Abstract
Key message
Lily R3-MYB transcription factors are involved in negative regulation to limit anthocyanin accumulation in lily flowers and leaves and create notable color patterns on ectopically expressed petunia flowers.
Abstract
In eudicots, both positive and negative regulators act to precisely regulate the level of anthocyanin accumulation. The R3-MYB transcription factor is among the main factors repressing anthocyanin biosynthesis. Although, in monocots, the positive regulators have been well characterized, the negative regulators have not been examined. Two R3-MYBs, LhR3MYB1 and LhR3MYB2, which were identified in lily transcriptomes, were characterized in this study to understand the regulatory mechanisms of anthocyanin biosynthesis. LhR3MYB1 and LhR3MYB2 had a C2 suppressor motif downstream of a single MYB repeat; the similar amino acid motif appears only in AtMYBL2 among the eudicot R3-MYB proteins. Stable and transient overexpression of LhR3MYB1 and LhR3MYB2 in tobacco plants showed suppression of anthocyanin biosynthesis by both; however, suppression by LhR3MYB2 was stronger than that by LhR3MYB1. In the lily plant, the LhR3MYB2 transcript was detected in leaves with light stimulus-induced anthocyanin accumulation and in pink tepals. Although LhR3MYB1 was expressed in some, but not all tepals, its expression was not linked to anthocyanin accumulation. In addition, LhR3MYB1 expression levels in the leaves remained unchanged by the light stimulus, and LhR3MYB1 transcripts predominantly accumulated in the ovaries, which did not accumulate anthocyanins. Thus, although LhR3MYB1 and LhR3MYB2 have an ability to repress anthocyanin accumulation, LhR3MYB2 is more strongly involved in the negative regulation to limit the accumulation than that by LhR3MYB1. In addition, the overexpression of LhR3MYB2 generated notable color patterns in petunia flowers; thus, the usefulness of the LhR3MYB genes for creating unique color patterns by genetic engineering is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins in many flowers and fruits are helpful in attracting pollinators and facilitating seed dispersion (Glover and Martin 2012). Anthocyanins also accumulate in vegetative organs in response to environmental stresses (Cui et al. 2014). In the vegetative organs, anthocyanins serve as a sunscreen to protect photosynthetic cells against intense light (Hughes et al. 2005; Albert et al. 2009) and may also scavenge reactive oxygen species under the conditions of photostress (Gould 2004). The anthocyanin biosynthesis pathway is well characterized and most of the genes encoding the anthocyanin biosynthesis enzymes have been isolated from many plant species (Tanaka and Ohmiya 2008). Anthocyanin biosynthesis is principally regulated at the transcriptional level of the biosynthesis genes and several transcription factors are involved in the regulation. Regulatory complexes composed of R2R3-MYB transcription factors, basic helix-loop-helix (bHLH) transcription factors, and WD40 proteins (i.e., MBW complexes) predominantly regulate the biosynthesis pathway (Lai et al. 2013; Xu et al. 2015). R2R3-MYB transcription factors are among the largest gene families specific to plants and are further classified into subgroups on the basis of the similarities and conserved motifs of amino acid sequences (Stracke et al. 2001). The subgroup 6 members of R2R3-MYB form the MBW complexes and positively regulate anthocyanin biosynthesis in many species (Lin-Wang et al. 2010; Matus et al. 2017). Single plant species often has a couple of subgroup 6 R2R3-MYB genes: Each of these genes has spatially and temporally distinct expression and causes restricted pigment deposition, resulting in color pattern formation. For example, in petunias, PhAN2 paints the entire petal region, while DEEP_PURPLE (PhDPL) and PURPLE_HAZE (PhPHZ) are responsible for the vein-associated pigmentation (venation) and light-induced pigment accumulation on the exposed petal surfaces (bud blush), respectively (Albert et al. 2011).
In eudicots, negative regulators are also involved in the anthocyanin biosynthesis to precisely modulate the pigment levels. Some R2R3-MYB transcription factors (e.g., FaMYB1 in strawberry and PhMYB27 in petunias), which form the MBW complexes with bHLH and WD40, negatively regulate the anthocyanin accumulation (Aharoni et al. 2001; Albert et al. 2014a). These R2R3-MYBs are included in the C2 repressor motif clade, which is divided into a subgroup 4 subclade and additional subclades, and share a motif called the C2 repressor motif or ethylene response factor-associated amphiphilic repression (EAR) motif (Cavallini et al. 2015).
An increasing number of R3-MYB transcription factors have been identified in plants, which possess only the R3 MYB domain. Some of R3-MYB proteins, e.g., CAPRICE and TRIPTYCHON in Arabidopsis, are involved in trichome and root hair formation (Wang and Chen 2014). Some of them, e.g., PhMYBx in petunias and AtMYBL2 in Arabidopsis, negatively regulate the anthocyanin biosynthesis (Dubos et al. 2008; Matsui et al. 2008; Albert et al. 2014a). The R3-MYB proteins maintain an amino acid sequence motif required for interaction with bHLH proteins and often disrupt the activity of the MBW protein complex by competing with the binding of R2R3-MYB to bHLH (Albert et al. 2014a; Wang and Chen 2014; Colanero et al. 2018). In addition, AtMYBL2 (R3-MYB) in Arabidopsis has a suppressor motif at the C terminal and directly represses the expression of anthocyanin biosynthesis genes (Matsui et al. 2008).
In eudicots, both positive and negative regulators are involved in anthocyanin biosynthesis and their balance determines the anthocyanin levels. For example, Atroviolacea (R3-MYB) in tomato (Colanero et al. 2018) and IlMYBL1 (R3-MYB) in Iochroma (Gates et al. 2018) prevent anthocyanin accumulation in fruits and flowers, respectively, and MlROI1 (R3-MYB) determines the color intensity of Mimulus flowers (Yuan et al. 2013). In monocots, many R2R3-MYB positive regulators have been clarified (Yamagishi et al. 2010b; Schwinn et al. 2016), but there is limited information about negative regulators; therefore, its evaluation will be necessary to understand the full regulatory mechanisms.
Lily is among the most valuable floricultural plants around the world. The genus Lilium consists of more than 90 species and is further classified into seven sections (Comber 1949). Interspecific hybridization is the principal breeding method in lily; Asiatic hybrid lilies (Lilium spp. division I, Leslie 1982) are derived from interspecific hybridization among species of the sections Sinomartagon and Daurolirion (a molecular phylogeny indicates these sections are closely related, Marasek-Ciolakowska et al. 2018). Species belonging to sections Archelirion have been used to develop Oriental hybrid lilies (Lilium spp. division VII, Leslie 1982). As lilies are a major floricultural crop, genetic evaluation of their agricultural traits is needed to improve the cultivar characteristics. However, lilies are non-model species and their genetic evaluation is not easy, because Lilium species have a huge genome (33–36 Gb, Bennett and Leitch 2011), often exhibit gametophytic self-incompatibility, and require 3–7 years between sowing and anthesis. In addition, mutant and tagged lines are yet to be developed. Stable transformation also remains a challenge.
