Abstract
Key message
Dual function of GhATAF1 in the responses to salinity stress and Verticillium dahliae infection in cotton.
Abstract
NAC (NAM/ATAF1/2/CUC2) is a large plant-specific transcription factor family that plays important roles in the response to abiotic stresses. We previously isolated a cotton NAC transcription factor gene, GhATAF1, which was up-regulated by ABA, cold and salt stresses and classified into AFAT1/2, a sub-family of NAC. Here, we report that GhATAF1 was also highly induced by MeJA, SA and Verticillium dahliae inoculation, which implied that GhATAF1 was involved not only in the response to abiotic stress but also in the response to biotic stress. GhATAF1 was localized in the nucleus and possessed transactivation activity. Overexpression of GhATAF1 enhanced cotton plant tolerance to salt stress by enhancing the expression of various stress-related genes, including the ABA response gene GhABI4; the transporter gene GhHKT1, involved in Na+/K+ homeostasis; and several stress-response genes (GhAVP1, GhRD22, GhDREB2A, GhLEA3, and GhLEA6). Additionally, overexpressing GhATAF1 increased cotton plant susceptibility to the fungal pathogens V. dahliae and Botrytis cinerea, coupled with the suppression of JA-mediated signaling and the activation of SA-mediated signaling. Our results suggested that GhATAF1, the cotton stress-responsive NAC transcription factor, plays important roles in the response to both abiotic stress and biotic stress by coordinating the phytohormone signaling networks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses and biotic stresses adversely affect the growth and productivity of plants. To survive, plants have evolved a multitude of mechanisms for adaptation to changing environmental conditions, such as drought, high salinity, and microbial pathogen infection. Signaling pathways dependent on the phytohormones abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) mediate the early response to abiotic and biotic stresses (Fujita et al. 2006; Glazebrook 2005). In recent years, increasing evidence has demonstrated that stress-related transcriptional factors function as important components in the crosstalk between abiotic and biotic stress signaling (Abe et al. 2003; Wu et al. 2009).
NAC (Petunia NAM, Arabidopsis ATAF1/2 and CUC2) proteins are plant-specific transcription factors that contain a conserved N-terminal DNA-binding domain (NAC-domain) and a variable C-terminal transcriptional domain (Ooka et al. 2003). Recent studies have demonstrated that many NAC proteins are involved in different aspects of plant developmental processes (Wang et al. 2011) as well as in abiotic stress responses, such as drought tolerance (Jeong et al. 2010; Tran et al. 2004), salt tolerance (Kim et al. 2008), or both drought and salt tolerance (Hu et al. 2006; Zheng et al. 2009) and in low-oxygen tolerance (Christianson et al. 2009). Meanwhile, studies also found that some NAC proteins participated in the response to pathogen inoculation (Delessert et al. 2005; Jensen et al. 2008; Wang et al. 2009; Yoshii et al. 2009), but few reports indicated that NAC proteins could be involved in both abiotic and biotic stress (Nakashima et al. 2007; Wu et al. 2009).
ATAF (Arabidopsis transcription activation factor) is a sub-family of NAC proteins in plants that consists of ATAF1, ATAF2, ANAC032, and ANAC102 in Arabidopsis. Functional studies have revealed that ATAF2 acts as a negative regulator in defense against the soil-borne fungal pathogen Fusarium oxysporum by repressing the expression of pathogenesis-related proteins in Arabidopsis (Delessert et al. 2005). Like ATAF2, ATAF1 was a negative regulator in defense responses against pathogens (Wang et al. 2009). Additionally, ATAF1 was also a negative regulator in ABA signaling, salt and oxidative stresses (Wu et al. 2009). In contrast to ATAF1, rice OsNAC6 was a positive regulator in drought, high-salt tolerance and defense against blast disease (Nakashima et al. 2007). Thus, the NAC proteins are bio-resources for the improvement of crop stress tolerance by genetic manipulation (Hu et al. 2006; Puranik et al. 2012).
Cotton (Gossypium spp.) is an important economic crop globally and is used as a source of natural fiber and edible oil. However, the yield and quality of cotton is restricted by many unfavorable environmental conditions, including abiotic and biotic stresses, even though cotton is a superior crop in salt and drought tolerance. In the last decade, several candidate genes in cotton were identified to be functionally related to drought and salt stresses (Luo et al. 2013; Zhou et al. 2014), and ectopic expression of the exogenous stress-related genes in cotton improved abiotic stress tolerance (Liu et al. 2014; Mittal et al. 2014).
