Abstract
Key message
VqERF114 regulates stilbene synthesis by interacting with VqMYB35.
Abstract
Resveratrol is a stilbene, an important class of secondary metabolites that accumulates in some plant species, including grapevine. In the plant, these are involved in the response to attack by plant pathogens and, as a component of the human diet, they offer a range of significant health benefits. Stilbene synthase (STS), the key enzyme responsible for resveratrol synthesis, has been characterised in a small number of plant species. However, the regulatory mechanisms for stilbene synthesis are uncertain. Here, an ERF family transcription factor from Chinese wild Vitis quinquangularis, VqERF114, was characterised as an indirect regulator of stilbene synthesis. A transient overexpression assay of VqERF114 in grapevine leaves led to increased STS expression and stilbene accumulation. However, VqERF114 did not bind to the promoters of VqSTSs but the MYB transcription factor, VqMYB35, did interact with VqERF114. This interaction was confirmed by a yeast two-hybrid assay and bimolecular fluorescence complementation. Furthermore, VqMYB35 showed activation effects on the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by binding directly to the MBS elements in their promoters. Co-overexpression of VqERF114 and VqMYB35 resulted in higher VqSTSs expression and more stilbene synthesis. These results demonstrate that VqERF114 regulates stilbene synthesis by interacting with VqMYB35.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Resveratrol is one of a number of natural polyphenols produced by secondary metabolism in some plant species. Resveratrol has so far been found in approximately 72 species, including in grapevine (Langcake and Pryce 1976), peanuts (Jennifer et al. 2002), sorghum (Yu et al. 2005), Scots pine (Rosemann et al. 1991), cranberry, blueberry and knotweed (Vastano et al. 2000). Resveratrol is a major stilbene-type phytoalexin that plays a role as an antimicrobial metabolite in plants (Hart 1981). In human nutrition, resveratrol offers a range of health benefits, having properties that include: anti-inflammatory, anticancer and anti-oxidative (Jang et al. 1997). Increasing attention is recently being paid to resveratrol, not only because its functions in plant defence but also because of its medical properties. In most stilbene-producing plants including grapevine, resveratrol is synthesised via the phenylpropanoid pathway. The phenylpropanoid pathway is a fully characterised metabolic pathway in plants. It produces all kinds of secondary metabolites including lignins, flavonoids and taxon-specific compounds, such as the stilbenes in grapevine (Chen et al. 2006). The full phenylpropanoid pathway contains two branches, the flavonoid branch and the stilbene branch. These two branches have three same upstream enzymes—phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-coumarate: CoA ligase (4CL). They are distinguished by the last step catalytic enzymes, chalcone synthases (CHSs) and stilbene synthases (STSs) (Vannozzi et al. 2012). For the CHSs, the substrates are p-coumaroyl-CoA and malonyl-CoA which result in the synthesis of the flavonoids, while STSs catalyse the same substrates to synthesise resveratrol. The key enzyme responsible for the synthesis of resveratrol is STS, which belongs to the chalcone synthase superfamily of type III polyketide synthases (Chong et al. 2009). In grapevine, an exhaustive characterisation of the STS multigenic family has been carried out with the PN40024 and PN ENTAV 115 genomes (Jaillon et al. 2007; Velasco et al. 2007), resulting in the identification of 48 putative STS gene sequences (Vannozzi et al. 2012). Of the two geometric isomers of resveratrol, trans-resveratrol is thought to be the more biologically active (Orallo 2006). In addition to resveratrol, more complex derivatives have also been found in grapevine, including trans-piceid (Waterhouse and Lamuela-Raventós 1994; Teguo et al. 1996; Romero-Pérez et al. 2001; Gatto et al. 2008), viniferins, pterostilbene (Langcake 1981), and piceatannol (Bavaresco et al. 2002). Resveratrol glucosyltransferase (RSGT) is the enzyme that synthesises stilbenoid glucoside from resveratrol (Hall and De Luca 2010).
China is one of the worlds’ major biodiversity centres for Vitis, so Chinese wild grapevines are very important germplasm resources. They also exhibit high levels of resistance to a number of key pathogens. The Chinese wild species Vitis quinquangularis, accession ‘Danfeng-2’ has attracted considerable interest because of its high resistance to pathogen invasion and the high levels of resveratrol found in its ripe berries (Wan et al. 2007; Shi et al. 2014; Zhou et al. 2015).
The AP2/ERF superfamily is a large group of transcription factors (TFs) in plants (Wessler 2005). This superfamily is identified by the existence of the AP2/ERF domain, which is composed of about 60–70 conserved amino acid residues and participates in DNA binding (Jofuku et al. 1994; Ohme-Takagi and Shinshi 1995). Based on the number of AP2 domains and on the presence of other domains, such as the B3 DNA-binding domain, the AP2/ERF superfamily is classified into three families: AP2, ERF and RAV (Sakuma et al. 2002). Thus, the AP2 family contains proteins with a tandem repetition of two AP2 domains (Okamuro et al. 1997), the ERF family proteins contain one AP2 domain (Nakano et al. 2006) and the RAV family proteins possess a single AP2 domain and an additional B3 domain (Kagaya et al. 1999). Within the ERF family, the AP2/ERF DNA-binding domain can be divided into DREB and ERF domains, based on the identity of residues at specific positions (Sakuma et al. 2002). The DREB subfamily (group A) proteins interact with an A/GCCGAC element (Stockinger et al. 1997), while the proteins of the ERF subfamily (group B) typically bind to an AGCCGCC element, called the GCC-box. GCC-box element is usually found in the promoters of genes in response to ethylene, pathogens and wounding (Ohme-Takagi and Shinshi 1995). These two subfamilies are further subdivided into six groups, respectively. Members of the ERF family are characterised and involved in many different functions, including the regulation of metabolism, response to biotic and abiotic stresses, hormonal signal transduction and developmental processes in numerous plant species (Nakano et al. 2006). The ERF transcription factor regulating the expression of STS has not yet been identified.