Flower color is an important characteristic that determines the commercial value of floricultural plants; thus, a lot of attention is paid to hues, intensity, and patterns of color. Color patterns include spots, stripes, venation, bud blush, and bicolor. Lily tepals mainly accumulate anthocyanins (cyanidin 3-O-rutinoside; Nørbæk and Kondo 1999) and carotenoids (Jeknić et al. 2012; Yamagishi et al. 2010a). Tepals of many Asiatic hybrid lily cultivars contain carotenoids and/or anthocyanins and those of Oriental hybrid lily cultivars mainly accumulate anthocyanins (Yamagishi 2013). Subgroup 6 R2R3-MYBs, in association with LhbHLH2 and LhWD40a, positively regulate anthocyanin pigmentation and color pattern development: LhMYB12 usually paints the entire tepal (Yamagishi et al. 2010b, 2012; Yamagishi 2011), while LhMYB12-Lat is responsible for splatter-type anthocyanin spot development (Yamagishi et al. 2014), LrMYB15 in L. regale regulates bud blush anthocyanin pigmentation (Yamagishi 2016), and LhMYB18 is involved in the large anthocyanin spot development (Yamagishi 2018). However, there is limited information about transcription factors that negatively regulate anthocyanin biosynthesis in lilies.
Genetic engineering is among the most important techniques in the breeding of floricultural plants (Azadi et al. 2016; Noman et al. 2017). Flower color can be modified by genetic engineering; e.g., flower color hues are altered by overexpression of flavonoid 3′,5′-hydroxylase genes and/or other genes encoding anthocyanin-modifying enzymes (Katsumoto et al. 2007; Hsu et al. 2017; Noda et al. 2017). As transcription factors regulate temporally and spatially restricted anthocyanin biosynthesis positively or negatively, the introduction of transcription factors should change the space and timing of anthocyanin accumulation. This should likely result in the creation of varying color intensities and patterns. However, only the genes encoding enzymes have been mostly used to date.
We previously sequenced the tepal transcriptomes of Asiatic hybrid lily ‘Lollypop’ and Oriental hybrid lily ‘Dizzy’, and identified LhR3MYB1, LhR3MYB2, and LhCAPRICE sequences in ‘Lollypop’ (Suzuki et al. 2016), and LhR3MYB2 sequence in ‘Dizzy’ (Yamagishi et al. 2018). Although the expression of LhR3MYB1 and LhR3MYB2 is examined in flowers (Suzuki et al. 2016; Yamagishi et al. 2018), their functions are not fully evaluated. In this study, stable and transient transformation of LhR3MYB1 and LhR3MYB2 in tobacco and petunia reveals their negative regulation of anthocyanin biosynthesis. Their expression profiles indicate their roles in lilies. In addition, the usefulness of R3-MYB genes to create unique color patterns by genetic engineering is discussed.
Materials and methods
Plant materials
The Asiatic hybrid lily (Lilium spp.) cultivars ‘Montreux’, ‘Centerfold’, ‘Grand Cru’, ‘Toronto’, ‘Connecticut King’, ‘Mirella’, ‘Navona’, ‘Gran Paradiso’, ‘Saija’, and ‘Blackout’, and the Oriental hybrid lily cultivars ‘Casablanca’ and ‘Sorbonne’ were grown in a greenhouse (unheated and natural photoperiod) at the experimental farm of Hokkaido University, Sapporo, Japan. Floral organs were collected 1 d before anthesis.
To analyze the effects of light exposure on the expression of the LhMYB6, R3-MYB, and anthocyanin biosynthesis genes, as well as on anthocyanin accumulation in the leaves, bulbs of Asiatic hybrid lily ‘Toronto’ were planted and grown in the dark for 15 days and then further grown at 23 °C in a 16 h light − 8 h dark photoperiod (under 35 µmol m−2 s−1 white light from fluorescent lamps). Leaves of light-treated lily were collected at 0, 12, 24, and 36 h after the start of light exposure.
Tobacco (Nicotiana tabacum ‘Xanthi NC’) and petunia (Petunia × hybrida ‘V26’) wild plants and their transformed plants were grown in a growth chamber at 24 °C under a 16 h light − 8 h dark photoperiod. Genotype of petunia ‘V26’ was AN2, an4, DPL, PHZ, AN1, and AN11 (Albert et al. 2011).
RNA isolation and cDNA synthesis
Total RNA was extracted from each organ (approximately 100 mg fresh weight) using NucleoSpin® RNA kit (MACHEREY–NAGEL GmbH & Co. KG, Düren, Germany). cDNA was synthesized using the PrimeScript® II 1st strand cDNA Synthesis Kit (Takara, Otsu, Japan) and poly (T)-adapter primer for sequencing, or the ReverTraAce® qPCR RT Master mix with gDNA Remover (Toyobo, Tokyo, Japan) for quantitative reverse transcription (qRT)-PCR analysis.
Gene sequencing
The unigene sequence of ‘Dizzy’ R3MYB2 (hereinafter, LhR3MYB2-Dizzy) contained a full open-reading frame, but unigene sequences of R3MYB1 and R3MYB2 in ‘Lollypop’ (hereinafter, LhR3MYB1-Lolly and LhR3MYB2-Lolly, respectively) lacked 3′ termini. Thus, 3′ rapid amplification of cDNA end PCR (RACE-PCR) was carried out for LhR3MYB1-Lolly and LhR3MYB2-Lolly using two sets of primers. To design the gene-specific primers (GSP) for qRT-PCR, the partial fragments of ‘Toronto’ LhMYB6 were amplified using primers designed from the LhMYB6 sequence in Asiatic hybrid lily ‘Montreux’ (Yamagishi et al. 2010b). All PCR primers used in this study are shown in the Supplementary Table S1. The PCR-amplified fragments were inserted in pGEM T-Easy vector (Promega, Tokyo, Japan) and sequenced. The amino acid sequences were aligned using the default parameters in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). A neighbor-joining tree construction and bootstrap analysis (1000 replicates) were performed using the default parameters in CLUSTAL X version 2.1 (Larkin et al. 2007).
DDBJ accession numbers of these sequenced genes are LC429593 (LhR3MYB1-Lolly), LC429594 (LhR3MYB2-Lolly), LC429595 (LhR3MYB2-Dizzy), LC429596 (LhMYB6-1 in ‘Toronto’), and LC429597 (LhMYB6-2 in ‘Toronto’).
