetic analyses of available strains, was also performed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Hazenia was described by Bold (1958) to accommodate a single species, Hazenia mirabilis (type species; type strain = UTEX 846), isolated from a pool near Nashville, Tennessee, USA. The occurrence of Hazenia sp. was mentioned by Broady (1979), who investigated a strain isolated from soil on Signy Island, maritime Antarctica. However, he refrained from drawing any taxonomic conclusions. Recently, we found another population of this genus in a freshwater habitat, as a component of the microvegetation colonizing the flat surface of the stones in shallow littorals of two stable shallow lakes on higher-lying leveled surfaces on James Ross Island, NE Antarctic Peninsula, Antarctica. These lakes originated after the deglaciation of volcanic mesas and they are among the oldest in the region (Nedbalová et al. 2013). Their community of microvegetation, dominated by the heterocytous cyanobacteria Calothrix elsteri and Hassallia andreassenii, participates in specific mosaic-like mucilaginous structures with limestone precipitations (Elster et al. 2009; Komárek et al. 2012). Hazenia grew on the surface of cyanobacterial mats in the form of green, macroscopic colonies.
Because Hazenia represents a relatively rare genus of green algae, we studied the morphology and life cycle of the Antarctic population, which we subsequently isolated into a monospecific culture (strain CCALA 986). The cultivation enabled the evaluation of the phylogenetic position of this alga and a comparison with related taxa.
Our results confirmed the existence of the genus Hazenia and enabled the revision of this generic unit, comprising three revised species. These species could be characterized by their ITS rDNA sequences, as well as by morphological, ecological, and phytogeographical traits.
Materials and methods
Natural material and isolation
Antarctic populations of Hazenia were collected in January 2009 in two shallow lakes near the Andreassen Point on the E coast of the deglaciated Ulu Peninsula, in the northern part of James Ross Island, NE Antarctic Peninsula (Fig. 1). Green Lake 1 (63°54′11.7″S, 57°46′49.9″W, altitude 65 m a.s.l., area 4,220 m2, max. depth 1.1 m, water volume about 1,000 m3, Fig. 2a) and Green Lake 2 (63°53′54.6″S, 57°46′33.8″W, altitude 40 m a.s.l., area 2,970 m2, max. depth 0.9 m, water volume about 2,200 m3, Fig. 2b) are fed by snow fields in their catchments and their water levels significantly decrease during summer months because of evaporation. The temperature of lake water can surpass 10 °C during clear summer days, and important diurnal fluctuations are probable, as was shown for another shallow lake on the Ulu Peninsula. Both lakes belong to the category of stable shallow lakes on higher-lying leveled surfaces. This type includes the oldest lakes in this region, with possible origins in the early Holocene. However, the exact ages of the two lakes under study are not known. Their bedrock is formed of neogene basaltic volcanic rocks, mostly of hyaloclastic breccias, tuffs, and subaerial basalts (Nedbalová et al. 2013).
In the middle of the summer seasons in 2008 and 2009, the lakes were ice free and the water with temperatures about 9–12 °C, saturated by oxygen (11–14 mg L−1), and with low mineral nutrient contents (oligotrophic character). Lake water pH was neutral to slightly alkaline (7.2–9.0) and the conductivity was 55 μS cm−1 (Green Lake 1) and 91 μS cm−1 (Green Lake 2). Further information about lake water chemistry can be found in Nedbalová et al. (2013).
Hazenia grows in mucilaginous, macroscopic, irregular colonies on the tops of low (up to 2 mm high) mosaic-like structures, which are of biogenic origin (Fig. 3). The main components of the active microphyte community in this locality are filamentous cyanobacteria Calothrix elsteri and Hassallia andreassenii, which are currently considered to be endemic to Antarctica (Komárek et al. 2012).