Among the biotic stress, Verticillium wilt in cotton is a devastating vascular disease caused by the soil-borne fungal pathogen V. dahliae. Although many G. barbadense germplasms are resistant to V. dahliae, the resistance is hardly transferred into upland cotton, which contributes approximately 95 % of the total cotton yield (Zhang et al. 2015). Previous analyses have identified sets of V. dahliae response signals, such as JA, SA, ET, and BR signaling and lignin and gossypol biosynthesis (Gao et al. 2013; Li et al. 2014; Xu et al. 2011). Although several individual genes, such as GhNDR1, GhMKK2, GbERF1, GbRLK, GbWRKY1, GbNRX1, and GhMLP28, were shown to be functionally related to Verticillium wilt resistance, only a few have been fully characterized (Gao et al. 2011; Guo et al. 2016; Jun et al. 2015; Li et al. 2014, 2016; Yang et al. 2015).
Previously, we reported the isolation of a cotton EST (DT549350) as a putative abiotic stress response gene using data-mining and expression pattern analysis (Zhu et al. 2011). In this study, we obtained the full length of GhATAF1 and demonstrated that this gene encoded a cotton NAC transcription factor and played important roles in the response to both abiotic stress and biotic stress via regulating the ABA, JA, and SA signaling networks.
Materials and methods
Plant materials, growth conditions and stress treatments
Upland cotton (Gossypium. hirsutum L. cv. YZ1) seedlings were grown in commercial sterilized soil or in Hoagland solutions under 16 h light/8 h dark conditions for 2–3 weeks. The cotton seedlings were dip-infected with suspensions of V. dahliae spores, and the roots were harvested at different time points after inoculation for RNA extraction (Xu et al. 2011). For treatments with MeJA and SA, 0.1 mM and 1 mM concentrations in Hoagland solutions were used, respectively. The roots were harvested at different time points after treatment for later RNA isolation. Four plants were harvested for each time point and treatment.
Expression analysis
Total RNAs from both control and stressed cotton tissues were isolated by a modified guanidine thiocyanate method (Zhu et al. 2005). For RT-PCR and qRT-PCR analyses, 3 µg total RNA/sample was used for cDNA biosynthesis with Superscript III reverse transcriptase (Invitrogen, San Diego, USA). The qRT-PCR reactions were performed using the 7500 Real Time PCR System (ABI, Foster City, USA) with SYBR green (Bio-Rad, USA). The GhUBQ7 (GenBank: DQ116441) gene from G. hirsutum was used as the endogenous reference gene (Tu et al. 2007). The relative transcript level was determined and normalized using the reference level and averaged over the three technical replicates. Primers for the RT-PCR and qRT-PCR (Supplemental Table 1) were designed according to the cDNA sequences using Primer Premier 5 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp).
Generation of transgenic cotton plants and Southern blot analysis
The full-length gene was amplified using the primers GhATAF1-OE-F and GhATAF1-OE-R and then cloned into pGWB417 (Shimane University) (Nakagawa et al. 2007). The vector was transformed into G. hirsutum YZ1 plants mediated by the Agrobacterium tumefaciens strain EHA105 (Jin et al. 2006).
Genomic DNA was extracted from the young leaves of wild-type (YZ1) and transgenic cotton plants using a DP305 plant genomic DNA kit (Tiangen Biotech, Beijing). Genomic DNAs were digested with HindIII, and the presence of the transgene was verified by the amplification of NPTII (Hao et al. 2012). The sequences of the NPT II forward primer and reverse primer were 5′-CGTAAAGCACGAGGAAGCG-3′ and 5′-GGCACAACAGACAATCGGC-3′, respectively.
Subcellular localization of GhATAF1 proteins
To construct the 35S::GFP-GhATAF1 vector, the ORF sequence of the GhATAF1 gene was cloned into the pGWB452 vector (the N-terminus of the green fluorescent protein gene controlled by the CaMV35S promoter) (Nakagawa et al. 2007). For transient expression, the plasmid DNA was used to transform onion epidermal cells using the particle bombardment method (Varagona et al. 1992). Then, the tissues were incubated on MS agar medium in dark conditions at 25 °C for 18 h. The subcellular localization of the GFP::GhATAF1 fusion protein in onion epidermal cells was observed using a fluorescence microscope (DM2500; Leica, Wetzlar, Germany).
Transcriptional activation activity of the GhATAF1 protein
For the transactivation assay, the full ORF and the N-terminus of GhATAF1 were obtained by PCR using the primers listed in Supplemental Table 1. The PCR products were fused in frame to the GAL4 DNA-binding domain in pDEST32 via a recombination reaction. The fusion proteins pDEST32-GhATAF1 and pDEST32-GhATAF1-ΔCT were used to transform the yeast strain AH109 with the reporter gene LEU2 according to the manufacturer’s protocols. The transformed stains were streaked on plates of SD/-Leu plus x-α-gal media.
Sensitivity analysis of transgenic cotton plants to SA and MeJA
To conduct the SA sensitivity assay, the 14-day-old cotton seedlings of wild-type and transgenic cotton plants were transferred to Hoagland solutions without or with SA (4 mM) for 6 h, after which the SA- and JA-response genes were analyzed by qRT-PCR. After treatment for 3–5 days, the wilt rate of the leaves and the survival rate of the plants were counted. To conduct the MeJA sensitivity assay, the surface-sterilized seeds of wild-type and transgenic cotton were germinated onto 1/2 MS medium and then were transferred to new 1/2 MS medium with or without MeJA (5 µM) and allowed to grow for 7 days before scoring.
