Abstract
Key message
Ds insertion in rice OsPS1-F gene results in semi-dwarf plants with reduced tiller number and grain yield, while genetic complementation with OsPS1-F rescued the mutant phenotype.
Abstract
Photosynthetic electron transport is regulated in the chloroplast thylakoid membrane by multi-protein complexes. Studies about photosynthetic machinery and its subunits in crop plants are necessary, because they could be crucial for yield enhancement in the long term. Here, we report the characterization of OsPS1-F (encoding Oryza sativa PHOTOSYSTEM 1-F subunit) using a single copy Ds insertion rice mutant line. The homozygous mutant (osps1-f) showed striking difference in growth and development compared to the wild type (WT), including, reduction in plant height, tiller number, grain yield as well as pale yellow leaf coloration. Chlorophyll concentration and electron transport rate were significantly reduced in the mutant compared to the WT. OsPS1-F gene was highly expressed in rice leaves compared to other tissues at different developmental stages tested. Upon complementation of the mutant with proUBI::OsPS1-F, the observed mutant phenotypes were rescued. Our results illustrate that OsPS1-F plays an important role in regulating proper growth and development of rice plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Productivity and yield in plants are regulated by photosynthesis. Understanding the molecular basis of this regulatory mechanism would help to improve productivity and yield in crop plants such as rice. Various studies have proposed different approaches to increase yield and productivity by altering different parameters of photosynthesis [e.g., improving photosynthetic efficiency (Ambavaram et al. 2014; Zhu et al. 2010), and by genetic manipulation of photoprotection (Kromdijk et al. 2016)]. However, the key players linking antenna function (light-harvesting) of photosystems and photosynthetic electron transport are not well studied in rice.
In higher plants, light reaction of photosynthesis is driven by two photosystems, namely, PSI and PSII (Ben-Shem et al. 2003; Chitnis 1996, 2001). Electron flow across these two photosystems (PSII to PSI) leads to NADPH and ATP generation, which are subsequently utilized to reduce carbon dioxide to carbohydrate. In higher plants, the core of PSI is composed of 14 polypeptides (PSI-A to L and PSI-N to O). All of them are encoded by nuclear genes except PSI-A to C, I, and J whose genes are localized in plastid DNA. PSI-C, D, E, and H are peripheral and found in the stromal side of the thylakoid membrane. In contrast, two large subunits (PSI-A and PSI-B) along with PS1-F polypeptide and others are on the lumenal side of the thylakoid (Busch and Hippler 2011; Chi et al. 2012; Chitnis 1996; Järvi et al. 2007). Different subunits present in PSI have distinct functions, which are coordinated in an organized manner for this photosystem to work. The functions of a few subunits of PSI have been reported in Arabidopsis (Haldrup et al. 2000; Ihnatowicz et al. 2004; Jensen et al. 2000). However, such studies are lacking in rice. A few studies showed the effect of loss of chlorophyll b molecules on regulation of photosynthesis in rice (Terao and Katoh 1996; Terao et al. 1996). In addition, various agronomic traits of rice have been reported to be related to the photosynthetic machinery (Ambavaram et al. 2014; Makino 2011; Murchie et al. 2002).
Psa-F is the ortholog of PS1-F in Chlamydomonas. Using Psa-F deficient mutant of the green alga, it has been shown that Psa-F regulates the interaction between plastocyanin and PSI without affecting its photoautotrophic growth (Farah et al. 1995). However, down-regulation of PS1-F was found to affect photoautotrophic growth in the dicotyledonous plant Arabidopsis (Haldrup et al. 2000). These findings suggest differences in the function of the gene between algae and higher plants. Rice being a monocotyledonous plant, its PS1-F may exhibit functional variation. Therefore, further studies are warranted to elucidate the function of PS1-F gene in rice.
Here, we report functional characterization of OsPS1-F and its role in regulating growth and development in rice. The importance of OsPS1-F was studied using a Ds insertion mutant line (osps1-f), which has reduced plant height, tiller number, and grain yield. The mutant plants have lower chlorophyll content and electron transport rate compared to the wild type (WT). Genetic complementation with proUBI::OsPS1-F rescued the developmental defects of the mutant. Hence, our findings demonstrate that OsPS1-F plays a critical role in regulating plant growth and development.