The MYB proteins are key factors in regulatory networks participating in metabolism, in development and in response to biotic and abiotic stresses (Dubos et al. 2010). The R2R3-MYB family members from different species possess DNA-binding specificities, with three main MYB binding types so far identified—type I: CNGTTR, type II: TNGTTR and type IIG/AC-elements: CCWAMC (Romero et al. 2010; Prouse and Campbell 2012). Several other variants have also been reported (Kelemen et al. 2015). In grapevine, the first survey of the R2R3-MYB family proteins characterised 108 genes in the × 8.4 genome draft sequence (Matus et al. 2008). Subsequently, a total of 134 MYB sequences have been retrieved in the CRIBI 12 × v1 genome accession (Wong et al. 2016). Recently, R2R3-MYB TFs have been reported to be involved in the regulation of the STS pathway in grapevine. VvMYB14 and VvMYB15 can enhance promoter activity of VvSTS29 and VvSTS41 (Höll et al. 2013), and VvMYB14 can bind to the VvSTS48 promoter (Fang et al. 2014). Moreover, another MYB transcription factor, VvMYB13, is characterised as playing a key role in regulating stilbene accumulation (Wong et al. 2016).
In our previous study, transcriptome analysis was performed on ‘Danfeng-2’ during the various stages of berry development to obtain specific TFs involved in stilbene synthetic pathway (PRJNA306731; GEO: GSE76256). Eighteen ERF TFs were found to be strongly co-expressed with STS genes. Expression analyses of 18 ERF genes co-expressed with stilbene synthases in V. quinquangularis accession ‘Danfeng-2’ in response to Uncinula necator were carried out, of which, VqERF114 (VIT_18s0072g00260) was found to be strongly co-expressed with STS genes and in response to U. necator. The functional study of VqERF114 involved in disease resistance will be explored in our further study. In this study, VqERF114 is identified as an activator of STS expression. However, it cannot directly bind to the promoters of VqSTSs. A yeast two-hybrid assay and a BiFC assay demonstrate that VqERF114 interacts physically with VqMYB35. VqMYB35 can promote the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by directly binding to their promoters. Furthermore, VqERF114 co-expresses with VqMYB35, resulting in higher expression of VqSTSs and in the enhanced accumulation of stilbene. These results demonstrate that VqERF114 is an indirect positive regulator of stilbene synthesis by interacting with VqMYB35 in grapevine.
Materials and methods
Plant materials
The Chinese wild grapevine species V. quinquangularis accession ‘Danfeng-2’ used in this study was cultivated in the grape germplasm collection at Northwest A&F University, Yangling, Shaanxi, China. The tobacco plants (Nicotiana benthamiana) used for subcellular localisation analysis and dual-luciferase assays were grown in the phytotron with a light/dark cycle of 16/8 h at 25 °C.
Gene isolation and sequence analysis
Total RNA was extracted from grape berries or leaves using a Plant RNA Kit (OMEGA). The PrimeScript™ RT reagent Kit (TaKaRa) was used to reverse transcribe the purified RNA into complementary DNA (cDNA) following the manufacture’s protocol. Full-length coding sequences (CDSs) of VqERF114, VqMYB35, VqMYB14 and VqMYB15 were cloned using the special primers listed in Table S1, using the V. vinifera cv. ‘Pinot Noir’ genome database P40024 as a reference. DNAMAN software was used to carry out amino-acid sequence alignment analyses of three ERF proteins, including VqERF114, VvERF114 (Gene ID: VIT_18s0072g00260) and AtABR1 (Gene ID: AT5G64750). The phylogenetic tree was constructed using MEGA 5.0 software to analyse the evolutionary relationships among the ERF subfamily proteins from grapevine and Arabidopsis.
Subcellular localisation
The VqERF114 and VqMYB35 CDSs without the stop codon were amplified and cloned into the KpnI–XbaI sites of the CaMV35S-GFP vector. The fusion construct was mobilised into the GV3101 strain of Agrobacterium tumefaciens and then transferred into tobacco leaves. Overnight grown bacterial cells were pelleted by centrifugation, resuspended with infiltration buffer (10 mM MES pH 5.8, 10 mM MgCl2, 200 μM acetosyringone) and incubated at 28 °C for 3 h. Tobacco leaves were transiently infiltrated with GV3101 carrying either the fusion construct or the control (CaMV35S-GFP), and then cultured in a growth chamber for 48 h. GFP fluorescence was visualised under a Zeiss confocal microscope LSM510 (Xie and Wang 2016).
Transcriptional activation assay in yeast
The full length of VqERF114 was amplified and inserted into the pGBKT7 vector (Clontech). The recombinant vector and the empty control vector (pGBKT7) were transferred into the yeast strain Y2HGold. In this assay, pGBKT7-53 vector co-transformed with pGADT7-T vector acted as positive control and pGBKT7-Lam vector co-transformed with pGADT7-T vector acted as negative control. The transformants were grown on SD/-Trp medium at 30 °C for 3–5 days. The yeast transformants were spotted on plates containing two media types: SD/-Trp + X-α-Gal and SD/-Trp + X-α-Gal + Aureobasidin A (200 ng mL−1). The transactivation activity was assessed by observing the growth of the yeast cells 2 days later.
Transient overexpression assay of VqERF114 and VqMYB35 in grapevine leaves
The A. tumefaciens GV3101-containing vector expressing VqERF114-GFP and VqMYB35-GFP fusion genes under the CaMV35S promoter was applied in the Agrobacterium-mediated transient assay. This assay was carried out as previously described by Xu et al. (2010) with minor modification. Leaves of V. quinquangularis accession ‘Danfeng-2’ were harvested 2 days after the treatment for quantitative RT-PCR and HPLC analysis.
qRT-PCR analysis
Agro-infiltrated leaves were collected to extract total RNA and 1 μg purified RNA was reversely transcribed into cDNA. Synthetic cDNA was diluted tenfold and used as template for qRT-PCR analysis. The PCR mix (20 μL) contained 10 μL of SYBR (novoprotein), 10 μM each primer, 1 μL of cDNA and sterile water. Three-step qRT-PCRs were carried out using the following procedure: 95 °C for 3 min, followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 15 s, extension at 72 °C for 15 s and a melt cycle with 0.5 °C increments (5 s) from 60 to 95 °C. Each experiment was carried out with three biological replicates, and three technical replicates were analysed for each biological sample. The expression of grapevine GAPDH was used to normalise gene transcript levels. The qRT-PCR primers of VqERF114, VqMYB35, PAL, STS, RSGT, VqSTS15, VqSTS28, VqSTS42/46 and GAPDH are described in Table S1.