Salt tolerance assays of transgenic cotton plants and measurement of the Na+ and K+ contents
For salt treatments, leaf discs (1 cm in diameter) of both transgenic and wilt-type cotton plants were incubated in solutions of various concentrations of NaCl (0, 0.4, and 0.8 M) for 3 days. The resistance of leaf discs to NaCl stress was compared by measuring the chlorophyll content using our previously described protocol (Munis et al. 2010). To further assess the effect of salinity on plant growth, the 14-day-old wild-type and transgenic cotton seedlings were transferred to Hoagland solutions containing 0.2 M NaCl. After treatment for 6 h, the stress-related genes were analyzed by RT-PCR, and after treatment for 3 days, the concentrations of Na+ and K+ were determined. The concentrations of Na+ and K+ in the shoots were determined by an atomic absorption spectrophotometer (Yang et al. 2014). The samples were dried at 80 °C for 3 days, digested in 65 % nitric acid in a MARS6 microwave (CEM) with a gradient of temperatures from 120 to 180 °C for 45 min, and diluted with deionized water. The Na+ and K+ contents of the samples were determined by inductively coupled plasma mass spectrometry (ICP-MS, Agilent 7700 series, USA).
Fungal pathogen cultivation and inoculation
A moderately aggressive Verticillium dahliae fungus, 1CD3-2, was incubated on PDA for 1 week and then was inoculated into Czapek broth on a shaker at 120 rpm at 25 °C for 3–4 days, until the concentration of spores reached ~108 spores ml−1. The suspension liquid was adjusted to 1 × 106 spores ml−1 with sterile distilled water for inoculation. The seedlings were infected with V. dahliae via root dip inoculation into a suspension of fungal spores for 1 min and were then returned to their original pots. Control plants were not inoculated but were otherwise treated and sampled with distilled water in the same way. The severity of the disease symptoms for each cotton seedling was scored using a 0–4 rating scale at 10 days post-inoculation (dpi), as described previously (Xu et al. 2012).
The Botrytis cinerea strain (stored at 4 °C) was transferred onto PDA medium, cultured for 5 days and then further incubated on fresh PDA medium for another 5 days at 25 °C. The discs of colonized agar (5 mm in diameter, made using a hole punch) were inoculated on the excised leaves of 3-week-old GhATAF1-overxpressing and wild-type plants at 25 °C and kept under a cover to maintain high humidity. Three days after inoculation, the average lesion size (diameter) was measured.
Results
GhATAF1 was induced by biotic stress, MeJA and SA treatments
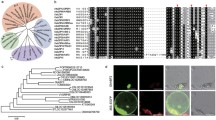
In our previous study, an EST (DT549350) was isolated as a candidate abiotic stress response gene in cotton that could be induced by ABA, salt, and cold treatments (Zhu et al. 2011). The full-length cDNA of the gene was obtained by RACE with primers designed according to the EST. The cDNA is 1236 bp in length and putatively encodes a polypeptide of 276 amino acid residues. The results of bioinformatics analysis showed that the protein, GhATAF1, was homologous to ATAF1 (AGI: At1g01720) with 59 % similarity and could be classified into the ATAF1/2 sub-family of the NAC family (Fig. 1 and Supplementary Fig. 1a). To identify the gene structure of GhATAF1 in the cotton genome, we amplified GhATAF1 from genomic DNA with gene-specific primers. According to the alignment between the cDNA and genomic DNA sequences, GhATAF1 consists of three exons and two introns (Supplementary Fig. 1b).
Alignment of GhATAF1 and its homologous NAC transcription factors in Arabidopsis. Alignment of protein sequences of GhATAF1 and other ATAF1 transcription factors from cotton and Arabidopsis. The identical amino acids are shown by stars and the conserved residues are shown by dots. The accession numbers of these known proteins in GenBank are as follows: GhNAC1: (ACI15347.1); ATAF1: (AEE27326.1); ATAF2: (AED91350.1); NAC032: (AEE35979.1); NAC102: (AED97798.1)
The results of qRT-PCR analysis indicated that GhATAF1 was dominantly expressed in cotton roots, leaves and stems, with low levels of expression in the ovules and fibers (Fig. 2a). To further investigate the involvement of GhATAF1 in disease responses, the gene expression was analyzed after treatment with the defense-related phytohormones methyl jasmonic acid (MeJA) and salicylic acid (SA) and after inoculation with V. dahliae. The expression of GhATAF1 was induced when cotton was inoculated with V. dahliae (Fig. 2b). Moreover, GhATAF1 was quickly up-regulated in cotton roots by the exogenous application of MeJA and SA, and SA activated the expression of GhATAF1 more highly and durably (Fig. 2c). These results implied that GhATAF1 was not only involved in the response to abiotic stress but also to biotic stress, possibly by regulating the phytohormone signaling network.