Gene expression
The expression levels of the genes were investigated by qRT-PCR or semi-qRT-PCR. qRT-PCR was carried out using the THUNDERBIRD® SYBR® qPCR Mix (Toyobo). Signals were monitored by the CFX Connect Real-Time system (Bio-Rad, Tokyo, Japan). The amount of Lilium hybrid POLYUBIQUITIN 4 (LhUBQ4), Nicotiana tabacum UBIQUITIN (NtUBQ), Petunia × hybrida UBIQUITIN (PhUBQ), or Renilla luciferase (Rluc) reporter gene (in the agroinfiltration assay) in each sample was used to normalize the amount of each target mRNA, using the formula, 2−∆Ct, where ∆Ct = Ct[target gene]–Ct[reference gene]. More than three biological replicates (details are shown in the legend of each figure) were used to calculate the mean values and standard error. The statistical differences were analyzed by t test using Microsoft Excel or Tukey’s honestly significant difference (Tukey’s HSD) test using R version 3.3.1 (https://www.R-project.org/).
For the semi-qRT-PCR, fragments of the LhR3MYB1, LhR3MYB2, and LhACTIN genes were amplified using cDNA from lily organs and electrophoresed on a 1.5% agarose gel.
Agroinfiltration in tobacco leaves
Full-length sequences of LhR3MYB1 and LhR3MYB2 in ‘Lollypop’ were PCR amplified using primers containing XbaI or SacI restriction sites. Then, intron-containing β-glucuronidase (iGUS) reporter gene in the binary vector pIG121 was exchanged to LhR3MYB1 or LhR3MYB2 using the restriction sites. iGUS, LhR3MYB1, and LhR3MYB2 were under the control of the constitutive cauliflower mosaic virus 35S promoter (35S).
An Agrobacterium-mediated transient assay (agroinfiltration) was performed using N. tabacum leaves. Agrobacterium harboring the 35S::LhR3MYB1 or 35S::LhR3MYB2 construct was infiltrated in tobacco leaves, together with Agrobacterium containing each of the 35S::LrMYB15, 35S::LhbHLH2, and nopaline synthase-promoter::Rluc–Lilium hybrid dihydroflavonol reductase (LhDFR)-promoter::iGUS construct (hereafter named LhDFRp::iGUS construct). Plasmid construction and the methods for agroinfiltration were described by Yamagishi (2016, 2018) and Suzuki et al. (2015), respectively. Three days after infiltration, leaf segments were harvested from six tobacco plants. Expression of iGUS, Rluc, N. tabacum DFR (NtDFR), N. tabacum anthocyanidin synthase (NtANS), and NtUBQ genes was evaluated by qRT-PCR.
Stable transformation of tobacco and petunia plants
Tobacco and petunia plants were transformed with the 35S::LhR3MYB1, 35S::LhR3MYB2, or 35S::iGUS constructs by Agrobacterium tumefaciens (EHA105)-mediated leaf disc procedure (Horsch et al. 1985). Transformed plants were selected in Linsmaier and Skoog (LS) medium containing kanamycin.
To evaluate flower color phenotypes in the progeny plants, two regenerated plants of petunia (R3MYB2-2 and R3MYB2-3) were crossed with petunia V26, mainly because anthers in these regenerated plants did not dehisce. The seeds were sown in an LS solid medium containing kanamycin and the seedlings that survived were transplanted to soil for cultivation.
Anthocyanin measurement
Pigments in leaves and flowers of petunias and lilies were extracted using 5% formic acid and total anthocyanin contents were measured by spectrophotometry (Yamagishi 2016). Segments from three leaves or petals from different leaves or flowers were analyzed.
Results
R3-MYB sequences in lilies
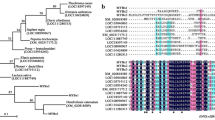
Full-length sequences of lily R3-MYB genes, which were initially found in the tepal transcriptomes, were determined by 3′ RACE PCR. Deduced amino acid sequence of LhR3MYB2-Lolly (148 amino acids) showed 73% (111/152) identity with that of LhR3MYB1-Lolly (152 amino acids), and 91% (134/148) identity with that of LhR3MYB2-Dizzy (148 amino acids). Amino acid sequence alignment of R3-MYB in lilies and other species (Fig. 1) revealed that a [D/E]Lx2[R/K]x3Lx6Lx3R motif was found within the R3 MYB repeat of the all R3-MYB proteins, required for interacting with R/B-like bHLH proteins (Zimmermann et al. 2004). Moreover, two short amino-acid motifs, C1 and C2 (or EAR), were found in some of the R3-MYB sequences, both of which are often observed in the C2 repressor motif clade R2R3-MYB sequences (Supplementary Fig. S1A). LhR3MYB1-Lolly, LhR3MYB2-Lolly, LhR3MYB2-Dizzy, and AtMYBL2 contained the C1 and C2 motifs (Matsui et al. 2008 indicate that AtMYBL2 has an EAR-like motif [DLNIGL], but this sequence is similar to the consensus sequence of the C2 motif [pdLNLD/ELxiG/S]), while, IlMYBL1, GtMYB1R1, and GtMYB1R9 included the C1 motif only and other R3-MYB sequences contained none of them.
Amino acid sequence alignment of R3-MYB transcription factors in lilies and eudicots. R3 repeat region is boxed. Letters in black background indicate the motif, (D/E)Lx2(K/R)x2Lx6Lx3R, required for binding to bHLH proteins. Residues consisting of C1, C2 (or EAR-like), and TLLLFR motifs are shown in red background. Gaps (−) are inserted to align amino acids. *Stop codon. Accession numbers of the sequences are shown in the legend of Supplementary Fig. S1. (Color figure online)
Because the amino-acid sequence lengths of the R3-MYB and R2R3-MYB proteins were very different, the phylogenetic trees of the R3-MYB and C2 repressor motif clade R2R3-MYB proteins were created using the amino acid sequences of either the whole region or the R3 repeat regions only (Supplementary Fig. S1 B and C). In both trees, LhR3MYB1-Lolly, LhR3MYB2-Lolly, and LhR3MYB2-Dizzy formed one cluster. R3-MYB sequences not containing the C1 and C2 motifs, including LhCAPRICE in Lollypop (Suzuki et al. 2016), formed one cluster that was distantly related to the lily LhR3MYB1 and LhR3MYB2 sequences. Among the C2 repressor motif clade R2R3-MYB proteins, the subclade proteins of subgroup 4 formed a single cluster, whereas the other proteins (FaMYB1, VvMYBC2-L1, and PhMYB27) exhibited relatively close relationship with the R3-MYB proteins.