Analysis of stilbene in grapevine leaves using HPLC
HPLC analysis of stilbene was carried out on an HPLC device (Waters) equipped with an Agilent Zorbax SB-C18 column (5 μm, 4.6 × 250 mm). Grapevine leaves harvested from transient expression assay were dried under vacuum for 24 h and then extracted by adding methanol (HPLC grade, 50 mL g−1) at 4 °C overnight. The extracts were filtered through a 0.22 μm membrane filter and analysed in an HPLC system. Chromatographic separation was carried out using a solvent system of ultrapure water and acetonitrile according to the procedure described in Xu et al. (2011). Stilbene level, expressed as μg g−1 dry weight, is the average of three replicates for each independent sample.
GUS activity assay
Three tandem copies of GCC-box were inserted into pC0380-GUS vector and fused with GUS reporter. pGCC-GUS was co-expressed with CaMV35S-VqERF114 in leaves of V. vinifera L. cv. Thompson seedless using Agrobacterium-mediated transient transformation. pMini35S-GUS, pGCC-GUS and pMini35S-GUS co-expressed with CaMV35S-VqERF114 served as controls. Grapevine leaves were collected for GUS staining and protein determination after culture for 2 days. The GUS activity assay was carried out according to the method described in Xu et al. (2010).
Yeast two-hybrid assay
The interaction between VqERF114 and VqMYB35 was investigated using the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech). The VqERF114 sequence was cloned into the pGBKT7 (BD) vector and the VqMYB35 sequence was cloned into the pGADT7 (AD) vector. BD and AD vectors were co-transferred into the yeast strain Y2HGold. Transformed cells were spread onto the following plates: SD/-Leu/-Trp medium and SD/-Ade/-His/-Leu/-Trp + X-α-Gal + AbA (400 ng mL−1) medium. The positive control and the negative control were the same as in the transcriptional activation assay.
Bimolecular fluorescence complementation assays
For the bimolecular fluorescence complementation (BiFC) assays, the coding sequences of VqERF114 and VqMYB35 without the stop codon were cloned into the pSPYNE and pSPYCE vectors, respectively. Tobacco protoplasts were co-transformed with the various plasmids using the polyethylene glycol (PEG) method (Yu et al. 2013). Protoplasts were cultured at room temperature for 24 h in the dark and observed using a Zeiss confocal microscope LSM510.
Promoter analysis
Genomic DNA of grape berries or leaves was isolated using the method of Xu et al. (2010). The promoters of the stilbene synthase genes (VqSTSs) were amplified using primers (Table S1) according to the homologous sequences. The Plant-CARE database was used to predict conserved cis-element motifs presented in each promoter (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Dual-luciferase assays
Dual-luciferase assays were carried out to measure transactivation activities of VqERF114 and VqMYB35 on the target promoter. The full-length cDNA of VqERF114 or VqMYB35 was inserted into pGreenII 62-SK vector and the promoters of VqSTSs were cloned into pGreenII 0800-LUC vector. All constructs were individually electroporated into Agrobacterium GV3101 and expressed transiently in tobacco leaves by A. tumefaciens-mediated infiltration. Agrobacterium cultures were resuspended with infiltration buffer to an OD600 of 0.4. The binding activity of the TFs on the promoter was assayed 3 days after infiltration and assessed by the ratio of enzyme activities of Firefly luciferase (LUC) and Renilla luciferase (REN) using Dual Luciferase Reporter Gene Assay Kit (Beyotime). The A. tumefaciens culture of TF (900 μL) was mixed with promoter (100 μL) to test the activity of a special TF on the promoter. The LUC/REN value of the empty vector 62-SK on the promoter was set as 1, as a calibrator. To determine the combined effect of two TFs on the promoter, 450 μL of each TF was mixed with 100 μL of promoter. The effect of the mixtures which contained each TF (450 μL) and empty vector 62-SK (450 μL) was also tested on the promoter as control. For each interaction between TF and promoter, three independent experiments were carried out, with three replicates in each experiment.
Yeast one-hybrid assay
The Matchmaker™ Gold Yeast One-Hybrid System (Clontech) was used to carry out the Y1H assay. Three tandem copies of GCC-box and the promoters of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 were inserted into pAbAi vector to build the pAbAi-bait and the constructs were integrated separately into the genome of the Y1HGold yeast strain. The core sequence of MBS element in the promoters of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 was mutated from CAGTTA to AAAAAA. The full length of VqMYB35 was cloned into pGADT7 to build the AD-prey vector. The minimal inhibitory concentrations of Aureobasidin A (AbA) for the bait strains were tested according to the system user manual. The AD-prey vectors were transferred into the bait strain and grown on SD/-Leu/AbA plate to confirm the interaction between TF and promoter.