Expression patterns of GhATAF1 in response to phytohormones and Verticillium dahliae infection. a Expression patterns of GhATAF1 in the root, stem, leaf, petal, anther, ovule and fiber of ‘YZ1’ (G. hirsutum). b qRT-PCR analysis of the expression patterns of GhATAF1 in cotton roots after inoculation with V. dahliae. Total RNAs were extracted from roots of ‘YZ1’ seedlings at the indicated time points after inoculation. c qRT-PCR analysis of GhATAF1 in ‘YZ1’ roots treated with or without MeJA (100 µM) and SA (1 mM) at the selected time points. The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates
Transcription factors are typically localized in the cell nucleus, where they perform transcriptional regulation roles. To examine the subcellular localization of GhATAF1, a fused 35S::GFP-GhATAF1 construct was constructed and introduced into onion epidermal cells via particle bombardment. Green fluorescence was predominately found in the nucleus, which suggested that GhATAF1 was a nuclear protein (Fig. 3a).
Subcellular localization and transactivation activity of the GhATAF1 protein. a The nuclear localization of the GhATAF1 protein in onion epidermal cells. GhATAF1 fused to GFP at the N-terminus under the control of the CaMV 35S promoter was transiently expressed in onion epidermal cells and analyzed by fluorescence microscopy in bright field (Bright), dark field (GFP), and combined views (Merge). b Transactivation analysis of pDEST32-GhATAF1 and pDEST32-GhATAF1-ΔCT. The transformants were streaked on SD-Leu plus 40 mg/L x-α-gal medium
A transactivation assay in the yeast strain AH109 using the full-length sequence of GhATAF1 fused to the DNA-binding domain of GAL4 produced strong transactivation activity, whereas the transactivation activity of the N-terminal region (1–158 aa) sequence of GhATAF1 fused to the GAL4 BD was abolished (Fig. 3b). These results indicated that the predicted C-terminus had transactivation activity.
GhATAF1 expression confers better salt tolerance in transgenic cotton plants
To characterize the function of GhATAF1 in response to multiple stresses, an overexpression construct of GhATAF1, pCaMV35S:GhATAF1-4MYC, was introduced into cotton. Two transgenic lines (OE-4, OE-5) with single copy insertions were identified through Southern blotting analysis and selected for further study (Supplemental Fig. 2). No differences were observed in the seedlings’ development between wild-type and GhATAF1-overexpressing cotton lines. Following 0.2 M NaCl treatment for 72 h, wild-type seedlings showed more severe wilt compared with GhATAF1-overexpressing cotton lines (Fig. 4a). In the presence of salt stress, the K+ content did not significantly change, but the Na+ content and the Na+/K+ ratio were significantly decreased in GhATAF1 transgenic lines compared to those in the wild-type lines (Fig. 4b–d). The chlorophyll content of the leaf discs was also measured to assess the cellular damage by salt stress. The chlorophyll contents in lines OE-4 and OE-5 were significantly higher than that in the wild-type line after treatment with 0.4 or 0.8 M NaCl (Fig. 4e, f).
GhATAF1-overexpressing cotton plants showed enhanced tolerance to salinity stress. a Performance of GhATAF1-overexpressing and wild-type cotton plants under NaCl treatment. b–d K+ concentration (b), Na+ concentration (c), Na+/K+ (d) in the shoots of GhATAF1-overexpressing and wild-type plants under normal or salt treatment conditions. The data represent the mean ± SD of three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and their corresponding wild-type (WT) plants at P < 0.05, t test. e Leaf disc assay for chlorophyll bleaching after incubation in water, 0.4 and 0.8 M NaCl solutions for 72 h at 25 °C. f Chlorophyll content determined by the leaf disc senescence assay for salinity tolerance of GhATAF1-overexpressing and wild-type cotton plants in (e). The data represent the mean ± SD of three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and the corresponding wild-type (WT) plants at P < 0.05, t test. g RT-PCR analysis of stress-related gene expression levels between GhATAF1-overexpressing and wild-type cotton plants under salt stress
The expression levels of stress-related genes were examined in GhATAF1-transgenic lines and wild-type plants grown under 0.2 M NaCl treatment or unstressed conditions. Compared to wild-type, the GhABI4, GhAVP1, GhHKT1, GhRD20, GhRD22, GhDREB2A, GhLEA3, and GhLEA6 gene expression levels were enhanced in GhATAF1-transgenic lines, while the expression of GhRD26, GhSOS1, GhSOS2, and GhNHX1 was not significantly different between wild-type and GhATAF1-transgenic lines (Fig. 4 g). Taken together, we concluded that GhATAF1 confers cotton tolerance to salt stress by up-regulating stress-related genes.