Transient expression of LhR3MYB1 and LhR3MYB2 in tobacco leaves
The function of LhR3MYB1 and LhR3MYB2 in the suppression of anthocyanin biosynthesis was verified using the agroinfiltration method (Fig. 2). The lily DFR-promoter driven iGUS gene was used as a reporter gene because subgroup 6 R2R3-MYB proteins in lilies had been shown to activate the lily DFR promoter in transient assays (Lai et al. 2012; Yamagishi 2018). LrMYB15, a subgroup 6 R2R3-MYB, positively regulates anthocyanin pigmentation (Yamagishi 2016). Overexpression of both LrMYB15 and LhbHLH2 stimulated the expression of lily DFR-promoter driven iGUS gene in tobacco leaves 3 days after infiltration, while the individual overexpression of LrMYB15 only mildly activated the reporter gene expression, and that of LhbHLH2 failed to activate it (Fig. 2a). The concomitant overexpression of the LhR3MYB2 protein with LrMYB15 and bHLH2 significantly reduced the activation of the LhDFR promoter by the MBW complexes; this expression was one-fifth of that observed in LrMYB15 and LrbHLH2 (Fig. 2b). LhR3MYB1 overexpression did not alter the expression level of the reporter gene. Overexpression of both LrMYB15 and LrbHLH2 induced the expression of NtDFR and NtANS, endogenous tobacco genes involved in the flavonoid biosynthetic pathway (Fig. 2c, e; Yamagishi 2016). Concomitant overexpression of LhR3MYB1 or LhR3MYB2, with LrMYB15 and bHLH2, reduced the expression levels of NtDFR and NtANS, compared to those in LrMYB15 and LrbHLH2 overexpression, but these suppression levels were higher in LhR3MYB2 than in LhR3MYB1 (Fig. 2d, f). This experiment was repeated twice and similar results were obtained (data not shown).
Relative transcription of iGUS, NtDFR, and NtANS in tobacco leaves infiltrated with Agrobacterium harboring 35S-promoter-driven transcription factor genes. In a, c, e, Agrobacterium harboring the 35S::LrMYB15 or 35S::LhbHLH2 construct was solely or simultaneously infiltrated into tobacco leaves, together with Agrobacterium containing the LhDFRp::iGUS reporter gene construct. In b, d, f, Agrobacterium harboring 35S::LhR3MYB1 or 35S::LhR3MYB2, as well as Agrobacterium, respectively, containing the 35S::LrMYB15, 35S::LhbHLH2, and LhDFRp::iGUS constructs, was infiltrated into tobacco leaves. The expression levels of iGUS (a, b), NtDFR (c, d), and NtANS (e, f) were estimated by qRT-PCR, and Rluc or NtUBQ was used to normalize the expression of the genes. Vertical bars indicate the standard error of data from six plants. Significance was compared to ‘No effector’ (a, c, e) or ‘MYB15 plus bHLH2’ (b, d, f) by t test. * or ** indicate significance at the 5 or 1% levels, respectively
Heterologous expression of LhR3MYB1 and LhR3MYB2 in tobacco plants
Involvement of LhR3MYB1 and LhR3MYB2 in anthocyanin biosynthesis was examined using stable tobacco transformants (Fig. 3). Tobacco plants were transformed either with construct 35S::LhR3MYB1 or 35S::LhR3MYB2, and two regenerated plants with white flowers were selected. The expression levels of NtDFR and NtANS were strongly down-regulated in the petal limbs of transgenic tobacco plants compared with those in wild-type (un-transformed) plants, whereas there was no clear difference in the expression levels of NtAN2 (R2R3-MYB, Pattanaik et al. 2010) between the transgenic and wild-type plants (Supplementary Fig. S2). These results indicate that LhR3MYB1 and LhR3MYB2 suppress the expression of anthocyanin biosynthesis genes without suppressing the expression of the endogenous regulatory gene, resulting in the formation of white flowers. Morphological changes other than the flower color, including hair formation, were not observed in these transformants.
Appearance and gene expression in flowers of tobacco plants transformed with 35S-driven LhR3MYB1 or LhR3MYB2 genes. The expression levels of transgenes (LhR3MYB1 and LhR3MYB2) and endogenous tobacco genes (NtDFR and NtANS) were investigated by qRT-PCR using cDNA from petal limbs collected at anthesis. NtUBQ was used to normalize the expression of the genes. WT: wild-type (un-transformed) tobacco plants. Vertical bars represent the standard error of the means of three flowers. The same letters above the columns indicate that the values are not statistically significant (p < 0.05) as per Tukey’s HSD test. Yellow bar 1 cm. (Color figure online)
Heterologous expression of LhR3MYB1 and LhR3MYB2 in petunia plants
The effects of LhR3MYB1 and LhR3MYB2 on anthocyanin biosynthesis were further confirmed using petunia plants. Petunia V26 plants were transformed with 35S::LhR3MYB1, 35S::LhR3MYB2, or 35S::iGUS constructs. Two of the three regenerated plants (R3MYB2-2 and R3MYB2-3) transformed with the 35S::LhR3MYB2 construct had pale flower colors (Fig. 4). The petal limbs of the R3MYB2-2 plant had many fine lines in a white background; blue pigment accumulated along the petal midribs and, between the midribs, thin blue lines appeared along the vascular bundles. Color intensities and patterns varied among flowers of a single transgenic plant of R3MYB2-2; some flowers developed more blue lines between midribs and some flowers had fewer lines. They also varied within a single flower, in which distinct regions of the limb were distinguished with different color intensities and/or irregular distribution of color pattern. In the R3MYB2-3 transgenic plant, blue thick lines along the midribs and fine lines in the vascular bundles developed in a blue background of limbs. Variations between flowers and within a flower were relatively low compared with that observed in the R3MYB2-2 plant. In one transformant (R3MYB2-1), the flower color had no changes. Morphological changes other than the flower color, including hair formation on the calyxes and flower stalks, were not observed in these transformants.
Appearance and gene expression in petunia flowers transformed with 35S-driven LhR3MYB2 or iGUS genes. In LhR3MYB2-2 and LhR3MYB2-3 plants, three flowers borne on a single regenerated plant are shown. Anthocyanin was extracted from the petal limbs. The expression levels of transgene (LhR3MYB2) and endogenous genes in petunia (PhDFR and PhANS) were investigated by qRT-PCR using cDNA from petal limbs collected at anthesis. PhUBQ was used to normalize the expression of the genes. Vertical bars represent the standard error of the means of three (anthocyanin) or five (genes) flowers. The same letters above the columns indicate that the values are not statistically significant (p < 0.05) as per Tukey’s HSD test. Yellow bar 2 cm. (Color figure online)
In petal limbs of R3MYB2-2 and R3MYB2-3, anthocyanin content and the expression levels of PhDFR and PhANS genes were significantly reduced compared with those in the iGUS-transformants (Fig. 4), while, in R3MYB2-1 transformant, the expression level of PhDFR and PhANS genes was similar to those in the iGUS-transformants. As the expression level of the LhR3MYB2 transgene was relatively low in the R3MYB2-1 plant compared to those in R3MYB2-2 and R3MYB2-3 plants, the anthocyanin content and the biosynthesis gene expression levels were likely to be negatively correlated with the expression levels of the transgene. The expression level of the petunia PhAN2 gene did not change significantly in the transformants (Supplementary Fig. S3).