Results
Identification and molecular characterisation of VqERF114
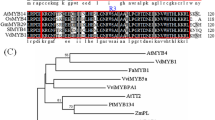
The cDNA clone of VqERF114 consists of a 1266 bp ORF predicted to encode a protein of 421 amino acids. Its predicted molecular weight and isoelectric point (pI) are 44.48 kDa and 6.526, respectively. There are 73 predicted ERF subfamily proteins in grapevine and 65 in Arabidopsis (Zhuang et al. 2009). Amino acid alignments show that VqERF114 contains a single AP2 DNA-binding domain, containing 58 amino acids from 201 to 258 aa. VqERF114 shows very high similarity to VvERF114 in V. vinifera (99% identity) (Fig. 1a). To determine the classification of VqERF114, a phylogenetic tree was built using the neighbour-joining method. This shows that VqERF114 is closely related to VvERF114 and that both belong to the ERF subfamily group B-4 (Fig. 1b). VqERF114 and green fluorescent protein (GFP) fusion construct were transiently expressed in tobacco leaves to determine the subcellular localisation of VqERF114. The signal of VqERF114–GFP was found exclusively in the nuclei of the epidermal cells whereas the GFP fluorescence in the control was observed throughout the cells (Fig. 1c). To test the transcriptional activity of VqERF114, pGBKT7-VqERF114 construct was produced and transferred into Y2HGold yeast cells. As with the positive control, yeast cells expressing VqERF114 grew well on SD/-Trp plates and showed AbA resistance (Fig. 1d), indicating that VqERF114 possesses transactivation ability in yeast cells. These results demonstrate that VqERF114 protein is nuclear localised, consistent with its putative role as a transcription factor.
Sequence alignment analysis, phylogenetic analysis and molecular characterisation of VqERF114. a Sequence alignment of the deduced VqERF114 protein with other ERF proteins including VvERF114 (Gene ID: VIT_18s0072g00260) and AtABR1 (Gene ID: AT5G64750). The consensus sequence is shown in black and the solid black line indicates the conserved AP2/ERF DNA-binding domain. Dashes show gaps in the alignment. b The phylogenetic analysis of VqERF114 and ERF subfamily proteins from Vitis vinifera and Arabidopsis thaliana. Multiple sequence alignment was carried out using ClustalW and the phylogenetic tree was constructed with MEGA 5.0 using a bootstrap test of phylogeny with the neighbour-joining method. c Subcellular localisation of VqERF114 in tobacco leaves. The fusion protein (VqERF114-GFP) and GFP control were transiently expressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens strain GV3101. GFP fluorescence was observed with a confocal microscope. Bars 50 μm. d Transcriptional activation of VqERF114 in yeast cells. The coding region of VqERF114 was cloned into the pGBKT7 (BD) vector to build the BD-VqERF114 construct. BD-VqERF114 together with the positive control (p53 + T-antigen), negative control (Lam + T-antigen) and empty vector (BD) were transferred into yeast cells. Yeast clones transformed with different constructs were grown on SD/-Trp plates at 30 °C for 3–5 days. Transcription activation was monitored according to the growth status of yeast cells and the X-α-Gal assay
Transient overexpression of VqERF114 in grapevine leaves
The expression of VqERF114 in infiltrated leaves was analysed using qRT-PCR 2 days after infiltration. The results show that the transcript levels were very much higher (8.9-fold) in VqERF114 overexpressed leaves than in the empty vector (EV) controls (Fig. 2a), confirming that VqERF114 was successfully overexpressed. The expressions of PAL and STSs were examined in the same leaves, and the transcript abundances were also increased by 5.6-fold and 10.9-fold, respectively (Fig. 2a). However, the transcript level of RSGT in the VqERF114-overexpressing leaves showed no significant difference from in the control EV (Fig. 2a). The stilbene levels in the transformed grapevine leaves were measured by HPLC. As shown in Fig. 2b, the contents of trans-resveratrol and trans-piceid increased after VqERF114 was overexpressed. Taken together, these results indicate that VqERF114 promotes the transcription of STSs and stilbene synthesis.
Transient overexpression of VqERF114 in grapevine leaves. a Relative expressions of VqERF114, PAL, STSs and RSGT in VqERF114 overexpressed grapevine leaves 2 days after infiltration. The expression was calculated relative to the average value in empty vector (EV) control leaves (set as 1). EV represents grapevine leaves treated with bacterial suspension carrying empty vector (CaMV35S-GFP). The GAPDH gene was used as internal control. Error bars indicate the SD from three independent experiments. b Concentrations of trans-resveratrol and trans-piceid in VqERF114 overexpressed grapevine leaves. The stilbene level, expressed as μg g−1 dry weight, is the average of three replicates for each independent sample. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05; **P < 0.01)
VqERF114 binds to GCC-box element
Proteins of the ERF subfamily specifically bind to an AGCCGCC element, called GCC-box, present in the promoters of genes (Ohme-Takagi and Shinshi 1995). Three tandem copies of GCC-box were inserted into pC0380-GUS vector and fused with GUS reporter. pGCC-GUS was co-expressed with CaMV35S-VqERF114 in grapevine leaves using Agrobacterium-mediated transient transformation. In GUS histochemical staining, no GUS activity was observed in pMini35S-GUS, pGCC-GUS or in pMini35S-GUS + CaMV35S-VqERF114. However, obvious GUS activity was observed in leaves co-infiltrated with pGCC-GUS and CaMV35S-GUS. The same result was obtained in the GUS protein determination assay. GUS activity appeared higher when pGCC-GUS co-expressed with CaMV35S-VqERF114 than in the control (Fig. 3a). Three tandem copies of GCC-box were inserted into pAbAi vector and transferred into Y1HGold. A yeast one-hybrid assay was carried out to confirm that VqERF114 binds specifically to the GCC-box element (Fig. 3b). It has been reported that the STS multigenic family contains 48 members and 33 members encoding complete ORFs (Vannozzi et al. 2012). Promoters of 13 full-length coding VqSTSs were amplified based on the homologous sequences, and the Plant-CARE database was used to predict the conserved cis-element motifs in each promoter. However, analysis of cis-element motifs indicates that promoters of these 13 VqSTSs had no GCC-box, but contained the MRE or MBS element, which can be recognised by MYB TFs (Fig. 3c). This led us to speculate that VqERF114 activates the expression of STSs which may occur via a transcription complex involving an additional (as yet unknown) regulator that can bind directly to the promoters of VqSTSs, such as MYB TFs.