Materials and methods
Plant materials and growth conditions and phenotype screens
The Ds insertion lines were generated in rice (Oryza sativa ssp japonica cv. Nipponbare (Kolesnik et al. 2004) and WT Nipponbare was used as control for all experiments). All the plants including WT, Ds insertion lines, and transgenic plants were grown in the greenhouse located in the National University of Singapore under natural light and temperature conditions. Phenotypic screening was done as reported in Ramamoorthy et al. (2011).
Ds copy number determination by Southern blot analysis
Genomic DNA preparation and Southern blot analyses were carried out according to Ramamoorthy et al. (2011). Around 5–6 µg of genomic DNA was digested with three different restriction enzymes (ClaI, EcoRI and HindIII) and fractioned on 1.0% agarose gel and transferred onto a Hybond-N+ membrane (GE Healthcare Life Sciences Cat. No. RPN119B) and fixed by UV-cross linking. This membrane was hybridized with digoxigenin (DIG)-labeled GUS probe in DIG Easy Hyb solution (Roche Cat. No. 11603558001) at 42 °C. Detection was carried out according to the manufacturer’s protocol using DIG Wash and Block Buffer Set and chemiluminescent substrate CDP-Star™ (Roche Cat. No. 11685627001). Hybridization signals were visualized by autoradiography.
Thermal asymmetric interlaced PCR (TAIL-PCR)
The FSTs of Ds insertion lines were obtained by TAIL-PCR (Liu and Whittier 1995) using the Ds specific primers and arbitrary degenerated (AD) primers. Ds terminal outward primers (at 5′ and 3′) were designed and AD primers were selected from Kolesnik et al. (2004). TAIL-PCR comprised of three steps, primary PCR products have nonspecific products in high amount and low level of desirable specific products. In subsequent secondary and tertiary PCR nonspecific primers got diluted and concentration specific products increased. Ds primer and AD primers ratio was (4 picomole/30 picomole) in PCR mix. Tertiary TAIL-PCR products were sequenced using Ds terminal primers and FSTs were obtained by BLAST search.
Molecular cloning of OsPS1-F and construct preparation for genetic transformation
The coding sequence of OsPS1-F was amplified by PCR using rice WT genomic DNA as template with gene or promoter-specific (for promoter cloning) primers using KAPA HiFi PCR Kit (Cat. No. KR0368). The primer sequences are listed in Online Resource 1. PCR was carried out as per manufacturer’s protocol. PCR products were purified by Promega Wizard® SV Gel and PCR Clean-Up System (Cat. No. A9282) and then cloned into pJET1.2/blunt vector provided with CloneJET PCR Cloning Kit (ThermoFisher Scientific, Cat. No. K1232). The cloned gene and promoter were confirmed by restriction digestion and Sanger sequencing. Selected clones were sub-cloned into appropriate destination vector pCAMBIA1300 Ti-derived binary vector (pCAMBIA; http://www.cambia.org.au) for Agrobacterium-mediated genetic transformation. The maize UBIQUITIN promoter-driven OsPS1-F construct was used for EX and complementation analyses. The Agrobacterium-mediated genetic transformation was carried out according to Hiei et al. (1994) and Ramamoorthy et al. (2011). OsPS1-F promoter-driven GUS gene construct was used for promoter GUS analysis. Cloning strategies and construct details are given as Online Resource 2.
RNA extraction and quantitative polymerase chain reaction (qPCR) analysis
Total RNA was isolated using the TRIZOL® Reagent (Life Technologies, Cat. No. 15596-026) from leaf samples of either young (6 weeks) or mature plants (2 months). One μg total RNA was reverse transcribed using Maxima First-Strand cDNA Synthesis Kit (ThermoFisher Scientific, Cat. No. K1671), as per the manufacturer’s protocol. The qPCR was carried out using OsPS1-F gene-specific primer pair and rice ACTIN1 primer pair was used for normalization (Online Resource 1). Three technical replicates were used for each independent qPCR reaction.