Hazenia samples collected in the field were transported to the laboratory in a frozen state. Isolation was performed on plates with agarized Z medium (Staub 1961). Plates were streaked with a small amount of field material and cultivated at ca. 18 °C in dim light (ca. 50 μmol m−2 s−1). After a few days of cultivation, algal colonies that developed were transferred separately to sterile agar plates. Unialgal Hazenia strains were cultured at 8 °C and a photon fluence rate of 80 μmol photons m−2 s−1. The strain was deposited in the CCALA culture collection of the Institute of Botany, Czech Academy of Sciences in Třeboň, Czech Republic, under the designation CCALA 986.
Additional cultures
In addition, three strains were obtained from two public culture collections, the Culture Collection of Algae, Georg-August University Göttingen, Germany (SAG), and the Culture Collection of Algae at the University of Texas at Austin, USA (UTEX), to allow their morphological and molecular comparisons with the newly isolated strain CCALA 986:
-
Hazenia mirabilis, strain SAG 1.87, designated as the type strain of the species and the genus Hazenia; isolated by Bold 1953.
-
Pseudendoclonium basiliense, strain SAG 466-1, designated as the type strain of the species; identical with UTEX 2593; isolated by Vischer 1923.
-
Pseudendoclonium printzii, strain SAG 467-1; isolated by Vischer 1924.
DNA isolation, polymerase chain reaction (PCR), and DNA sequencing
Total genomic DNA was isolated using the InstaGene matrix (Bio-Rad, Hercules, CA, USA) as described in Škaloud et al. (2012). Sequences of the small subunit (SSU) 18S ribosomal DNA (rDNA) and the internal transcribed spacer (ITS) region were obtained by PCR amplification using an XP thermal cycler (Bioer, Tokyo, Japan). Each 20-μL PCR contained 13.1 μL of sterile Milli-Q water (Millipore, Billerica, MA, USA), 2 μL of AmpliTaq Gold® 360 Buffer 10× (Life Technologies, Carlsbad, CA, USA), 2.2 μL of MgCl2 (25 mM), 0.4 μL of dNTP mix (10 mM), 0.25 μL of each primer (25 nM), 0.6 μL of 360 GC Enhancer (Life Technologies), 0.2 μL of AmpliTaq Gold® 360 DNA Polymerase, and 1 μL of DNA (10 ng μL−1). The SSU rDNA gene was amplified using the primers 18S-F (5′-AAC CTG GTT GAT CCT GCC AGT-3′) and 18S-R (5′-TGA TCC TTC TGC AGG TTC ACC TAC G-3′; Katana et al. 2001). The ITS rDNA region was amplified using the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′; White et al. 1990). The amplifications of the SSU rDNA and ITS markers started with an initial denaturation at 94 °C for 4 min, followed by 35 cycles of denaturing at 94 °C for 1 min, annealing at 52/50 °C for 1 min, and elongation at 72 °C for 2/1.5 min, with a final extension at 72 °C for 10 min, respectively. The PCR products were stained with bromophenol blue loading dye, quantified on 1 % agarose gel, stained with ethidium bromide, and cleaned with the JETQUICK PCR Purification Kit (Genomed, Löhne, Germany). The purified amplification products were sequenced using an Applied Biosystems (Foster City, CA, USA) automated sequencer (ABI 3730xl) at Macrogen Corp. in Seoul, Korea. Sequencing reads were assembled and edited using the SeqAssem programme (Hepperle 2004). The sequences are available in the EMBL Nucleotide Sequence Database under accession numbers HF570951-HF570955.