Transcription factor VqERF114 binds directly to GCC-box element. a GUS activity assay in transiently transformed grapevine leaves. Three tandem copies of GCC-box were inserted into pC0380-GUS vector and fused with GUS reporter. pGCC-GUS was co-expressed with CaMV35S-VqERF114 using Agrobacterium-mediated transient transformation. pMini35S-GUS, pGCC-GUS and pMini35S-GUS co-expressed with CaMV35S-VqERF114 served as controls. Grapevine leaves were collected for GUS staining and protein determination after culture for 2 days. Statistical significance was determined by Student’s two-tailed t test (**P < 0.01). b Yeast one-hybrid assay of the interaction between VqERF114 and GCC-box. Three tandem copies of GCC-box were inserted into pAbAi vector. 3 × GCC-box-pAbAi vector was transferred into yeast Y1HGold strain and interaction was confirmed on an SD/-Leu + AbA medium. cCis-element motifs presented in each STS promoter are showed by different shapes. None of the promoters of VqSTSs contain the GCC-box element and 13 STS promoters contain MRE or MBS element which can be recognised by the MYB transcription factor
Identification of VqMYB35 as a VqERF114 interacting protein
It has been reported that the R2R3-MYB TFs VvMYB14 and VvMYB15 can regulate the expression of STS in grapevine (Höll et al. 2013). All MYB TFs co-expressed with STS according to our previous study were amplified and cloned into pGADT7 vector. The yeast two-hybrid system was carried out to confirm the potential interactions between VqERF114 and VqMYBs co-expressed with STS (data not shown). Among all VqMYBs, one MYB was found to interact with VqERF114, showing 98% amino acid identity with VvMYB35 (Gene ID: VIT_14s0066g02180). This was designated VqMYB35. However, VqERF114 could not interact with VqMYB14 or VqMYB15 (Fig. 4a). A BiFC assay was carried out using a tobacco protoplast transient expression system to further confirm the interaction between VqERF114 and VqMYB35. Fluorescence was observed in the nuclei only when pSPYNE-VqERF114 was co-expressed with pSPYCE-VqMYB35, while negative controls produced no detectable fluorescence (Fig. 4b). Overexpression of VqERF114 could enhance the transcription of VqMYB35 as it has been proven via qRT-PCR analysis (Fig. 4c). These results suggest that VqERF114 interacts with VqMYB35 in planta and positively regulates the transcription of VqMYB35.
VqERF114 interacts with the R2R3-MYB transcription factor VqMYB35 and positively regulates its expression. a Interaction of VqERF114 with VqMYB35 in yeast two-hybrid assay. pGADT7-VqMYB35 and pGBKT7-VqERF114 plasmids were co-transferred into yeast Y2H Gold cells. The empty pGADT7 vector co-transformed with pGBKT7-VqERF114 was used as control. Transformed cells were spread onto the SD/-Ade/-His/-Leu/-Trp + X-α-Gal + AbA medium for the confirmation assay. b BiFC assays were carried out to confirm the interaction between VqERF114 and VqMYB35 in tobacco protoplasts. Plasmids carrying pSPYNE-VqERF114 and pSPYCE-VqMYB35 were transferred into protoplasts with the combinations indicated. Bars 10 μm. c Overexpression of VqERF114 increased the transcript of VqMYB35. The expression of VqMYB35 was calculated using qRT-PCR. The GAPDH gene was used as internal control. Error bars indicate the SD from three independent experiments. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05)
VqMYB35 activates the expressions of VqSTSs by directly targeting their promoters
Analysis of cis-element motifs indicates that there were 13 promoters of VqSTSs containing MRE or MBS elements, which can be recognised by MYB transcription factor (Fig. 3c). Dual-luciferase assays were carried out in tobacco leaves to detect the effects of VqMYB35 on the promoter expression of 13 VqSTSs. VqSTSs promoters were inserted into the pGreenII 0800-LUC vector and co-infiltrated into tobacco leaves with VqMYB35 (Fig. 5a). As illustrated in Fig. 5b, VqMYB35 showed a significant activation effect on the VqSTS15, VqSTS28, VqSTS42 and VqSTS46 promoters, while the remaining members showed very limited effects. In qRT-PCR analyses, the transcripts of VqMYB35, VqERF114, VqSTSs, VqSTS15, VqSTS28 and VqSTS42/46 were increased in VqMYB35-overexpressing grapevine leaves compared with the EV control (Fig. 5c). This confirms that VqMYB35 positively regulates the transcription of VqERF114. Subcellular localisation analysis revealed that VqMYB35 is also localised in the nuclei (Fig. 5d). Analysis of conserved cis-element motifs demonstrates that these four promoters contain single MBS element (Fig. 6a). The yeast one-hybrid assay confirmed that VqMYB35 can bind directly to the VqSTS15, VqSTS28, VqSTS42 and VqSTS46 promoters (Fig. 6b). However, the binding activity disappeared when the MBS core sequence CAGTTA was mutated to AAAAAA (mMBS) (Fig. 6c). These results illustrate that the MBS element in the VqSTS15, VqSTS28, VqSTS42 and VqSTS46 promoters can be recognised by VqMYB35. This indicates that VqMYB35 activates the expressions of VqSTSs by directly targeting their promoters via the MBS element.