Chlorophyll content, F v/F m ratio, and electron transport rate measurements
The chlorophyll concentration was measured in the fully expanded leaves by a non-disruptive method using SPAD-502m (Konica-Minolta, Japan). About 50 readings were taken for each plant and the mean and standard error of data are presented. In addition, chlorophyll content was measured as described previously (Arnon 1949). Different leaf samples (100 mg each) were homogenized in liquid nitrogen and extracted with 1 ml of 80% (v/v) acetone. The absorbance was measured using DU®730 Life Science UV/Vis Spectrophotometer (Beckman Coulter, USA) at 645 and 663 nm. Four samples were measured for each line and three measurements were taken for each sample and the average values were used to calculate the chlorophyll a, chlorophyll b, chlorophyll a/b ratio, and total chlorophyll contents (Arnon 1949). The chlorophyll fluorescence and electron transport rates were measured by a non-disruptive method using a portable chlorophyll fluorometer PAM-210 (Walz, Germany) with the default setting of slow kinetics mode. The 2-month-old plants were shifted to Percival (Percival Scientific Inc, USA) plant growth chamber (10 h light/14 h dark, 30 °C day/25 °C night temperature and 55% of relative humidity) and dark adapted for overnight and measurements were taken at several timepoints after an hour of illumination. The F v/F m ratio and the electron transport rate were shown as mean and standard error. Relative electron transport rate was calculated using this formula ETR = Yield (ΦPSII) × PAR × 0.5 × ETR Factor (0.84) by PAM-210. Here, yield is PSII effective quantum yield at actinic light, and PAR is photosynthetically active radiation. The factor 0.5 is taking into account that about 50% of all absorbed quanta reach PSII. The ETR factor is defining that the fraction of incident light estimated to be absorbed by the sample default value is 0.84.
Amino acid sequence alignment and phylogenetic analysis of OsPS1-F
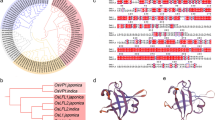
The derived amino acid sequence alignment and phylogenetic trees were constructed using selected sequences from algae, dicotyledonous, and monocotyledonous plants. Phylogeny.fr online software (http://www.phylogeny.fr/version2_cgi/simple_phylogeny.cgi) was used with one click mode, where MUSCLE, Gblocks, PhyML, and TreeDyn programs were used for multiple alignment, curation, tree building, and tree rendering, respectively (Dereeper et al. 2008).
Statistical analyses
All statistical analyses were performed by two-tailed t test using Microsoft® Office Excel 2010 and data with P value < 0.05 are shown as significant.
Results
osps1-f is a novel semi-dwarf Ds insertion mutant
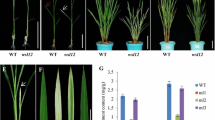
A large pool of Ds insertion lines using Ac/Ds transposable element system was generated previously (Jiang et al. 2007; Kolesnik et al. 2004). Based on a phenotypic screen, a mutant showed that light green leaves, reduced plant height, tiller number, and grain yield compared to Nipponbare WT rice was selected for further study (Fig. 1). The height difference was observed throughout the growth stages from seedling to mature plant (Fig. 1a, b). At the beginning of tillering stage, osps1-f mutant plant height was about 30 cm (60% of WT), and at mature stage, it was two-thirds of the WT (Fig. 1b, d). In addition, osps1-f had significantly lower tiller number, with only one tiller observed per plant until booting stage (Fig. 1a), but at maturity, one or two more tillers were produced (Fig. 1b, e). Panicle length, floret, and grain number were also significantly reduced in osps1-f compared to the WT (Fig. 1c, Online Resource 3).
Phenotypic characterization of osps1-f. Comparison of height between WT and osps1-f plant at (a) young and (b) mature stage (c) main panicle from WT (top) and osps1-f (bottom). Comparison of plant height (d) and tiller number (e) of WT and osps1-f (significantly different from the WT as per Student’s t test **P < 0.01, n = 10). Error bars denote standard deviation, scale bar 10 cm
Ds element was inserted into photosystem I subunit F
For further molecular analysis, Ds flanking sequence of mutant was obtained by thermal asymmetric interlaced (TAIL)-PCR. The flanking sequence analysis showed that Ds is inserted in the 5′-UTR of the gene encoding PSI-F (OsPS1-F for Oryza sativa PHOTOSYSTEM 1-F subunit, Fig. 2a), located in rice chromosome 3. The Ds insertion position was further confirmed by PCR analysis of seven different mutant siblings and WT as a control. The PCR result showed that the Ds element was present in the above-mentioned position and the mutant was homozygous in nature (Fig. 2b). A genomic Southern blot analysis showed that the selected osps1-f mutant had a single copy of Ds insertion in its genome (Fig. 2c). Quantitative PCR (qPCR) carried out using primer pair for the coding sequence of OsPS1-F with total RNA from leaves demonstrated that negligible amount of transcript levels were detected in the osps1-f mutant compared to the WT (Fig. 2d).