Sequence alignment, model selection, and phylogenetic analyses
The newly determined sequences were aligned to other 18S/ITS rDNA sequences from the DDBJ/EMBL/GenBank database. Two different alignments were constructed for the phylogenetic analyses: (1) an SSU rDNA alignment of 52 sequences selected to encompass all lineages within Ulvales/Ulotrichales and (2) an ITS rDNA alignment of 35 Ulotrichalean sequences. The sequences were aligned using MAFFT v. 6 software (Katoh et al. 2002) under the Q-INS-i strategy. The alignment of ITS2 sequences was guided by the consensus secondary structure model of ITS2 in the Ulvales (Caisová et al. 2011). The most appropriate substitution models were estimated using the Akaike information criterion with PAUP/MrModeltest 1.0b (Nylander 2004). The GTR + I + G substitution model was estimated for both SSU rDNA gene and ITS rDNA regions.
The phylogenetic trees were inferred with Bayesian inference (BI) using MrBayes v. 3.1 (Ronquist and Huelsenbeck 2003). Two parallel Markov chain Monte Carlo (MCMC) runs were carried out for four million generations each with one cold and three heated chains. Trees and parameters were sampled every 100 generations. Convergence of the two cold chains was checked and “burn-in” was determined using the “sump” command. Bootstrap analyses were performed by maximum likelihood (ML) and weighted parsimony (wMP) criteria in GARLI v. 0.951 (Zwickl 2006) and PAUP* v. 4.0b10 (Swofford 2002), respectively. ML analyses consisted of rapid heuristic searches (100 pseudo-replicates) using automatic termination (genthreshfortopoterm command set to 100,000). The wMP bootstrapping (1,000 replications) was performed using heuristic searches with 100 random sequence addition replicates, tree bisection reconnection swapping, random addition of sequences (maxtrees set to 10,000 for each replicate), and gap characters treated as a fifth character state. Character weights were assigned using the rescaled consistency index on a scale of 0–1,000. New weights were based on the mean of the fit values for each character over all of the trees in memory.
Results
Morphological observations
Our populations of Hazenia (later described as Hazenia broadyi spec. nova) are currently known only from two localities in the littoral zones of two Antarctic lakes, Green Lake 1 and Green Lake 2, on James Ross Island in the Weddell Sea. They occurred there in dense populations (Fig. 3), commonly colonizing the surfaces of littoral stones lying in the flooded spray zone, particularly on the top of flexuous, low ridges formed by the cyanobacteria Calothrix elsteri and Hassallia andreassenii.
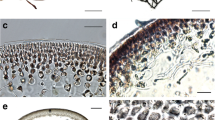
Hazenia broadyi grew in macroscopic, up to 3 mm high, green colonies, with very dense, relatively short, and abundant, irregularly branched filaments (Figs. 3, 4, 5d–f). The branches were short, mostly unilateral, often repeatedly divaricated, and narrowed and rounded at the ends. The cells were irregular in outline, usually slightly elongated, rounded at the ends, and 3.4–15.0 μm in diameter. Terminal cells were usually 3.2–5.4 μm wide and up to 2.5× longer than wide, and slightly narrowed and rounded at the top. More or less pseudoparenchymatous groups of cells developed in the centers and near the bases of colonies, whereas densely arranged filaments were produced peripherally. Reproduction took place by fragmentation of thalli or individual filaments and probably by dormant, larger, and irregular cells developing inside colonies at the ends of branches (Fig. 6). These cells develop from the normal vegetative cells (they are not endogenic, arise neither from zoospores nor from aplanospores), and their chloroplast is more compact and starch granules usually accumulate in the cell volume. In natural material, they sometimes form spherical dormant cells at the ends of branches. Less frequently, the enlarged cells can also develop in old laboratory cultures. They probably represent a kind of resting spores; however, their exact nature needs further study.
The cytomorphology of cells is specific for this taxon. The massive, in principal parietal, irregular chloroplast, occupies the large part of the cell volume and is characterized by slightly irregular wavy margin. In the chloroplast, the pyrenoid-like bodies are visible in many cases, but not with a special starch envelop and concentrated granules around. Numerous rounded granules and oil droplets, which finally fuse in large storage granules and fill a large part of cell volume, occur in old material and especially under culture conditions.