VqMYB35 is localised in the nucleus and activates the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46. a Diagrams of the reporter and effector vectors used in dual-luciferase assays. b VqMYB35 activates the promoters of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 in dual-luciferase assays. The LUC/REN value of the empty vector 62-SK on each promoter was set as 1, as a calibrator. Each value represents the mean ± SD of three independent experiments, with three replicates in each experiment. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05). c Overexpression of VqMYB35 promotes the expressions of VqERF114, VqSTSs, VqSTS15, VqSTS28 and VqSTS42/46. The expression was calculated relative to the average value in empty vector (EV) control leaves (set as 1). EV represents grapevine leaves treated with bacterial suspension carrying empty vector (CaMV35S-GFP). The GAPDH gene was used as internal control. Error bars indicate the SD from three independent experiments. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05; **P < 0.01). d Subcellular localisation of VqMYB35 in tobacco leaves. The fusion protein (VqMYB35-GFP) and GFP control were transiently expressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens strain GV3101. GFP fluorescence was observed with a confocal microscope. Bars 50 μm
VqMYB35 directly targets the promoters of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by binding to the MBS element. a Schematic representation of promoter structure of four STS genes. Black diamonds indicate MBS element (CAGTTA) and white diamonds indicate mutant MBS element (AAAAAA). b Yeast one-hybrid assay of the interaction between VqMYB35 and four STS promoters. c Yeast one-hybrid assay of the interaction between VqMYB35 and four mutant STS promoters. The minimal inhibitory concentrations of Aureobasidin A (AbA) for the bait strains were tested on SD/-Ura medium in the presence of AbA and interaction was confirmed on SD/-Leu medium in the presence of AbA
VqERF114 is an indirect regulator of VqSTSs by cooperating with VqMYB35
When the combined effects of VqERF114 and VqMYB35 were analysed in the dual-luciferase assays, at least a synergistic 3.5-fold activation of the VqSTS15, VqSTS28, VqSTS42 and VqSTS46 promoters was observed, compared with the activation by either VqERF114 or VqMYB35, which alone promoted trans-activation at most 1.3- and 2-fold, respectively (Fig. 7a). Additionally, transient overexpression of both genes (VqERF114/VqMYB35) in grapevine leaves resulted in higher expressions of VqSTSs, VqSTS15, VqSTS28 and VqSTS42/46 transcripts (Fig. 7b) and higher stilbene synthesis (Fig. 7c) than overexpression of either VqERF114 or VqMYB35. These results indicate that the VqERF114–VqMYB35 transcription complex regulates stilbene synthesis by activating the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46.
Combined effects of VqERF114/VqMYB35 on the promoter activation of VqSTSs, the STS expression and stilbene synthesis. a VqERF114 co-expressed with VqMYB35 in dual-luciferase assays shows more significant activation of four STS promoters. The LUC/REN value of the empty vector 62-SK on each promoter was set as 1, as a calibrator. Each value represents the mean ± SD of three independent experiments, with three replicates in each experiment. b qRT-PCR analysis of combined effects of VqERF114/VqMYB35 on the STS expression. The GAPDH gene was used as internal control. Error bars indicate the SD from three independent experiments. c Combined effects of VqERF114/VqMYB35 on the synthesis of trans-resveratrol and trans-piceid. The stilbene level, expressed as μg g−1 dry weight, was the average of three replicates for each independent sample. Statistical significance was determined by Student’s two-tailed t test (*P < 0.05; **P < 0.01; ***P < 0.001)
Discussion
AP2/ERF TFs exist widely in the plant kingdom. These proteins play role in the regulation of primary and secondary metabolism, responses to environmental stimuli and in the processes of growth and development. The participation of ERF TFs in the control of secondary metabolism derives mostly from plants producing metabolites for pharmaceutical application (Licausi et al. 2013). For example, ORCA2 and ORCA3 in Catharanthus roseus regulate the expression of strictosidine synthase, and participate in the synthesis of terpenoid indole alkaloids (Van Der Fits and Memelink 2001). In tobacco, the AP2/ERF factor ORC1 is characterised as regulating the nicotine synthesis (De Boer et al. 2011). The expression of artemisin synthetic genes is controlled by AaERF1 and AaERF2 in Artemisia annua (Yu et al. 2012). Moreover, TcERF12 and TcERF15 regulate taxol synthesis in Taxus chinensis, by acting as repressor and activator, respectively, of the tasy gene (Zhang et al. 2015). In Newhall sweet orange (Citrus sinensis Osbeck), CitERF71 can regulate the synthesis of E-geraniol by binding directly with the CitTPS16 promoter (Li et al. 2017). The production of phenolic acids is positively regulated by SmERF115 in Salvia miltiorrhiza (Sun et al. 2019), while MdERF1B controls anthocyanin and proanthocyanidin synthesis in apple (Zhang et al. 2018a). In grapevine, resveratrol is a major secondary metabolite that plays important role in pathogen resistance. To identify new regulators of secondary metabolism in grapevine, multi-omics studies incorporating systems biological approaches have been carried out by Wong and Matus (2017). The phenylpropanoid regulatory network in the grape berry was generated, and positive co-expression correlations between AP2/ERF TFs and STS genes were demonstrated (Wong and Matus 2017). Furthermore, VvERF114, the homologue of VqERF114 in V. vinifera, is identified as part of the grapevine STS regulatory network (Wong et al. 2016). Vannozzi et al. (2018) integrated two subnetworks built from RNA-Seq data and microarray, and obtained the gene co-expression network (GCN) of grapevine TFs and STS. In this GCN, VvERF114 was also highly represented in the microarray interaction module (Vannozzi et al. 2018). As shown in Fig. 2, VqERF114 positively co-expressed with STS and facilitated the accumulation of stilbene. Our own experimental results further confirm the GCN of grapevine STS and VqERF114. Proteins of the ERF subfamily specifically bind to the GCC-box element, which is often found in the promoters of genes (Ohme-Takagi and Shinshi 1995). However, none of the promoters of VqSTSs contain the GCC-box element (Fig. 3c). This result led us to speculate that VqERF114 may regulate the expression of VqSTSs in an indirect manner, rather than directly binding to the promoters of VqSTSs.