Molecular characterization of osps1-f. a Genomic organization of the osps1-f showing the location of Ds insertion in the 5′-UTR, 70 nucleotides upstream from the start codon ATG. Arrows show the position of the primers used for the characterization of the insertion mutant. b Zygosity determination of seven different siblings of osps1-f and WT by PCR using three primers, first one 3′ border primer (3′B) of Ds, second primer was designed before the 5′ of insertion (LP), and third was after the 3′ of insertion (RP). I and II denote amplification with primer pairs 3′B, LP and LP, RP, respectively. c Determination of Ds copy number by Southern blot hybridization using genomic DNA of osps1-f digested with three different restriction enzymes (HindIII, EcoRI and ClaI). d qPCR analysis with cDNAs from six different homozygous osps1-f seedlings and a WT seedling. Error bars denote standard deviation
Genetic complementation of osps1-f mutant and ectopic expression of OsPS1-F
To confirm that observed mutant phenotype was due to OsPS1-F disruption, osps1-f mutant was complemented with proUBI::OsPS1-F. Ten independent transgenic lines were generated and all of them showed different degrees of rescue of the mutant phenotype. We selected one line that showed full phenotype rescue (Fig. 3a) and with ~ 3:1 segregation ratio (WT-like:mutant, indicating single copy transgene insertion) in the next two generations (data not shown). Furthermore, qPCR analysis showed that this line has elevated level of OsPS1-F transcripts (Fig. 3b), which confirmed that the mutant phenotype was due to loss of function of OsPS1-F. To test whether ectopic expression (EX) of OsPS1-F will lead to increased photosynthetic rates, proUBI::OsPS1-F was introduced into the WT background. Out of 20 lines generated, three independent lines were selected (with high, medium, and low transgene expression levels) for further analysis. The EX lines generated (with elevated transcript level of OsPS1-F) did not exhibit any significant changes in phenotype compared to the untransformed WT (Fig. 3c, d).
Analyses of OsPS1-F complementation (CO) lines and ectopic expression (EX) lines. a Phenotype of 2-month-old WT, osps1-f and CO32 line, b qPCR expression analysis of OsPS1-F in WT, osps1-f and CO32 line, c Phenotypes of 2-month-old WT, and EX lines, and d qPCR expression analysis of OsPS1-F in WT and EX lines. Error bars denote standard deviation (significantly different from the WT as per Student’s t test **P < 0.01), scale bar 10 cm
OsPS1-F was strongly expressed in leaves compared to other WT tissues
OsPS1-F expression was compared at different developmental stages using young and mature leaves, young and mature panicles, young and mature roots, and 2-week-old WT seedlings. Our qPCR results showed that the expression of OsPS1-F was higher in young and mature leaves than in other tissues at different developmental stages (Fig. 4a). To further validate this result, we generated transgenic plants harboring proOsPS1-F::GUS gene construct. The GUS staining analysis of T0 plantlets revealed that OsPS1-F was highly expressed in the leaves, which correlates with its important function in photosynthesis (Fig. 4a, b).
Spatiotemporal expression of OsPS1-F in rice. a Expression pattern of OsPS1-F in various tissues (YL young leave, ML mature leave, YP young panicle, MP mature panicle, YR young root, MR mature root, SL seedlings). b Expression patterns of OsPS1-F revealed by GUS staining of proOsPS1-F::GUS seedlings (1–3) and control (C)
osps1-f mutant showed reduced chlorophyll content, F v/F m ratio, and electron transport rate
The light green coloration of leaves in mutant plants suggests reduced chlorophyll content in these plants. Indeed, chlorophyll concentration was significantly reduced (by about 35%) in fully expanded leaves of osps1-f mutant compared to WT (Fig. 5a). The complementation and EX transgenic lines showed no significant difference in chlorophyll level compared to WT by the non-disruptive method (Fig. 5a). Similarly, measurement of chlorophyll content by acetone extraction revealed that total chlorophyll and chlorophyll a/b ratios were significantly lower in the osps1-f mutant and there was no significant difference in complementation and EX lines compared to WT (Fig. 5b; Table 1). Furthermore, chlorophyll fluorescence measured using PAM210 showed that the maximum quantum yield (F v/F m) of osps1-f mutant was significantly lower (0.68) than that of the WT (0.74) (Fig. 6a) and the representative slow kinetics graphs of chlorophyll fluorescence are given in the Online Resource 4. In addition, the osps1-f mutant showed about 40% reduction in electron transport rate, whereas an increase was seen in the complementation and EX lines (P value < 0.01) compared to the WT (Fig. 6b).