Morphological differences exist between the natural populations (Figs. 4, 5d–f) and specimens, cultured on agar plates (Fig. 5a–c). The basic morphology of filaments and cytology are very similar, but compact colonies found in natural habitats (Fig. 3e–f) were never observed in culture. The colonies on agar are usually composed of agglomerations of irregular, rounded cells (Fig. 5a), which only in peripheral and older parts grow into the elongated and narrowed cells (Fig. 5b–c). This development of growth form was also recorded in natural populations (Fig. 5d–f).
Phylogenetic analyses
An alignment 1,725 bp in length of 52 SSU rDNA sequences was used to infer the phylogenetic placement of H. broadyi within the Ulvophycean lineage of Ulvales/Ulotrichales (Fig. 7). The Bayesian analysis resolved both orders as reciprocally monophyletic, with moderate statistical support. H. broadyi was inferred within Ulotrichales, in a close relation to the morphologically similar H. mirabilis and Pseudendoclonium basiliense. Although both taxa showed a very high level of SSU rDNA similarity (99.8 %, corresponding to three nucleotide changes), only H. mirabilis formed a monophyletic clade with H. broadyi. Our phylogenetic reconstruction provides evidence that the genus Pseudendoclonium as currently circumscribed is polyphyletic, with the type species, P. submarinum, being inferred in the family Kornmanniaceae within Ulvales.
Phylogenetic analysis of the orders Ulotrichales and Ulvales based on 18S ribosomal DNA sequences. The tree was inferred using MrBayes with the GTR + G + I nucleotide substitution model. Values at the nodes correspond to Bayesian posterior probabilities, maximum likelihood, and maximum-parsimony bootstrap percentages, respectively. Full statistical support (1.00/100/100) is marked with an asterisk. Thick branches represent nodes receiving the highest posterior probability support (1.00). Sequences determined in this study are given in bold. Species traditionally affiliated with the genus Pseudendoclonium are marked by arrows (type species marked by filled arrow). The scale bar shows the estimated number of substitutions per site
To better resolve the placement of H. broadyi in the Ulotrichales, we also performed phylogenetic analysis using ITS rDNA data (Fig. 8), including newly obtained sequences for H. mirabilis (SAG 1.87) and P. basiliense (SAG 466-1). The alignment of 35 ITS rDNA sequences was 529 bp in length. Phylogenetic analysis of these sequences resolved three currently recognized families in the highly supported core Ulotrichales (Gayraliaceae, Gomontiaceae, and Monostromataceae), plus a monophyletic, moderately supported lineage of Hazenia and two P. basiliense isolates. Although all families of core Ulotrichales were monophyletic with high support, the relationships between the Hazenia lineage and these families were poorly resolved.
Phylogenetic analysis of Ulotrichales based on internal transcribed spacer ribosomal DNA sequences. The tree was inferred using MrBayes with the GTR + G nucleotide substitution model. Values at the nodes correspond to Bayesian posterior probabilities, maximum likelihood, and maximum-parsimony bootstrap percentages, respectively. Thick branches represent nodes receiving the highest posterior probability support (1.00). Sequences determined in this study are given in bold. The scale bar shows the estimated number of substitutions per site
Based on 18S rRNA gene sequencing combined with ITS, we concluded that the genus Hazenia comprises three defined species, the type H. mirabilis, our populations from Antarctica (which we describe as a new species H. broadyi), and also (as a separate species) the alga described as P. basiliense. The latter must be excluded from the genus Pseudendoclonium and recombined into the genus Hazenia, according to the molecular analyses. The taxonomic position of the fourth published Hazenia taxon, Hazenia sp. from Signy Island (Broady 1979), must be resolved in the future.
Taxonomic review
-
Hazenia Bold, Amer. J. Botany 45(10): 742, 1958 (Gayraliaceae, Ulotrichales)
Diagnostic features:
Position in the phylogenetic tree, morphology of thallus, specific type of branching, life cycle, ecology.