Overexpression of GhATAF1 in cotton decreased resistance to fungal pathogens
Leaves from GhATAF1-overexpressing cotton lines and wild-type plants were inoculated with Botrytis cinerea, and the lesion size was measured 3 days after inoculation. The lesion size in GhATAF1-overexpressing lines was larger than that in wild-type plants (Fig. 5a, b). To analyze V. dahliae resistance responses in wild-type and GhATAF1-overexpressing plants, 3-week-old seedlings were inoculated with a V. dahlia strain, ‘1CD3-2’. The results showed that GhATAF1-overexpressing lines presented more wilting and de-colored leaves than did wild-type plants 10 days after inoculation (Fig. 5c), and the disease index of GhATAF1-overexpressing plants was higher than that of wild-type plants (Fig. 5d). Meanwhile, we also analyzed the expression of genes participating in the JA and SA signaling pathways in cotton roots through RT-PCR. The results showed that the expression of genes involved in JA biosynthesis (GhLOX1) and the JA response (GhMYC2, GhPR3, and GhPR4) were suppressed in GhATAF1-overexpressing lines compared with the wild-type plants, while the transcripts of genes involved in the SA signaling pathway, such as GhNDR1-1, GhNPR1, GhPR1, and GhPR5, were up-regulated in GhATAF1-overexpressing lines compared with wild-type plants (Fig. 5e). These results indicated that GhATAF1 was a negative regulator in the resistance of cotton to B. cinere and V. dahliae by suppressing JA-mediated and activating SA-mediated defense signaling.
Reduced resistance of GhATAF1-overexpressing cotton plants to fungal pathogens. a Phenotypes of GhATAF1-overexpressing and wild-type cotton leaves after inoculation with B. cinerea for 3 days. b The size of the lesions on the leaves indicated in (a). Error bars indicate the standard deviation of ten lesions. Asterisks indicate statistically significant differences between transgenic lines and their corresponding wild-type (WT) plants at P < 0.05, t test. c The phenotypes of GhATAF1-overexpressing and wild-type cotton plants after inoculation with V. dahliae strain ‘1CD3-2’ for 10 days. d Disease index of the plants indicated in (c). e RT-PCR analysis of the expression levels of JA and SA signaling-related and PR genes in GhATAF1-overexpressing and wild-type cotton plants after inoculation with V. dahliae strain ‘1CD3-2’ for 24 h
GhATAF1-overexpressing plants exhibited reduced JA responses and increased SA responses
JA and SA are important phytohormones that respond to fungal pathogens (Dong 1998). Because GhATAF1 was induced by MeJA and SA (Fig. 2c), it might function as a regulator in the JA and SA responses. To further verify whether GhATAF1 could negatively affect JA signaling, JA-mediated growth (hypocotyl length and fresh weight) inhibition was compared between GhATAF1-overexpressing and wild-type plants. The results demonstrated that the overexpression of GhATAF1 significantly promoted cotton growth (hypocotyl elongation) under 5 µM MeJA treatment conditions compared with wild-type plants (Fig. 6a, b). Meanwhile, the fresh weights of the two GhATAF1-overexpressing lines were significantly higher than the wild-type plants (Fig. 6c).
GhATAF1-overexpressing plants exhibited reduced JA responses and an activated SA response. a Phenotypes of GhATAF1-overexpressing and wild-type cotton seedlings grown on 1/2 MS medium supplied without (Mock) or with 5 µM MeJA for 7 days. b, c Hypocotyl length (b) and fresh weight (c) of GhATAF1-overexpressing and wild-type seedlings grown on 1/2 MS medium containing the indicated concentrations of MeJA. Error bars indicate the standard deviation of over 15 seedlings. Asterisks indicate statistically significant differences between transgenic lines and the corresponding wild-type (WT) plants at P < 0.05, t test. d The phenotypes of GhATAF1-overexpressing and wild-type seedlings 4 days after treatment without (Mock) or with 4 mM SA. e Relative survival ratio of GhATAF1-overexpressing and wild-type seedlings treated with 4 mM SA. The values are the mean ± SD from three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and the corresponding wild-type (WT) plants at P < 0.05, t test. f qRT-PCR analysis of the GhATAF1, GhJAZ1, GhPDF1.2, GhWRKY70, GhPR1, and GhPR3 expression levels 3 h after treatment with or without MeJA and SA. GhUBQ7 was used as an internal control to normalize expression levels. The data represent the mean ± SD of three technical replicates
GhATAF1 could also be highly induced by SA (Fig. 2c), which was an antagonist to JA in the response to pathogens. To investigate the SA sensitivity, 3-week-old seedlings of wild-type and GhATAF1-overexpressing lines were transferred into Hoagland solutions with or without 4 mM SA for 5 days. The survival rate of wild-type plants was significantly higher than the survival rates of the two GhATAF1-overexpressing lines under SA treatment (Fig. 6d, e), suggesting that GhATAF1 is responsible for activating SA signaling.