Chlorophyll content and chlorophyll a/b ratio in mutant and transgenic lines. a Chlorophyll content of different rice lines. b Chlorophyll a/b ratio in different rice lines [WT, osps1-f, complementation (CO) lines and ectopic expression (EX) lines]. Error bars denote standard error (**significantly different as per Student’s t test P < 0.01)
F v/F m ratio of different rice lines. a F v/F m ratio and electron transport rate in mutant and transgenic lines. b Electron transport rate of different rice lines [WT, osps1-f, complementation (CO) lines, ectopic expression (EX) lines]. Error bars denote standard error (significantly different as per Student’s t test *P < 0.05, **P < 0.01)
Discussion
Photosynthetic machinery is highly complex, which makes it one of the important components that may be manipulated by plant biotechnology for crop improvement. It consists of two important multi-component photosystems, namely, PSI and PSII among others (Ben-Shem et al. 2003; Chitnis 1996, 2001). PS1-F encodes 1 of the 14 polypeptides forming the core of PSI. It has been reported that Psa-F polypeptide (PS1-F ortholog) is involved in the docking of plastocyanin to the PSI reaction center and hence in electron transport chain in Chlamydomonas (Farah et al. 1995). Based on the amino acid sequence analysis of PS1-F and its orthologs from algae and higher plants, it was observed that PS1-F is highly conserved among plants. At the same time, it is less conserved between algae and higher plants (Online Resource 5), which suggests that there might be functional divergence between the genes from the algae and higher plants. This opens up the possibility of functional differences among divergent groups of higher plants such as dicotyledonous versus monocotyledonous species. Such studies are facilitated by functional genomics and analysis of selected mutants. Findings from such studies can be useful for crop improvement efforts.
Insertional mutagenesis using T-DNA or transposons (Jiang et al. 2007) is an important strategy used for gene function analysis. In this study, we have investigated the role of a photosynthetic machinery subunit, OsPS1-F, using osps1-f rice mutant generated by Ds insertion. The resultant mutant showed several growth defects in plant height, tiller number, and grain yield (Fig. 1, Online Resource 3), suggesting that the gene is critical for maintaining growth and development in rice. Our findings of severely compromised OsPS1-F expression (Fig. 2d) in the osps1-f mutant combined with the fact that the phenotypic defects could be rescued by complementing the mutant with OsPS1-F (Fig. 3a, b, Online Resource 3) confirmed that the observed phenotypes are caused by the loss of function of the tagged gene. Similar results were reported earlier in Arabidopsis when reduction of PS1-F transcript levels was caused by antisense suppression (Haldrup et al. 2000). The same study reported that the Arabidopsis plants could not survive beyond the seedling stage when the gene expression level was below 3% that of WT. However, in our study, despite insignificant levels of expression of the gene in osps1-f mutant, the plants survived, albeit with severe reduction in their growth and biomass accumulation compared to the WT. There were no adverse or positive effects on plant phenotype in the EX lines, which appeared similar to the normal WT plants (Fig. 3c, d, Online Resource 3). This could be due to the fact that ectopic expression or overexpression of a subunit from multi subunit protein complex might not be adequate for altering the phenotype (Semple et al. 2008). Likewise, ectopic expression of one gene from a multi-gene family also might not produce any obvious phenotypes in rice, for example, SU(VAR)3–9 homolog (SUVH) genes SDG703, SDG704, SDG709, SDG713, SDG714, SDG715, SDG726, and SDG728 (Ding et al. 2007; Qin et al. 2010), and YAB3 (Dai et al. 2007). In some cases, such gene alterations showed no visible changes in phenotype unless induced by salt treatment (Liu et al. 2014). Similarly, it is not unreasonable to speculate that overexpressing one subunit of a specific protein complex is not enough to produce any noticeable enhancement of growth and development. However, it is important to analyze the functions of various components of such complexes or multi-gene families, which might help to reveal a combination that can lead to pronounced enhancement in plant growth in the long term.