Flower color appearances were further evaluated in the progeny plants derived from the crosses of R3MYB2-2 × V26 or R3MYB2-3 × V26 (Supplementary Fig. S4). Altered color patterns, which were observed in the regenerated plants, also appeared in the crossed progeny plants. However, the color of the petal background in the plants from the R3MYB2-2 × V26 cross was darker than in the regenerated plant of R3MYB2-2, most likely because V26 was used as a parent. The intensity and patterns were not the same among the three progeny plants, but were similar among the three flowers in a single progeny plant. Thus, the variations between flowers and within single flowers observed in the regenerated plants of R3MYB2-2 and R3MYB2-3 were reduced in the progenies.
Four transgenic plants expressing LhR3MYB1 transgene were regenerated; however, their flower color appearance did not change (Supplementary Fig. S5). The anthocyanin content was similar to that in the iGUS-transformants, and the expression levels of the PhDFR and PhANS genes were not significantly different from those in the iGUS transformants.
Expression of the LhR3MYB1 and LhR3MYB2 in lily organs
Expression of LhR3MYB1 and LhR3MYB2 in lily organs was verified using semi-quantitative RT-PCR (Fig. 5a–e). Tepals of ‘Montreux’ and ‘Toronto’ were fully pigmented, while ‘Centerfold’ and ‘Grand Cru’ developed a brush mark pattern and large anthocyanin spots, respectively, at the basal tepal region (the latter two cultivars had no anthocyanin accumulation in the upper tepal region). ‘Casablanca’ is a white-tepal cultivar. Relatively, high expression of LhR3MYB1 was detected in the ovaries of ‘Montreux’, ‘Centerfold’, ‘Grand Cru’, ‘Toronto’, and ‘Casablanca’. The ovary color was green and did not accumulate anthocyanin pigments at this stage (1 d before anthesis). Weak LhR3MYB1 signals appeared in the tepals and leaves of ‘Montreux’ and the filaments of ‘Casablanca’. The LhR3MYB2 signal was detected in the tepals of ‘Montreux’ and ‘Toronto’, and only in the basal tepal region of ‘Centerfold’ and ‘Grand Cru’, but not in the white ‘Casablanca’ tepals. The major signal of LhR3MYB2 was not detected in any of the organs other than the tepals.
Expression of LhR3MYB1, LhR3MYB2, and LhACTIN genes in organs of Asiatic hybrid lily cultivars ‘Montreux’ (a), ‘Centerfold’ (b), ‘Grand Cru’ (c), ‘Toronto’ (d), an Oriental hybrid lily cultivar ‘Casablanca’ (e), and in the tepals (f) of Asiatic hybrid lily cultivars ‘Connecticut King’, ‘Mirella’, ‘Navona’, ‘Gran Paradiso’, ‘Saija’, ‘Blackout’, and an Oriental hybrid lily cultivar ‘Sorbonne’. The number of PCR cycles is shown in parentheses. Note that ‘Montreux’, ‘Gran Paradiso’, ‘Saija’, ‘Blackout’, and ‘Sorbonne’ accumulate anthocyanins in their entire tepal region. ‘Centerfold’ and ‘Grand Cru’ developed color patterns and accumulated anthocyanin at the basal region of tepals. ‘Casablanca’, ‘Connecticut King’, ‘Mirella’, and ‘Navona’ did not accumulate anthocyanin pigments
Correlation of LhR3MYB1 and LhR3MYB2 expression with tepal color was further verified using seven lily cultivars (Fig. 5f). ‘Gran Paradiso’, ‘Saija’, ‘Blackout’, and ‘Sorbonne’ accumulated anthocyanins in their tepals, but ‘Connecticut King’, ‘Mirella’, and ‘Navona’ did not. LhR3MYB2 expression was detected in ‘Gran Paradiso’, ‘Saija’, ‘Blackout’, and ‘Sorbonne’, indicating that LhR3MYB2 expresses only in the anthocyanin-pigmented tepals. While LhR3MYB1 expression was detected in ‘Connecticut King’, ‘Navona’, ‘Gran Paradiso’, ‘Saija’, and ‘Blackout’, indicating that its expression in tepals was independent of the presence or absence of anthocyanins.
Light induced expression of the LhR3MYB1 and LhR3MYB2 in leaves
When etiolated lily shoots are exposed to light, they rapidly accumulate anthocyanins (Nakatsuka et al. 2009) for protecting photosynthetic cells against light. Thus, the effects of light exposure on LhR3MYB1 and LhR3MYB2 expression were investigated using young etiolated leaves of ‘Toronto’ (Fig. 6). During 36 h of light treatment, anthocyanin content increased rapidly. The expression of LhMYB6, a subgroup 6 R2R3-MYB positively regulating anthocyanin biosynthesis in vegetative organs (Yamagishi et al. 2010b), and of anthocyanin biosynthesis genes of LhDFR and LhANS increased and peaked at 24 h after light exposure. A similar expression change occurred for LhR3MYB2; with its expression being highest at 24 h after light exposure. However, LhR3MYB1 expression remained unchanged during the 36 h of light treatment. These indicate that LhR3MYB2 is among the main factors regulating anthocyanin levels in the leaves.
Effect of light on leaf appearance, anthocyanin accumulation, and expression of LhMYB6, LhR3MYB1, LhR3MYB2, LhDFR, and LhANS genes in leaves of ‘Toronto’. Etiolated lily shoots were exposed to light, and changes in color, anthocyanin content, and gene expression in leaves were evaluated at 0, 12, 24, and 36 h after the start of light exposure. Vertical bars represent the standard error of the means of four leaves. The same letters above the columns indicate that the values are not statistically significant (p < 0.05) as per Tukey’s HSD test. White bar 1 cm
Discussion
LhR3MYB1 and LhR3MYB2 suppress anthocyanin biosynthesis
MYB transcription factors consist of a very large family in plants and are classified into four major groups according to the number of MYB repeats: 4R-MYB, 3R-MYB, R2R3-MYB, and MYB-related. The protein members of the MYB-related group possess either a single or a partial MYB repeat and are further classified into several subgroups, including R3-MYB (Dubos et al. 2010). Functions of R3-MYB proteins have been well studied in eudicots, but R3-MYBs that repress anthocyanin biosynthesis have not yet been demonstrated outside of the eudicots (Albert et al. 2014b). In the present study, we evaluated R3-MYB proteins in monocot lilies. The sequences of three R3-MYB proteins, LhR3MYB1-Lolly, LhR3MYB2-Lolly, and LhR3MYB2-Dizzy, were compared: On the basis of the same length and high identity (91%) in the amino acid sequences of LhR3MYB2-Lolly and LhR3MYB2-Dizzy, they were considered as orthologous proteins. In the stable tobacco transformants, overexpression of LhR3MYB1-Lolly and LhR3MYB2-Lolly generated flower phenotypes that were deficient in anthocyanin. LhR3MYB2 led to a pale colored anthocyanin of petunia flowers. In addition, LhR3MYB1-Lolly and LhR3MYB2-Lolly suppressed the expression of endogenous anthocyanin biosynthesis genes NtDFR and NtANS in transient and stable expression assays using tobacco plants. These results strongly support that lily R3MYB1 and R3MYB2 act as functional anthocyanin repressors. The roles similar to lily R3-MYB have been reported in AtMYBL2 (Dubos et al. 2008; Matsui et al. 2008) and AtCAPRICE (Nemie-Feyissa et al. 2014) in Arabidopsis, MlROI1 in Mimulus (Yuan et al. 2013), GtMYB1R1 and GtMYB1R9 in gentian (Nakatsuka et al. 2013), PhMYBx in petunia (Albert et al. 2014a), and IlMYBL1 in Iochroma (Gates et al. 2018).