To further validate the function of GhATAF1 in the JA and SA signaling response, the expression of GhJAZ1, GhPDF1.2, GhPR1, GhPR3, and GhWRKY70 in transgenic lines and wild-type plants was examined. Whether the plants were treated with MeJA or SA, the transcripts of GhJAZ1 and GhPR3 were increased, but fewer GhJAZ1 and GhPR3 transcripts were found in GhATAF1-overexpressing lines than in the wild-type (Fig. 6f). The GhPDF1.2 transcripts were induced by MeJA, but fewer transcripts of GhPDF1.2 were found in GhATAF1-overexpressing lines than in the wild-type (Fig. 6f). In contrast to these JA-related genes, the transcript levels of SA-related genes (GhPR1 and GhWRKY70) were more up-regulated in GhATAF1-overexpressing plants than in wild-type plants (Fig. 6f). All of these results indicated that the overexpression of GhATAF1 in cotton repressed JA signaling and activated SA signaling.
Discussion
NAC proteins are a large family of plant-specific transcription factors that are involved in a wild range of growth and stress responses. Although a number of NAC transcription factors have been isolated in cotton, their biological functions were not analyzed in detail (Shah et al. 2013; Shang et al. 2013). ATAF1/2, a sub-family of NAC, plays an important role in the response to abiotic stresses. Although several reports have indicated that ATAF1 TFs are involved in the biotic and abiotic stress responses, the molecular mechanism of ATAF1 TF-mediated regulation of multifarious stresses is not well documented (Wu et al. 2009). In this study, we isolated and characterized GhATAF1, a cotton ATAF1/2 sub-family NAC transcription factor. In addition to induction by salt, cold and ABA (Zhu et al. 2011), the transcription levels of GhATAF1 were also induced by V. dahliae infection, MeJA and SA (Fig. 2). Based on the analysis of overexpressing GhATAF1 in cotton, we here propose a function for the cotton NAC gene GhATAF1 in modulating the cellular network of the phytohormone-mediated abiotic and biotic stress responses.
Overexpression of GhATAF1 enhanced salt tolerance by up-regulating GhHKT1 and stress response genes
To date, numerous stress-responsive NACs have been reported to have key functions in tolerance to abiotic stresses such as salinity and drought. For example, overexpression of RD26, SNAC1, OsNAC6, TaNAC69, or PeNAC1 increases salt tolerance in transgenic plants by regulating various types of downstream targets, such as stress-related transcriptional factors, ROS-related enzymes or Na+ transporters (Fujita et al. 2004; Hu et al. 2006; Nakashima et al. 2007; Xue et al. 2011). However, overexpression of Arabidopsis ATAF1 increases sensitivity to salt and oxidative stress (Wu et al. 2009). Na+ disequilibrium is a primary consequence of salt stress, and maintaining a low Na+/K+ ratio in the cytoplasm is important for plant salt resistance. The SOS regulatory pathway, NHX1 (vacuolar Na+/H+ exchanger) and HKT1 (K+/Na+ co-transporter) are the most important regulators for maintaining a low cytoplasmic concentration of Na+ under salt stress (Zhu 2003). In our experiments, GhATAF1-overexpressing cotton plants exhibited improved salt tolerance (Fig. 4). Additionally, we detected the expression of Na+ transporters and found that the expression of GhSOS1, GhSOS2, and GhNHX1 in GhATAF1-overexpressing plants was not increased significantly compared with the wild-type under either normal or salt stress conditions. This indicated that the improved salt tolerance of GhATAF1-overexpressing plants occurred through different pathways rather than SOS and NHX1. Interestingly, the expression of GhHKT1 was significantly increased in GhATAF1-overexpressing cotton plants compared to wild-type plants under salt stress condition, and the Na+ content and Na+/K+ ratio in the shoots of GhATAF1-overexpressing plants were lower than those in wild-type plants (Fig. 4). In addition, more accumulation of stress response genes, such as GhABI4, GhAVP1, GhLEA3, GhLEA6, GhRD22, and GhDREB2A, was found in GhATAF1-overexpressing lines than in wild-type plants under salt stress (Fig. 4). ABA plays a key role in the abiotic stress response in plants, and ABI4 is a key regulator of ABA signaling and plastid signals derived from reactive oxygen species (ROS) (Hauser et al. 2011). Overexpression of AVP1 (a vacuolar H+-pyrophosphatase) was reported to enhance plant salt and drought tolerance substantially (Pasapula et al. 2011). LEA3 and LEA6 belong to the late embryogenesis abundant genes, and RD22 and DREB2A are stress response genes that are involved in the ABA response, dehydration and salt stress (Hundertmark and Hincha 2008; Mowla et al. 2006; Nakashima et al. 2000). Thus, GhATAF1 conferred salt tolerance by regulating various stress response genes, including some transporters involved in Na+/K+ homeostasis and stress response genes that appear to function in ABA signaling and damage limitation or repair. Further studies are necessary to determine whether the involvement of GhATAF1 in response to drought and osmotic stresses shares a similar molecular basis to the response to salt stress.