-
Hazenia mirabilis Bold, Amer. J. Botany 45(10): 742, 1958.
Diagnosis:
Colonies macroscopic, tuft- to cushion-like, mucilaginous, 3 mm high and in diameter, irregularly spheroidal, growing on stones, sometimes slightly lime-encrusted. Filaments richly, irregularly, and usually unilaterally repeatedly branched, relatively short, uni- to multiseriate, consisting of prostrate pseudoparenchymatous and erect systems, forming loose to pseudoparenchymatous and dense fascicles. Short branches narrowed toward ends and terminated by blunt tips. Cells very irregular, globose, barrel-shaped or cylindrical, 3.4–15.0 μm in diameter or somewhat irregularly inflated; at ends usually elongated and slightly narrowed, conically rounded, 3.2–5.4 μm wide and up to 2.5× longer than wide. Cell contains a prominent parietal chloroplast with indistinct pyrenoid. Reproduction by fragmentation of filaments and probably by larger, irregular cells with green content.
Type:
BRNM-HY/CCALA 986, deposited in: Herbarium of the Moravian Museum Brno, typical morphology (icona typica): shown in Figs. 4, 5; type strain: CCALA 986.
Habitat:
On surfaces of flat stones in the littoral zones of Antarctic lakes; original locality (locus classicus): Antarctica, James Ross Island, NE Antarctic Peninsula, E coast, near Andreassen Point (Green Lake 2).
Distribution:
Thus far known only from two lakes (Green Lake 1 and Green Lake 2) near Andreassen Point on James Ross Island.
Etymology:
The species is dedicated to Paul Broady (Christchurch, New Zealand), the prominent specialist in freshwater and terrestrial algae in Antarctica.
-
Hazenia basiliensis (Vischer) comb. nova (basionym: Pseudendoclonium basiliense Vischer, Verh. Schweiz. Naturf. Ges. 107(2): 204, 1926).
-
Hazenia sp. in Broady, Brit. Antarct. Surv., Sci. Rep. 98: 89–90, 1979.
Discussion
James Ross Island is situated in the transitory zone between maritime and continental Antarctica with harsher climatic conditions than the Western Antarctic Peninsula (Øvstedal and Lewis-Smith 2001). The discovery of filamentous, branched green algae in this region is therefore an important novel finding from the ecological and evolutionary points of view.
The species newly described in this paper, Hazenia broadyi, must be considered endemic to Antarctica, based on the current data. In lakes on James Ross Island, a high level of endemism has been reported for cyanobacteria and pennate diatoms that form the major part of the littoral biomass (Komárek et al. 2008, 2012; Kopalová et al. in press). Overall, striking discontinuities and high degrees of regional endemism have been demonstrated for various taxonomic groups of Antarctic biota, including lichens and many invertebrates (Convey et al. 2008). Studies focusing on the origins of terrestrial life in Antarctica, with its extensive glaciation history (Convey et al. 2008), suggested the possibility that organisms survived on land, especially during the last glacial maxima (Huybrechts 1993; Denton and Hughes 2002). New and more precise knowledge of glaciations, together with the increasing amount of molecular data from various taxonomic groups, is changing our understanding of the ability of living organisms to persist during extensive glaciations of Antarctica. However, the dispersal of microalgae to Antarctica has been long discussed (De Wever et al. 2009; Rybalka et al. 2009). Reintroduction or persistence of microalgal species during cold climate cycles connected with the cleaning action of extensive ice cover may be confirmed by molecular studies of strains collected in Antarctica and by assessing their relationships with populations on surrounding islands and continents.