Lily R3-MYB proteins were somewhat different from those of eudicots from a sequence point of view. LhR3MYB1 and LhR3MYB2 in lilies and the R3-MYB proteins in eudicots possess a conserved six-amino-acid motif required for interactions with R-like bHLH proteins (Zimmermann et al. 2004) in the R3 repeat and, thus, are likely to bind with the bHLH transcription factors. This motif strongly suggests that LhR3MYB1 and LhR3MYB2 proteins are capable of repressing anthocyanin gene expression by competing with R2R3-MYB proteins that positively regulate anthocyanin biosynthesis for interactions with bHLH proteins, as postulated from studies on AtCAPRICE and PhMYBx (Simon et al. 2007; Albert et al. 2014a). In the tobacco and petunia transformants, expression of the biosynthesis genes was suppressed without affecting the AN2 expression levels. These results are consistent with that the LhR3MYB1 and LhR3MYB2 proteins should compete with the binding of R2R3-MYB to bHLH. In addition, LhR3MYB1 and LhR3MYB2 had a conserved C2 motif; the similar amino acid motif (EAR-like motif) appears only in AtMYBL2 among the eudicot R3-MYB proteins. The C2 repressor motif in the C2 repressor motif clade R2R3-MYBs directly represses the expression of target genes (Jin et al. 2000; Albert et al. 2014a; Cavallini et al. 2015). It is possible that LhR3MYB1 and LhR3MYB2 actively repress anthocyanin biosynthesis. Additional studies are needed to confirm the functions of the C2 motif in lily R3-MYB, because the C2 (EAR-like) motif in AtMYBL2 is not involved in the suppression of anthocyanin biosynthesis. Instead, a TLLLFR motif in AtMYBL2 is responsible for the repressive activity (Matsui et al. 2008). The TLLLFR motif is not found in other R3-MYB proteins including in lilies.
The C1 motif was found in LhR3MYB1 and LhR3MYB2. Function of the C1 motif is unknown but it often appears in the R2R3-MYB proteins regulating benzenoid/phenylpropanoid biosynthesis (Colquhoun et al. 2011; Cavallini et al. 2015), including ODORANT1 in petunia and lily that regulates benzenoid volatile production (Verdonk et al. 2005; Yoshida et al. 2018). R3-MYBs of AtMYBL2, IlMYBL1, GtMYB1R1, and GtMYB1R9, repressing anthocyanin biosynthesis in eudicots, also possess the C1 motif.
When LhR3MYB1 and LhR3MYB2 were ectopically expressed in tobacco leaves concomitantly with LrMYB15 and LrbHLH2, these R3-MYB genes reduced the expression levels of endogenous anthocyanin genes, although their suppression was higher by LhR3MYB2 than by LhR3MYB1. In the same transient assay, LhR3MYB2 suppressed the expression level of the lily DFR promoter-driven iGUS gene but LhR3MYB1 could not. In addition, overexpression of LhR3MYB2 altered the color phenotype in petunia plants but that of LhR3MYB1 did not. Thus, it is likely that the magnitude suppression of gene expression is higher in LhR3MYB2 than in LhR3MYB1. The strength of suppression also varied among the R3-MYB proteins in eudicots: Atroviolacea in tomato (Colanero et al. 2018) and IlMYBL1 in Iochroma (Gates et al. 2018) strongly suppressed anthocyanin accumulation resulting in anthocyanin deficient fruits and flowers, respectively, although other R3-MYB proteins in eudicots cannot create anthocyaninless phenotypes.
The overexpression of LhR3MYB1 led to an anthocyanin deficient phenotype in tobacco plants but did not alter the anthocyanin color in petunia flowers, indicating that the formation of white flowers by suppressing anthocyanin biosynthesis is likely to be difficult in petunia compared to tobacco plants. Petunia V26 develops deep purple flowers that accumulate large amounts of pigments and the expression levels of the biosynthesis genes should be high. Thus, LhR3MYB1 with relatively weak suppression activity is unable to suppress the anthocyanin accumulation in petunia flowers.
Roles of R3-MYB in lily plants
Expression of LhR3MYB2 gene in lily tepals has been analysed previously. Asiatic hybrid lily ‘Lollypop’ develops bicolor tepals with a pink upper half and white lower half. The expression levels of LhR3MYB2-Lolly (shown as c24227_g2 in Suzuki et al. 2016) in ‘Lollypop’ are tenfold higher in the red-pigmented region than in the white region, and the change in expression during tepal development is similar to those of the anthocyanin biosynthesis genes (Suzuki et al. 2016). Oriental hybrid lily ‘Dizzy’ forms red stripes along the tepal midribs in white background, and the expression signal (fragments per kilobase of exon per million mapped fragments [FPKM] values) of LhR3MYB2-Dizzy is detected only in the red stripe region (Yamagishi et al. 2018). In the present study, LhR3MYB2 expressed dominantly in the anthocyanin-pigmented tepals and tepal regions. So, LhR3MYB2 is likely to provide negative regulation to limit the accumulation of anthocyanins in lily flowers, as seen with GtMYB1R1 and GtMYB1R9 in gentian (Nakatsuka et al. 2013), MlROI1 in Mimulus (Yuan et al. 2013), and PhMYBx in petunia (Albert et al. 2011). Expression of PhMYBx is under the control of the MBW complex and, thus, its expression is high when R2R3-MYB positive regulators are expressed (Albert et al. 2014a).