Recently, an increasing number of TFs have been identified as being involved in the regulation of STS expression in grapevine. Members of R2R3-MYB TFs, such as VvMYB13, VvMYB14 and VvMYB15, have been characterised as regulators of stilbene synthesis (Höll et al. 2013; Fang et al. 2014; Wong et al. 2016). Besides MYB TFs, some WRKY TFs have been identified as involved in control of the stilbene synthetic pathway. For example, VvWRKY24 seems to be the sole activator of the VvSTS29 promoter and VvWRKY3 functions through a combined effect with VvMYB14 (Vannozzi et al. 2018). Moreover, VvWRKY8 represses the expressions of VvSTS15 and VvSTS21 through direct interaction with VvMYB14 (Jiang et al. 2018). With the increasing numbers of TFs identified as regulators of STS genes, the regulatory network of stilbene synthesis in the grapevine is becoming increasingly clear. Here, VqMYB35 is demonstrated to activate the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by directly targeting the MBS element in their promoters (Figs. 5, 6). In Arabidopsis, AtMYB35 (TDF1) is identified as playing a key role in anther development and in tapetal function for microspore maturation (Dubos et al. 2010; Zhu et al. 2010). Meanwhile, AoMYB35 is a candidate for the sex-determining genes in Asparagus officinalis (Tsugama et al. 2017). Our study discovers a novel function for VqMYB35 in regulating the expression of STS genes in grapevine.
Transcriptional regulation by TFs is a complex process and most plant TFs play a role in a combined manner. The TFs can bind to DNA and regulate the expression of the corresponding gene or cooperate by protein–protein interactions by forming protein complexes (Bemer et al. 2017). A meaningful number of interfamily TF interactions have recently been recognised. These help us to explore specific and dynamic gene expression. Cross-family TF interactions between AP2/ERF and MYB have been identified by employing the publicly available BioGRID database (Bemer et al. 2017). In Chinese wild V. quinquangularis accession ‘Danfeng-2’, the interaction between VqERF114 and VqMYB35 was confirmed using the yeast two-hybrid assay and the BiFC assay (Fig. 4). Moreover, VqERF114 and VqMYB35 can positively regulate the transcription of one another (Figs. 4c, 5c). Recent research has provided evidence for interactions between ERF and MYB TFs in regulating secondary metabolism. In pear, PyERF3 is reported to interact with PyMYB114 to regulate fruit anthocyanin synthesis (Yao et al. 2017), while Pp4ERF24 and Pp12ERF96 can interact with PpMYB114 to control fruit coloration (Ni et al. 2019). Besides fruit colour, the ERF-MYB transcription complex can also affect fruit fragrance by controlling the synthesis of volatile compounds. For example, FaERF9 and FaMYB98 can form transcription complex to activate the expression of quinone oxidoreductase and enhance furaneol synthesis in strawberry (Zhang et al. 2018b). It has been demonstrated that VqERF114 can interact with VqMYB35 (Fig. 4a, b) and VqMYB35 can activate the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by directly binding to the MBS element in their promoters (Figs. 5, 6). These findings allow us to speculate a regulatory mechanism in which VqERF114 regulates STS expression and resveratrol production as part of a complex with VqMYB35. As shown in Fig. 7, co-overexpression of VqERF114 and VqMYB35 resulted in much higher accumulations of STS transcripts and stilbene. These results provide evidence that VqERF114 controls the expression of STS indirectly by interacting with VqMYB35.
All the results presented here indicate that VqERF114 participates indirectly in stilbene synthesis by interacting with VqMYB35, and VqMYB35 then enhances the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by targeting their promoters. These results confirm this, previously uncharacterised, regulatory model of STS expression which is regulated by ERF transcription factor (Fig. 8). This work offers new ideas regarding the complex regulatory network controlling the synthesis of stilbene.
Hypothetical model for resveratrol synthesis regulated by VqERF114/VqMYB35 transcription complex. VqERF114 interacts with VqMYB35 and activates VqMYB35 transcription. Meanwhile, VqMYB35 increases the expression of VqERF114. VqMYB35 also promotes the expressions of VqSTS15, VqSTS28, VqSTS42 and VqSTS46 by binding to the MBS element in their promoters and increases resveratrol synthesis. Hence, VqERF114 indirectly regulates STS expression and resveratrol synthesis by interacting with VqMYB35. Co-overexpression of VqERF114 and VqMYB35 results in much higher accumulations of STS transcripts and stilbene
References
Bavaresco L, Fregoni M, Trevisan M, Mattivi F, Vrhovsek U, Falchetti R (2002) The occurrence of the stilbene piceatannol in grapes. Vitis 41:133–136
Bemer M, van Dijk AD, Immink RG, Angenent GC (2017) Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci 22:66–80
Chen JY, Wen PF, Kong WF, Pan QH, Zhan JC, Li JM, Wan SB, Huang WD (2006) Effect of salicylic acid on phenylpropanoids and phenylalanine ammonia-lyase in harvested grape berries. Postharvest Biol Technol 40:64–72
Chong J, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177:143–155
De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A (2011) APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J 66:1053–1065
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Fang L, Hou Y, Wang L, Xin H, Wang N, Li S (2014) Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep 33:1629–1640
Gatto P, Vrhovsek U, Muth J, Segala C, Romualdi C, Fontana P, Pruefer D, Stefanini M, Moser C, Mattivi F (2008) Ripening and genotype control stilbene accumulation in healthy grapes. J Agric Food Chem 56:11773–11785
Hall D, De Luca V (2010) Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J 49:579–591
Hart JH (1981) Role of phytostilbenes in decay and disease resistance. Annu Rev Phytopathol 19:437–458
Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25:4135–4149
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM (1997) Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275:218–220
Jennifer B, Takao Y, Hiroshi A, Lean MEJ, Alan C (2002) Plant foods and herbal sources of resveratrol. J Agric Food Chem 50:3337–3340
Jiang J, Xi H, Dai Z, Lecourieux F, Yuan L, Liu X, Patra B, Wei Y, Li S, Wang L (2018) VvWRKY8 represses stilbene synthase gene through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J Exp Bot 70:715–729
Jofuku KD, Den Boer B, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27:470–478