Our study contributes another fragment to the mosaic of the understanding of freshwater filamentous, branched, green algae distributions in the Antarctic. These algae have been sparsely recorded from this region. To our knowledge, there are only a few records from shallow lentic wetlands in maritime Antarctica and on sub-Antarctic Islands. Bonaventura et al. (2006) recorded Stigeoclonium sp. in lentic water bodies at Hope Bay, Antarctic Peninsula. Pseudendoclonium sp. and Stigeoclonium sp. have been recorded by Komárek and Komárek (1999) in the Arctowski Station region on King George Island (South Shetlands). The only Antarctic record of a species from the genus Hazenia comes from Signy Island, South Orkney Islands (Broady 1979). On sub-Antarctic Islands with relatively rich vegetation of vascular plants, branched green algae have been found more frequently (see Prescott 1979).
Phylogenetic analyses confirmed our generic determination of the population from James Ross Island; it was genetically allied to the type strain H. mirabilis. However, the ITS rDNA phylogeny, a detailed morphological investigation, as well as ecological criteria indicated that the population should be considered a separate taxon, which we describe as H. broadyi. The genus Hazenia is clearly defined and confirmed based on modern taxonomic methods using molecular data. Interestingly, the type strain of P. basiliense (the type strain SAG 466–1, distant from the type species of the genus, P. submarinum), was in close affinity with both Hazenia taxa, indicating that its generic classification was wrong. Its position within Hazenia was also confirmed based on morphological observations. The genus Pseudendoclonium is thus apparently polyphyletic and has representatives in disparate phylogenetic clusters, of which one, including the type of Pseudendoclonium based on P. submarinum, must be classified separately from Hazenia.
Hazenia belongs to the order Ulotrichales, family Gayraliaceae, according to the modern classification. Modern classifications of green algae based on molecular data often contradict traditional systematics (cf. Pröschold and Leliaert 2007). According to our results, the genera of filamentous, branched, green algae can be characterized by selected morphological features after detailed revision and synthesis of morphological and molecular data.
The only previous record of Hazenia sp. from the Antarctic region was published by Broady (1979) (Signy Island, South Orkney Islands), but this population was not taxonomically defined. Moreover, neither its ecology nor its morphology corresponds to the populations from James Ross Island. Therefore, these Antarctic populations could not be unified in a single species, and the taxonomic position of the Signy Island population (Broady 1979) must be resolved in the future. However, more precise and dense field studies in various geographical areas across the Antarctic region, together with detailed molecular and taxonomic evaluations, are necessary to understand the distribution of green algae in Antarctica in more detail.
References
Bold H (1958) Three new chlorophycean algae. Am J Bot 45:737–743
Bonaventura SM, Vinocur A, Allende L, Pizarro H (2006) Algal structure of the littoral epilithon in lentic water bodies at Hope Bay, Antarctic Peninsula. Polar Biol 29:668–680
Broady P (1979) The terrestrial algae of Signy Island, South Orkney Islands. Br Antarct Surv Sci Rep 98:1–117
Caisová L, Marin B, Melkonian M (2011) A close-up view on ITS2 evolution and speciation—a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol Biol 11:262
Convey P, Gibson JAE, Hillenbrand CD, Hodgson AD, Pigh PJA, Smellie JL, Stevens MI (2008) Antarctic terrestrial life—challenging the history of the frozen continent? Biol Rev 83:103–117
De Wever A, Leliaert F, Verleyen E, Vanormelingen P, Van der Gucht K, Hodgson DA, Sabbe K, Vyverman W (2009) Hidden levels of phylodiversity in Antarctic green algae: further evidence for the existence of glacial refugia. Proc R Soc B 276:3591–3599