GhATAF1 is involved in biotic stress by suppressing JA-responsive genes but activating SA-induced genes
Previous studies indicated that several NAC proteins were involved in various plant-pathogen interactions by activating or suppressing pathogenesis-related genes. ATAF1 was induced by MeJA, but not SA, and positively regulated penetration resistance towards the biotrophic fungus Blumeria graminis f. sp. hordei by attenuating ABA signaling (Jensen et al. 2008) but negatively regulated resistance to the necrotrophic pathogens B. cinerea, Alternaria brassicicola and the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 by regulating the expression of defense genes (Wang et al. 2009; Wu et al. 2009). Overexpression of ATAF2 resulted in increased susceptibility to the necrotrophic soil-borne fungal pathogen Fusarium oxysporum due to the repression of PR genes (Delessert et al. 2005). In this study, GhATAF1 was rapidly induced by SA and MeJA and responded to inoculation with V. dahliae (Fig. 2). Overexpression of GhATAF1 decreased the resistance to necrotrophic pathogens B. cinerea and V. dahliae in cotton and activated the expression of SA-responsive genes (GhNDR1-1, GhNPR1, GhPR1, and GhPR5) but suppressed the expression of JA-responsive genes (GhLOX1, GhMYC2, GhPR3, and GhPR4) after inoculation with V. dahlia (Fig. 5). These results suggest that GhATAF1 acts as a negative regulator in cotton resistance to V. dahlia, possibly by repressing JA signaling.
Although the molecular basis of the interaction between cotton and V. dahliae is still poorly understood, evidence has suggested that JA signaling is required in Ve1-mediated resistance to V. dahliae in tomato and Arabidopsis (Fradin et al. 2011). Furthermore, JA signaling was also activated in cotton following V. dahliae infection, and it positively contributed to cotton resistance to V. dahliae (Gao et al. 2013; Li et al. 2014). As shown in Fig. 6, the overexpression of GhATAF1 decreased MeJA-mediated seedling growth (hypocotyl elongation and fresh weight) inhibition, and the expression levels of GhJAZ1, GhPDF1.2, and GhPR3 (marker genes induced by JA) were significantly lower in GhATAF1-overexpressing plants than in wild-type plants. The results demonstrated that GhATAF1 repressed the JA responses. Signaling crosstalk between SA and JA commonly manifests as a reciprocal antagonism (Thaler et al. 2012). In contrast to the response to JA signaling, GhATAF1-overexpression plants displayed an increased sensitivity to SA, and the expression levels of defense-related genes associated with SA-mediated defense responses (GhPR1 and GhWRKY70) were higher in GhATAF1-overexpressing plants than in wild-type plants after treatment with SA (Fig. 6). WRKY70 is a node of convergence for JA-mediated and SA-mediated signals in plant defense (Li et al. 2004), and JAZs are the repressor of JA and play key roles in the crosstalk between JA and other hormone signaling pathways (Kazan and Manners 2012). These results demonstrated that GhATAF1 acts as a key regulator in the antagonistic interaction between the JA and SA signaling pathways, possibly by regulating the expression of GhJAZ1 and GhWRKY70.
To date, some NAC TFs are involved in crosstalk between abiotic and biotic stress signaling. ATAF1 was a negative regulator in defense responses against pathogens and in ABA signaling, salt and oxidative tolerance but a positive regulator in drought tolerance (Wang et al. 2009; Wu et al. 2009). Rice OsNAC6 was a positive regulator in drought, high-salt tolerance and resistance against blast disease (Nakashima et al. 2007). In this study, we regarded GhATAF1 as a new convergence point of the interaction of ABA-, JA-, and SA-dependent responses to abiotic and biotic stresses. Although the molecular basis of the hormone crosstalk in the transgenic plants has not been completely resolved in this report, our results suggested that GhATAF1 can influence the phytohormonal signaling networks in response to multiple stresses by regulating the crucial regulators of phytohormones and stress-related genes. Further characterization of the direct target genes of GhATAF1 and the proteins interacting with GhATAF1 will expand our understanding of the functions of the GhATAF1 in the crosstalk of the biotic and abiotic stress responses.