In many lily cultivars, sprouting shoots often accumulate anthocyanins, visible as dark red color. The anthocyanin pigments gradually disappear as the shoots grow resulting in green shoots (Nakatsuka et al. 2009). Anthocyanin in the sprouting shoots would protect photosynthetic cells against sunlight until sufficient amounts of photosynthetic pigments accumulate. The accumulation of anthocyanin for photoprotection is usually balanced against light-harvesting requirements for photosynthesis (Albert et al. 2009). In this study, etiolated ‘Toronto’ shoots were exposed to light to mimic the event occurred in the sprouting shoots, and changes related to anthocyanin biosynthesis were monitored. These shoots accumulated anthocyanins rapidly, and the changes in the expression of LhMYB6, LhDFR, LhANS, and LhR3MYB2 were well correlated with the increase in anthocyanin levels. Thus, LhR3MYB2 may be involved in fine tuning the anthocyanin levels in leaves and balancing the inductive effects of LhMYB6. A similar function in vegetative tissues has been reported in petunia PhMYBx, the expression of which increases under high light conditions (Albert et al. 2011, 2014a).
Expression profile of LhR3MYB1 was different from that of LhR3MYB2. In our previous study, the expression level of LhR3MYB1 (shown as c24227_g1 in Suzuki et al. 2016) in ‘Lollypop’ was higher in the pigmented region than in the white region, but the difference was relatively small compared to that of LhR3MYB2 (Suzuki et al. 2016). In ‘Dizzy’, LhR3MYB1 sequence was not identified in the tepal transcriptome (Yamagishi et al. 2018), suggesting that LhR3MYB1 expression level may be very low in Dizzy tepals. In this study, LhR3MYB1 expression was detected in tepals of some lily cultivars but was not correlated to the presence or absence of anthocyanins. In addition, LhR3MYB1 expression in the sprouting buds remained unchanged during the 36 h of light treatment. Thus, expression of LhR3MYB1 was not linked to the anthocyanin accumulation. Compared with LhR3MYB2, contribution of LhR3MYB1 to the negative regulation of anthocyanin biosynthesis is not high, although the color change in tobacco transformants indicates that LhR3MYB1 has the ability to suppress anthocyanin biosynthesis.
The highest expression of LhR3MYB1 was found in the ovaries of all cultivars: Expression profiles of LhR3MYB1 and LhR3MYB2 differed in the various lily organs. In eudicots, expression sites of R3-MYB proteins are diverse; AtMYBL2 expresses mainly in the vegetative organs (Dubos et al. 2008), while MlROI1, GtMYB1R1, GtMYB1R9, IlMYBL1 are expressed in the flowers (Nakatsuka et al. 2013; Yuan et al. 2013; Gates et al. 2018), and Atroviolacea in fruits (Colanero et al. 2018). As LhR3MYB1 is predominantly expressed in the ovaries (which do not accumulate anthocyanin), it is presumed that LhR3MYB1 has an unknown role in the ovaries, in addition to the suppression of anthocyanin biosynthesis. R3-MYB proteins in eudicots are often multifunctional (Wang and Chen 2014), e.g., AtCAPRICE in Arabidopsis is involved in the regulation of epidermal cell patterning and in the down-regulation of anthocyanin synthesis (Wang et al. 2008; Nemie-Feyissa et al. 2014).
The roles of the R3-MYB proteins in lily floral and vegetative tissues are summarized as follows. LhR3MYB1 and LhR3MYB2 are involved in the negative regulation to limit the production of anthocyanins once activated by endogenous or exogenous stimuli through the expression of the R2R3-MYB positive regulators. Thus, the features of R3-MYB proteins involved in the regulatory network of anthocyanin biosynthesis are similar in both monocots and eudicots and are thus likely to be widespread in higher plants.
Pattern development in petunia V26 flowers
Creating new flower color patterns is valuable for practical reasons because modified color patterns in the floral organs increase their commercial value (Nakayama 2014). In the present study, petunia V26 plants transformed with LhR3MYB2 had notable flower color phenotypes: Dark pigmentation appeared along the petal midribs and vascular bundles in white or pale blue backgrounds. The color pattern in the transformants is somewhat similar to a venation pattern in petunia but not the same; pigmented lines on vascular bundles seem clearer in the venation, while many small and slimly fragmented lines were observed in the flowers of transformed plants.
A range of distinct phenotypes and color patterns were observed among the flowers of a single transgenic line and within single flowers as well. Such a variation in flower coloration is often observed in petunia transformants (Chu et al. 2014; Cavallini et al. 2015), due to epigenetic effects such as DNA methylation of transgenes (Elomaa et al. 1995; Meyer et al. 1992). These variations gradually diminish in selfed and crossed progenies (Meyer et al. 1992; Oud et al. 1995). In this study, the level of the variation was reduced in the first crossed progeny plants.
Petunia flowers develop a variety of anthocyanin color patterns, which are mainly caused by the insertion of transposable elements (Spelt et al. 2000; Quattrocchio et al. 2006), post-transcriptional gene silencing of the chalcone synthase gene (Koseki et al. 2005; Morita et al. 2012), and spatially distinct expression of R2R3MYB positive regulators (e.g., DPL and PHZ, Albert et al. 2011). In addition, the suppression of endogenous flavonoid biosynthesis genes and the expression of heterologous flavonoid biosynthesis genes alter color intensities and patterns of petunia flowers (Meyer et al. 1992; Tsuda et al. 2004). In the present study, LhR3MYB2 overexpression created the tiny stripe color pattern in petunia flowers; to the best of our knowledge, such a phenotype has not been reported previously. As genetic engineering is one of the important methods to modify flower colors in floricultural plants (Zhang et al. 2014), LhR3MYB2 is a strong candidate for altering color intensities and creating novel floral color patterns.
Author contribution statement
MY designed research and conducted experiments. MS, KM, and MY analyzed data. MS and MY wrote the manuscript. All authors read and approved the manuscript.