Although OsPS1-F is not directly involved in the chlorophyll fluorescence, because there was no significant change observed when OsPS1-F was ectopically expressed, it appears to have an indirect influence on photosynthetic electron transport rate. Earlier observations in Arabidopsis indicated that PS1-F plays a major role in the later steps of photosynthesis such as electron transport (Farah et al. 1995; Haldrup et al. 2000). Likewise, the PSII electron transport rate was significantly reduced in osps1-f mutant (Fig. 6b) and which is consistent with the earlier findings in Arabidopsis and algae (Farah et al. 1995; Haldrup et al. 2000). In addition, a significant reduction in chlorophyll a/b ratio in osps1-f mutant (Fig. 5) indicates a reduced PSI/PSII ratio which is consistent with the restricted PSI electron transport in the PsaF-suppressed plants (Haldrup et al. 2000). This could be the primary reason for the reduced growth of plants observed in our study. Alternatively, the lack of PS1-F may cause a structural alteration in PSI that might be brought about due to changes in the protein complex within PSI, which will be ultimately manifested as reduced overall plant biomass. Collectively, these data suggest that disruption of OsPS1-F gene expression could affect the PSII function as well, which can explain the reduction in the overall antenna function as was also reported in Chlamydomonas (Farah et al. 1995) and higher plants (Haldrup et al. 2000). The significant increase in the PSII electron transport rate (P value < 0.05 by t test) in the EX lines that showed higher OsPS1-F expression (Figs. 3d, 6b) further supports this. In addition, this indicates that increasing the photosynthetic electron transport rate alone is not adequate to elicit growth enhancement. As OsPS1-F is a part of the multi subunit protein complex, there may also be some unknown feedback regulation to control the functions of different subunits.
In conclusion, our results suggest that OsPS1-F modulates chlorophyll content and photosynthetic electron transport rate. Along with the previous studies, these findings indicate that PS1-F is one of the major factors that affect plant growth and development, which has a profound influence on crop productivity. Further investigations with other PSI protein subunits may lead to a better understanding of the function of this protein complex.
Author contribution statement
RR and BV planned and carried out the experiments and data analysis and wrote the manuscript draft. SR provided the mutant line and contributed to manuscript writing. PK conceptualized the project and contributed to data analysis and manuscript preparation. All authors read and approved the final manuscript.
Abbreviations
- OsPS1-F:
-
Oryza sativa PHOTOSYSTEM 1-F subunit
- TAIL-PCR:
-
Thermal Asymmetric Interlaced PCR
- Ac/Ds :
-
Activator/Dissociation transposable elements
- F m :
-
Maximum yield of fluorescence
- F v :
-
Variable fluorescence
- F v/F m :
-
Maximal photochemical efficiency of PSII
References
Ambavaram MM et al (2014) Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat Commun 5:5302. https://doi.org/10.1038/ncomms6302
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol 24:1–15. https://doi.org/10.1104/pp.24.1.1
Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426:630–635. https://doi.org/10.1038/nature02200
Busch A, Hippler M (2011) The structure and function of eukaryotic photosystem I. Biochim Biophys Acta 1807:864–877. https://doi.org/10.1016/j.bbabio.2010.09.009
Chi W, Ma J, Zhang L (2012) Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos Trans R Soc Lond B Biol Sci 367:3420–3429. https://doi.org/10.1098/rstb.2012.0065
Chitnis PR (1996) Photosystem I. Plant Physiol 111:661–669. https://doi.org/10.1104/pp.111.3.661
Chitnis PR (2001) Photosystem I: function and Physiology. Annu Rev Plant Physiol Plant Mol Biol 52:593–626. https://doi.org/10.1146/annurev.arplant.52.1.593
Dai M, Hu Y, Zhao Y, Zhou DX (2007) Regulatory networks involving YABBY genes in rice shoot development. Plant Signal Behav 2:399–400. https://doi.org/10.1104/pp.107.095737
Dereeper A et al (2008) Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465-W469. https://doi.org/10.1093/nar/gkn180