Author contribution statement
X. Zhang, X. He and L. Zhu conceived and designed the experiments. X. He performed most of the experiments. L. Xu and W. Guo participated in the experiments. X. He analyzed the data and drafted the manuscript; X. Zhang and L. Zhu revised the manuscript.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149:1724–1738
Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43:745–757
Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1:316–323
Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GCM, Joosten MHAJ, Thomma BPHJ (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156:2255–2265
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66:293–305
Gao W, Long L, Zhu LF, Xu L, Gao WH, Sun LQ, Liu LL, Zhang XL (2013) Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol Cell Proteomics 12:3690–3703
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Guo W, Jin L, Miao Y, He X, Hu Q, Guo K, Zhu L, Zhang X (2016) An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol Biol. doi:10.1007/s11103-016-0467-6
Hao J, Tu L, Hu H, Tan J, Deng F, Tang W, Nie Y, Zhang X (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63:6267–6281
Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21:R346–R355
Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom 9:1–22
Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF (2008) Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J 56:867–880
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H, et al (2006) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plan 50:519–524
Jun Z, Zhang Z, Gao Y, Zhou L, Fang L, Chen X, Ning Z, Chen T, Guo W, Zhang T (2015) Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci Rep 5:15048
Kazan K, Manners JM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17:22–31
Kim SG, Lee AK, Yoon HK, Park CM (2008) A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 55:77–88
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li C, He X, Luo X, Xu L, Liu L, Min L, Jin L, Zhu L, Zhang X (2014) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166:2179–2194
Li YB, Han LB, Wang HY, Zhang J, Sun ST, Feng DQ, Yang CL, Sun YD, Zhong NQ, Xia GX (2016) The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. doi:10.1104/pp.15.01930
Liu G, Li X, Jin S, Liu X, Zhu L, Nie Y, Zhang X (2014) Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 9:e86895
Luo X, Wu J, Li Y, Nan Z, Guo X, Wang Y, Zhang A, Wang Z, Xia G, Tian Y (2013) Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS One 8:e54002
Mittal A, Gampala SS, Ritchie GL, Payton P, Burke JJ, Rock CD (2014) Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J 12:578–589
Mowla SB, Cuypers A, Driscoll SP, Kiddle G, Thomson J, Foyer CH, Theodoulou FL (2006) Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J 48:743–756
Munis MF, Tu L, Deng F, Tan J, Xu L, Xu S, Long L, Zhang X (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commun 393:38–44
Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71:2095–2100
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol 42:657–665
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, Chen J, Qiu X, Zhu L, Zhang X, Auld D, Blumwald E, Zhang H, Gaxiola R, Payton P (2011) Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol J 9:88–99
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Shah ST, Pang C, Fan S, Song M, Arain S, Yu S (2013) Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene 531:220–234
Shang H, Li W, Zou C, Yuan Y (2013) Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol 55:663–676
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Tu L, Zhang X, Liu D, Jin S, Cao J, Zhu L, Deng F, Tan J, Zhang C (2007) Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin Sci Bull 52:3110–3117
Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4:1213–1227
Wang X, Basnayake BM, Zhang H, Li G, Li W, Virk N, Mengiste T, Song F (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant Microbe Interact 22:1227–1238
Wang H, Zhao Q, Chen F, Wang M, Dixon RA (2011) NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 68:1104–1114
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62:5607–5621
Xu F, Yang L, Zhang J, Guo X, Zhang X, Li G (2012) Prevalence of the defoliating pathotype of Verticillium dahliae on cotton in central china and virulence on selected cotton cultivars. J Phytopathol 160:369–376
Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yang M, Zhang Y, Zhang L, Hu J, Zhang X, Lu K, Dong H, Wang D, Zhao FJ, Huang CF, Lian X (2014) OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J Exp Bot 65:4849–4861
Yang CL, Liang S, Wang HY, Han LB, Wang FX, Cheng HQ, Wu XM, Qu ZL, Wu JH, Xia GX (2015) Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol Plant 8:399–411
Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T (2009) Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. Plant J 57:615–625
Zhang J, Yu J, Pei W, Li X, Said J, Song M, Sanogo S (2015) Genetic analysis of Verticillium wilt resistance in a backcross inbred line population and a meta-analysis of quantitative trait loci for disease resistance in cotton. BMC Genom 16:577
Zheng X, Chen B, Lu G, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zhou T, Yang X, Wang L, Xu J, Zhang X (2014) GhTZF1 regulates drought stress responses and delays leaf senescence by inhibiting reactive oxygen species accumulation in transgenic Arabidopsis. Plant Mol Biol 85:163–177
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zhu L, Tu L, Zeng F, Liu D, Zhang X (2005) An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agron Sin 31:1657–1659
Zhu LF, He X, Yuan DJ, Xu L, Xu L, Tu LL, Shen GX, Zhang H, Zhang XL (2011) Genome-wide identification of genes responsive to ABA and cold/salt stresses in Gossypium hirsutum by data-mining and expression pattern analysis. Agric Sci China 10:499–508
Acknowledgments
This work supported by the Program of Introducing Talents of Discipline to Universities in China (Grant No. B14032) and the National Cotton Agricultural Research System (Grant no. CARS-18-09). Technical assistance from Professor Xingming Lian (Huazhong Agricultural University, China) for Na+ and K+ quantification is especially appreciated.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Additional information
Communicated by Y.-T. Lu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, X., Zhu, L., Xu, L. et al. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep 35, 2167–2179 (2016). https://doi.org/10.1007/s00299-016-2027-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2027-6