Kelemen Z, Sebastian A, Xu W, Grain D, Salsac F, Avon A, Berger N, Tran J, Dubreucq B, Lurin C (2015) Analysis of the DNA-binding activities of the Arabidopsis R2R3-MYB transcription factor family by one-hybrid experiments in yeast. PLoS One 10:e0141044
Langcake P (1981) Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, ε-viniferin, α-viniferin and pterostilbene. Physiol Plant Pathol 18:213–226
Langcake P, Pryce R (1976) The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol Plant Pathol 9:77–86
Li X, Xu Y, Shen S, Yin X, Klee H, Zhang B, Chen K (2017) Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J Exp Bot 68:4929–4938
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199:639–649
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Ni J, Bai S, Zhao Y, Qian M, Tao R, Yin L, Gao L, Teng Y (2019) Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol Biol 99:67–78
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci 94:7076–7081
Orallo F (2006) Comparative studies of the antioxidant effects of cis- and trans-resveratrol. Curr Med Chem 13:87–98
Prouse MB, Campbell MM (2012) The interaction between MYB proteins and their target DNA binding sites. BBA Gene Regul Mech 1819:67–77
Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz AJ (2010) More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14:273–284
Romero-Pérez AI, Lamuela-Raventós RM, Andrés-Lacueva C, Torre-Boronat MC, La De (2001) Method for the quantitative extraction of resveratrol and piceid isomers in grape berry skins. Effect of powdery mildew on the stilbene content. J Agric Food Chem 49:210–215
Rosemann D, Heller W, Sandermann H (1991) Biochemical plant responses to ozone: II. Induction of stilbene biosynthesis in Scots Pine (Pinus sylvestris L.) seedlings. Plant Physiol 97:1280–1286
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Shi J, He M, Cao J, Wang H, Ding J, Jiao Y, Li R, He J, Wang D, Wang Y (2014) The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiol Biochem 74:24–32
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Sun M, Shi M, Wang Y, Huang Q, Yuan T, Wang Q, Wang C, Zhou W, Kai G (2019) The AP2/ERF transcription factor SmERF115 positively regulates the biosynthesis of phenolic acids in Salvia miltiorrhiza. J Exp Bot 70:243–254
Teguo PW, Decendit A, Vercauteren JG, Merillon JM (1996) Trans-resveratrol-3-O-β-glucoside (piceid) in cell suspension cultures of Vitis Vinifera. Phytochemistry 42:1591–1593
Tsugama D, Matsuyama K, Ide M, Hayashi M, Fujino K, Masuda K (2017) A putative MYB35 ortholog is a candidate for the sex-determining genes in Asparagus officinalis. Sci Rep 7:41497. https://doi.org/10.1038/srep41497
Van Der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J 25:43–53
Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M (2012) Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12:130. https://doi.org/10.1186/1471-2229-12-130
Vannozzi A, Wong DCJ, Höll J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M (2018) Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol 59:1043–1059
Vastano BC, Chen Y, Zhu N, Ho C-T, Zhou Z, Rosen RT (2000) Isolation and identification of stilbenes in two varieties of Polygonum cuspidatum. J Agric Food Chem 48:253–256
Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2:e1326
Wan Y, Schwaninger H, He P, Wang Y (2007) Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 46:132–136
Waterhouse AL, Lamuela-Raventós RM (1994) The occurrence of piceid, a stilbene glucoside, in grape berries. Phytochemistry 37:571–573
Wessler SR (2005) Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci 10:54–56
Wong DCJ, Matus JT (2017) Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: recent applications and future opportunities. Front Plant Sci 8:505. https://doi.org/10.3389/fpls.2017.00505
Wong DCJ, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT (2016) A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res 23:451–466
Xie X, Wang Y (2016) VqDUF642, a gene isolated from the Chinese grape Vitis quinquangularis, is involved in berry development and pathogen resistance. Planta 244:1075–1094
Xu W, Yu Y, Ding J, Hua Z, Wang Y (2010) Characterization of a novel stilbene synthase promoter involved in pathogen-and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 231:475–487
Xu W, Yu Y, Zhou Q, Ding J, Dai L, Xie X, Xu Y, Zhang C, Wang Y (2011) Expression pattern, genomic structure, and promoter analysis of the gene encoding stilbene synthase from Chinese wild Vitis pseudoreticulata. J Exp Bot 62:2745–2761
Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, Wu J (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J Cell Mol Biol 92:437–451
Yu CKY, Springob K, Schmidt J, Nicholson RL, Chu IK, Yip WK, Lo C (2005) A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol 138:393–401
Yu Z, Li J, Yang C, Hu W, Wang L, Chen X (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5:353–365
Yu Y, Xu W, Wang J, Wang L, Yao W, Yang Y, Xu Y, Ma F, Du Y, Wang Y (2013) The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol 200:834–846
Zhang M, Li S, Nie L, Chen Q, Xu X, Yu L, Fu C (2015) Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol Biol 89:463–473
Zhang J, Xu H, Wang N, Jiang S, Fang H, Zhang Z, Yang G, Wang Y, Su M, Xu L, Chen X (2018a) The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol Biol 98:205–218
Zhang Y, Yin X, Xiao Y, Zhang Z, Li S, Liu X, Zhang B, Yang X, Grierson D, Jiang G, Klee HJ, Chen K (2018b) An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol 178:189–201
Zhou Q, Du Y, Cheng S, Li R, Zhang J, Wang Y (2015) Resveratrol derivatives in four tissues of six wild Chinese grapevine species. N Z J Crop Hortic Sci 43:204–213
Zhu J, Chen H, Li H, Gao J-F, Jiang H, Wang C, Guan Y-F, Yang Z-N (2010) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55:266–277
Zhuang J, Peng R-H, Cheng Z-M, Zhang J, Cai B, Zhang Z, Gao F, Zhu B, Fu X-Y, Jin X-F, Chen J-M, Qiao Y-S, Xiong A-S, Yao Q-H (2009) Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci Hortic 123:73–81
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 31672129). The research was carried out by the Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China. The authors specifically thank Dr. Alexander (Sandy) Lang from RESCRIPT Co. (New Zealand) for useful comments and language editing, which have greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
YW designed and initiated this study. LW carried out the experiments and analysed the results. LW wrote the manuscript and YW revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Qiaochun Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Wang, Y. Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep 38, 1347–1360 (2019). https://doi.org/10.1007/s00299-019-02456-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02456-4