Ding Y et al (2007) SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19:9–22. https://doi.org/10.1105/tpc.106.048124
Farah J, Rappaport F, Choquet Y, Joliot P, Rochaix J-D (1995) Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J 14:4976. https://doi.org/10.1002/j.1460-2075.1995.tb00180.x
Haldrup A, Simpson DJ, Scheller HV (2000) Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J Biol Chem 275:31211–31218. https://doi.org/10.1074/jbc.M002933200
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282. https://doi.org/10.1046/j.1365-313X.1994.6020271.x
Ihnatowicz A et al (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37:839–852. https://doi.org/10.1111/j.1365-313X.2004.02011.x
Järvi S, Gollan PJ, Aro E-M (2007) Understanding the roles of the thylakoid lumen in photosynthesis regulation. Current challenges in photosynthesis: from natural to artificial. Front Plant Sci 4:31. https://doi.org/10.3389/fpls.2013.00434
Jensen PE, Gilpin M, Knoetzel J, Scheller HV (2000) The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J Biol Chem 275:24701–24708. https://doi.org/10.1074/jbc.M000550200
Jiang S-Y, Bachmann D, La H, Ma Z, Venkatesh PN, Ramamoorthy R, Ramachandran S (2007) Ds insertion mutagenesis as an efficient tool to produce diverse variations for rice breeding. Plant Mol Biol 65:385–402. https://doi.org/10.1007/s11103-007-9233-0
Kolesnik T et al (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37:301–314. https://doi.org/10.1046/j.1365-313X.2003.01948.x
Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354:857–861. https://doi.org/10.1126/science.aai8878
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681. https://doi.org/10.1016/0888-7543(95)80010-J
Liu G, Li X, Jin S, Liu X, Zhu L, Nie Y, Zhang X (2014) Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 9:e86895. https://doi.org/10.1371/journal.pone.0086895
Makino A (2011) Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol 155:125–129. https://doi.org/10.1104/pp.110.165076
Murchie EH, Yang J, Hubbart S, Horton P, Peng S (2002) Are there associations between grain-filling rate and photosynthesis in the flag leaves of field-grown rice? J Exp Bot 53:2217–2224. https://doi.org/10.1093/jxb/erf064
Qin F-J, Sun Q-W, Huang L-M, Chen X-S, Zhou D-X (2010) Rice SUVH histone methyltransferase genes display specific functions in chromatin modification retrotransposon repression. Mol Plant 3:773–782. https://doi.org/10.1093/mp/ssq030
Ramamoorthy R, Jiang S-Y, Ramachandran S (2011) Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PLoS One 6:e28069. https://doi.org/10.1371/journal.pone.0028069
Semple JI, Vavouri T, Lehner B (2008) A simple principle concerning the robustness of protein complex activity to changes in gene expression. BMC Syst Biol 2:1 https://doi.org/10.1186/1752-0509-2-1
Terao T, Katoh S (1996) Antenna sizes of photosystem I and photosystem II in chlorophyll b-deficient mutants of rice. Evidence for an antenna function of photosystem II centers that are inactive in electron transport. Plant Cell Physiol 37:307–312. https://doi.org/10.1093/oxfordjournals.pcp.a028947
Terao T, Sonoike K, Yamazaki J-y, Kamimura Y, Katoh S (1996) Stoichiometries of photosystem I and photosystem II in rice mutants differently deficient in chlorophyll b. Plant Cell Physiol 37:299–306. https://doi.org/10.1093/oxfordjournals.pcp.a028946
Zhu X-G, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61:235–261. https://doi.org/10.1146/annurev-arplant-042809-112206
Acknowledgements
This work was supported by research funding from the National Research Foundation, Prime Minister’s Office, Singapore under its Competitive Research Programme (CRP award no. NRF-CRP 7-2010-02). We thank Drs. Pannaga Krishnamurthy, Pratibha Ravindran, and Jiang ShuYe for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Prakash Lakshmanan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ramamoorthy, R., Vishal, B., Ramachandran, S. et al. The OsPS1-F gene regulates growth and development in rice by modulating photosynthetic electron transport rate. Plant Cell Rep 37, 377–385 (2018). https://doi.org/10.1007/s00299-017-2235-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2235-8