Denton GH, Hughes TH (2002) Reconstruction the Antarctic ice sheet at the last glacial maximum. Quat Sci Rev 21:193–202
Elster J, Nedbalová L, Komárek J, Vodrážka R (2009) Biogenic calcite structures in Green Lake, James Ross Island, Antarctica. In: Barták M, Hájek J, Váczi P (eds) Structure and function of Antarctic terrestrial ecosystems, Brno, October 22–23, 2009, pp 38–40
Hepperle D (2004) SeqAssem©. A sequence analysis tool, contig assembler and trace data visualization tool for molecular sequences. (http://www.sequentix.de)
Huybrechts P (1993) Glaciological modelling of the late Cenozoic east Antarctic ice sheet: stability or dynamism? Geogr Ann Ser A Phys Geogr 75:221–238
Katana A, Kwiatowski J, Spalik K, Zakrys B, Szalacha E, Szymanska H (2001) Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J Phycol 37:443–451
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Komárek O, Komárek J (1999) Diversity of freshwater and terrestrial habitats and their oxyphototroph microflora in the Arctowski Station region, South Shetlands Islands. Pol Polar Res 20:259–282
Komárek J, Elster J, Komárek O (2008) Diversity of cyanobacterial microflora of the northern part of James Ross Island, NW Weddell Sea, Antarctica. Polar Biol 31:853–865
Komárek J, Nedbalová L, Hauer T (2012) Phylogenetic position and taxonomy of three heterocytous cyanobacteria dominating the littoral of deglaciated lakes, James Ross Island, Antarctica. Polar Biol 35:759–774
Kopalová K, Nedbalová L, Nývlt D, Elster J, van de Vijver B Diversity and ecology of the freshwater diatom communities from Ulu Peninsula (James Ross Island, NE Antarctic Peninsula). Polar Biol. doi:10.1007/s00300-013-1317-5 (in press)
Nedbalová L, Nývlt D, Kopáček J, Šobr M, Elster J (2013) Freshwater lakes of Ulu Peninsula, James Ross Island, northeast Antarctic Peninsula: origin, geomorphology and physical and chemical limnology. Antarct Sci 25:358–372
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. (http://www.abc.se/~nylander/)
Øvstedal DO, Lewis-Smith RI (2001) Lichens of Antarctica and South Georgia. A guide to their identification and ecology. Cambridge University Press, Cambridge
Prescott GW (1979) A contribution to a bibliography of Antarctic and Subantarctic algae. Bibl Phycol 45:1–312
Pröschold T, Leliaert F (2007) Systematics of the green algae: conflict of classic and modern approaches. In: Brodie L, Lewis J (eds) Unravelling the algae: the past, present, and future of algal systematics (Systematics Association Special Volumes). CRC Press, Boca Raton, FL, pp 123–153
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rybalka N, Andersen RA, Kostikov I, Mohr KI, Massalski A, Olech M, Friedl T (2009) Testing for endemism, genotypic diversity and species concept in Antarctic terrestrial microalgae of the Tribonemataceae (Stramenophiles, Xanthophyceae). Environ Microbiol 11:554–565
Škaloud P, Šťastný J, Nemjová K, Mazalová P, Poulíčková A, Neustupa J (2012) Molecular phylogeny of baculiform desmid taxa (Zygnematophyceae). Plant Syst Evol 298:1281–1292
Staub R (1961) Untersuchungen an der Blaualge Oscillatoria rubescens DC. Schweiz Z Hydrol 23:83–198
Swofford DL (2002) PAUP 4.0b10: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, University of Texas at Austin
Acknowledgments
The study was supported by the Ministry of Education, Youth and Sport of the Czech Republic (long-term research development grant RVO67985939, projects KONTAKT ME 945 and CzechPolar LM2010009). The authors would like to thank to the scientific infrastructure of the Czech Antarctic Station “J.G. Mendel” and its crew for their support. We are also very grateful to Mrs. Jana Šnokhousová and Mrs. Dana Švehlová for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Škaloud, P., Nedbalová, L., Elster, J. et al. A curious occurrence of Hazenia broadyi spec. nova in Antarctica and the review of the genus Hazenia (Ulotrichales, Chlorophyceae). Polar Biol 36, 1281–1291 (2013). https://doi.org/10.1007/s00300-013-1347-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-013-1347-z