References
Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28:319–332
Albert NW, Lewis DH, Zhang H, Irving LJ, Jameson PE, Davies KM (2009) Light-induced vegetative anthocyanin pigmentation in Petunia. J Exp Bot 60:2191–2202
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014a) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980
Albert NW, Davies KM, Schwinn KE (2014b) Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal Behav 9:e29526
Azadi P, Bagheri H, Nalousi AM, Nazari F, Chandler SF (2016) Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol Adv 34:1073–1090
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107:467–590
Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167:1448–1470
Chu Y, Pan J, Wu A, Cai R, Chen H (2014) Molecular cloning and functional characterization of dihydroflavonol-4-reductase gene from Calibrachoa hybrida. Sci Hortic 165:398–403
Colanero S, Perata P, Gonzali S (2018) The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front Plant Sci 9:830
Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG (2011) PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H. J Exp Bot 62:1133–1143
Comber HF (1949) A new classification of the genus Lilium. In: Lily yearbook, vol 13. The Royal Horticultural Society, London, pp 85–105
Cui L, Shan J, Shi M, Gao J, Lin H (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Dubos C, Le GJ, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul J-M, Alessandro A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Elomaa P, Helariutta Y, Kotilainen M, Teeri TH, Griesbach RJ, Seppänen P (1995) Transgene inactivation in Petunia hybrida is influenced by the properties of the foreign gene. Mol Gen Gen 248:649–656
Gates DJ, Olson BJSC, Clemente TE, Smith SD (2018) A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol 217:1346–1356
Glover BJ, Martin C (2012) Anthocyanins. Curr Biol 22:R147–R150
Gould KS (2004) Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004:314–320
Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Hsu YH, Tagami T, Matsunaga K, Okuyama M, Suzuki T, Noda N, Suzuki M, Shimura H (2017) Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus. Plant J 89:325–337
Hughes NM, Neufeld HS, Burkey KO (2005) Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol 168:575–587
Jeknić Z, Morré JT, Jeknić S, Jevremović S, Subotić A, Chen THH (2012) Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from tiger lily (Lilium lancifolium Thunb. ‘Splendens’). Plant Cell Physiol 53:1899–1912
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Noriko N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao G, Nehra NS, Lu C, Dyson BK, Tsuda S, Ashikari T, Kusumi T, Mason JG, Tanaka Y (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48:1589–1600
Koseki M, Goto K, Masuta C, Kanazawa A (2005) The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol 46:1879–1883
Lai Y, Shimoyamada Y, Nakayama M, Yamagishi M (2012) Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci 193–194:136–147
Lai Y, Li H, Yamagishi M (2013) A review of target gene specificity of flavonoid R2R3-MYB transcription factors and a discussion of factors contributing to the target gene selectivity. Front Biol 8:577–598
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X Version 2.0. Bioinformatics 23:2947–2948
Leslie AC (1982) The international lily register, 3rd edn. The Royal Horticultural Society, London
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Marasek-Ciolakowska A, Nishikawa T, Shea DJ, Keiichi Okazaki K (2018) Breeding of lilies and tulips—interspecific hybridization and genetic background. Breed Sci 68:35–52
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Matus JT, Cavallini E, Loyola R, Höll J, Finezzo L, Santo SD, Vialet S, Commisso M, Roman F, Schubert A, Alcalde JA, Bogs J, Ageorges A, Tornielli GB, Arce-Johnson P (2017) A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J 91:220–236
Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H (1992) Endogenous and environmental factors influence 35S promoter methylation of a maize Al gene construct in transgenic petunia and its colour phenotype. Mol Gen Genet 231:345–352
Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M (2012) Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J 70:739–749
Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N (2009) Light-induced expression of basic helix–loop–helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic 121:84–91
Nakatsuka T, Yamada E, Saito M, Fujita K, Nishihara M (2013) Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep 32:1925–1937
Nakayama M (2014) A research strategy to understand the mechanisms that govern flower color pattern formation. JARQ Jpn Agric Res Q 48:271–277
Nemie-Feyissa D, Olafsdottir SM, Heidari B, Lillo C (2014) Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98:34–40
Noda N, Yoshioka S, Kishimoto S, Nakayama M, Douzono M, Tanaka Y, Aida R (2017) Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci Adv 3:e1602785
Noman A, Aqeel M, Deng J, Khalid N, Sanaullah T, Shuilin H (2017) Biotechnological advancements for improving floral attributes in ornamental plants. Front Plant Sci 8:530
Nørbæk R, Kondo T (1999) Anthocyanins from flowers of Lilium (Liliaceae). Phytochem 50:1181–1184
Oud JSN, Schneiders H, Kool AJ, van Grinsven MQJM (1995) Breeding of transgenic orange Petunia hybrida varieties. Euphytica 85:403–409
Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L (2010) Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231:1061–1076
Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18:1274–1291
Schwinn KE, Ngo H, Kenel F, Brummell DA, Albert NW, McCallum JA, Pither-Joyce M, Crowhurst RN, Eady C, Davies KM (2016) The Onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front Plant Sci 7:1865
Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311:566–578
Spelt C, Quattrocchio F, Mol J, Koes R (2000) anthocyanin1 of petunia encodes a basic-helix loop helix protein that directly activates structural anthocyanin genes. Plant Cell 12:1619–1631
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Suzuki K, Tasaki K, Yamagishi M (2015) Two distinct spontaneous mutations involved in white flower development in Lilium speciosum. Mol Breed 35:193
Suzuki K, Suzuki T, Nakatsuka T, Dohra H, Yamagishi M, Matsuyama K, Matsuura H (2016) RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom 17:611
Tanaka Y, Ohmiya A (2008) Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotechnol 19:190–197
Tsuda S, Fukui Y, Nakamura N, Katsumoto Y, Yonekura-Sakakibara K, Fukuchi-Mizutani K, Ohira K, Ueyama Y, Ohkawa H, Holton TA, Kusumi T, Tanaka T (2004) Flower color modification of Petunia hybrida commercial varieties by metabolic engineering. Plant Biotech 21:377–386
Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17(5):1612–1624
Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5:133
Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen J (2008) Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol 8:81
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci 20:176–185
Yamagishi M (2011) Oriental hybrid lily Sorbonne homologue of LhMYB12 regulates anthocyanin biosyntheses in flower tepals and tepal spots. Mol Breed 28:381–389
Yamagishi M (2013) How genes paint lily flowers: regulation of colouration and pigmentation patterning. Sci Hortic 163:27–36
Yamagishi M (2016) A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol Breed 36:3
Yamagishi M (2018) Involvement of a LhMYB18 transcription factor in large anthocyanin spot formation on the flower tepals of the Asiatic hybrid lily (Lilium spp.) cultivar ‘Grand Cru’. Mol Breed 38:60
Yamagishi M, Kishimoto S, Nakayama M (2010a) Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed 129:100–107
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010b) Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol 51:463–474
Yamagishi M, Yoshida Y, Nakayama M (2012) The transcription factor LhMYB12 determines anthocyanin pigmentation in the tepals of Asiatic hybrid lilies (Lilium spp.) and regulates pigment quantity. Mol Breed 30:913–925
Yamagishi M, Toda S, Tasaki K (2014) The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol 201:1009–1020
Yamagishi M, Uchiyama H, Handa T (2018) Floral pigmentation pattern in Oriental hybrid lily (Lilium spp.) cultivar ‘Dizzy’ is caused by transcriptional regulation of anthocyanin biosynthesis genes. J Plant Physiol 228:85–91
Yoshida K, Oyama-Okubo N, Yamagishi M (2018) An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Mol Breed 38:144
Yuan Y-W, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD Jr. (2013) Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194:255–263
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Acknowledgements
This work was supported by a Grant-In-Aid for Scientific Research (No. 15H04447) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sakai, M., Yamagishi, M. & Matsuyama, K. Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep 38, 609–622 (2019). https://doi.org/10.1007/s00299-019-02391-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02391-4