Abstract
Key message
Bacterial phosphite oxidoreductase gene and chemical phosphite can be used as a selection system for Agrobacterium -mediated maize transformation.
Abstract
Application of phosphite (Phi) on plants can interfere the plant metabolic system leading to stunted growth and lethality. On the other hand, ectopic expression of the ptxD gene in tobacco and Arabidopsis allowed plants to grow in media with Phi as the sole phosphorous source. The phosphite oxidoreductase (PTXD) enzyme catalyzes the conversion of Phi into phosphate (Pi) that can then be metabolized by plants and utilized as their essential phosphorous source. Here we assess an alternative selectable marker based on a bacterial ptxD gene for Agrobacterium-mediated maize transformation. We compared the transformation frequencies of maize using either the ptxD/Phi selection system or a standard herbicide bar/bialaphos selection system. Two maize genotypes, a transformation amenable hybrid Hi II and an inbred B104, were tested. Transgene presence, insertion copy numbers, and ptxD transcript levels were analyzed and compared. This work demonstrates that the ptxD/Phi selection system can be used for Agrobacterium-mediated maize transformation of both type I and type II callus culture and achieve a comparable frequency as that of the herbicide bar/bialaphos selection system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Global maize production is increasing annually and reached around 1 billion metric tons in 2013 (http://faostat3.fao.org/). Because of its importance as a crop for food, feed, fiber and fuel, the improvement of maize agronomic traits such as yield and pathogen resistance is central to ensuring productivity of this crop. Crop improvement has been accomplished by using conventional breeding, which can take several decades to establish a new variety or by genetic engineering. Genetic transformation has been utilized as a method for targeted crop improvement allowing specific traits to be introduced into the maize genome (Moose and Mumm 2008). Agrobacterium-mediated transformation is a tool that has been developed for more than 25 years and offers advantages compared to any other genetic transformation method in terms of transgene stability, low copy transgene integration, and the length of the DNA segments that can be introduced into the plant genome (Frame et al. 2002; Ishida et al. 2007).
During genetic transformation, plant tissues are grown on media containing a selection agent that kills non-transformed cells, allowing only transformed cells to survive (Miki and McHugh 2004). Choice of selectable marker is a key factor in the successful production of transgenic plants. Typically the selectable marker gene is placed along with the gene of interest in the DNA vector construct. Antibiotic (hygromycin and kanamycin) and herbicide (bialaphos and glyphosate) resistance genes have been widely utilized as selectable markers (Block et al. 1987; Gordon-Kamm et al. 1990; van den Elzen et al. 1985). However, some plant species have natural resistance to antibiotics. For example, orchids are naturally resistant to hygromycin and kanamycin (Knapp et al. 2000). On the other hand, many plant species are sensitive to herbicides such as bialaphos or glyphosate. In fact a number of commercial genetically modified (GM) crops are either bialaphos resistant (LibertyLink®) or glyphosate resistant (Roundup Ready®) (Breyer et al. 2014). Although herbicide resistant genes are effective in achieving plant transformation, intellectual property restrictions can limit the utilization of such genes by the public sector. Recently, concern over possible horizontal gene transfer of herbicide resistance traits to wild relative species has also encouraged researchers to seek alternatives (Ramessar et al. 2007).
One of the systems that has been developed is positive selection in which the selectable marker supports growth of transformed cells instead of killing non-transformed cells. The positive selection system based on Escherichia coli phosphomannose isomerase (PMI) has been reported in a number of plants including maize, rice, tobacco, and cabbage (Joersbo and Okkels 1996; Lucca et al. 2001; Min et al. 2007). The PMI enzyme reversibly converts mannose-6-phosphate to fructose-6-phosphate allowing the identification of transgenic events on media containing mannose (Negrotto et al. 2000; Reed et al. 2001). Higher transformation frequency compared to antibiotic selection using this system has been reported (Joersbo and Okkels 1996; Min et al. 2007).
A recent study showed the utilization of phosphite oxidoreductase (ptxD) gene derived from Pseudomonas stutzeri WM88 as a positive selectable marker for plant transformation (López-Arredondo and Herrera-Estrella 2013). The PTXD catalyzes the conversion of phosphite (Phi), a non-metabolizable form of phosphorus (P), into inorganic phosphate (Pi) that can be readily used by plant cells as P source. Phi is a structural analogue of Pi and has been reported to act as an alternative fungicide that can inhibit the growth of oomycete (McDonald et al. 2001). Plants can only metabolize Pi. Selection of transgenic events using the ptxD gene allows transformed cells to utilize Phi as a phosphorous (P) source and survive on media containing Phi. A selectable marker system based on ptxD/Phi has been reported in tobacco and Arabidopsis (López-Arredondo and Herrera-Estrella 2013) and more recently in yeast transformation (Kanda et al. 2014) with some advantages in term of cost and safety compared to antibiotic selection.
In this current study, PTXD was utilized as a selectable marker for Agrobacterium-mediated maize transformation. Maize Hi II hybrid genotype and B104 inbred line have been shown to be amenable for the routine Agrobacterium-mediated transformation protocol (Frame et al. 2002, 2006; Zhao et al. 2002). Selection of transformed cells was performed on media containing monopotassium phosphite (KH2PO3) and compared to bialaphos selection. The bar/bialaphos is a selection system routinely used for maize transformation. The ptxD/Phi system resulted in positive selection of transgenic events from Hi II and B104. Molecular analyses further confirmed the expression of ptxD gene in all transgenic events selected with Phi. Our results indicate that ptxD/Phi system can be effectively used for Agrobacterium-mediated maize transformation.
Materials and methods
DNA construct
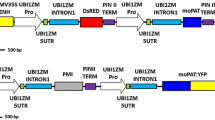
The T-DNA region harboring the A522 plasmid (11,171 bp) containing both the ptxD (Lopez-Arredondo and Herrera-Estrella 2012) and the bar (White et al. 1990) genes was used in the experiment as shown in Fig. 1. The plasmid is based on the Gateway binary vector pB7WG2 (Karimi et al. 2002), that contains the bar gene under the control of the nos promoter and terminator (Bevan et al. 1983), with no intron included, as selectable marker and the spectinomycin resistance (Sm/SpR) gene for selection in Agrobacterium and E. coli. The ptxD gene was codon optimized to be expressed in maize and regulated by the maize ubiquitin gene ubi1 promoter with its first intron (Christensen et al. 1992) and the CaMV 35S terminator (Odell et al. 1985). The DNA construct was transformed into Agrobacterium tumefasciens EHA101 (Hood et al. 1986) and utilized to infect maize immature embryos.
T-DNA region of A522 construct expressing the ptxD gene. Pubi + int, maize ubi-1 promoter and its first intron; T35S, CaMV 35S terminator; Pnos-bar-Tnos, bar gene expression regulated driven by Agrobacterium nopaline synthase gene promoter (Pnos) and terminator (Tnos); LB, Agrobacterium T-DNA left border; RB, Agrobacterium T-DNA right border; Hind III, restriction enzyme; kb, kilobases
Plant material
Maize Hi II hybrid (Armstrong et al. 1991) and B104 inbred (Hallauer et al. 1997) genotypes were used in the experiments. Greenhouse grown maize ears were kindly provided by the Plant Transformation Facility of Iowa State University. Immature zygotic embryos (1.5–2.0 and 1.8–2.0 mm) were dissected from Hi II and B104 maize ears, respectively.
Media preparation
N6-based (Chu 1975) and MS-based (Murashige and Skoog 1962) media for Hi II and B104, respectively, were prepared as previously described by Frame et al. (2011). The media formulations for infection, resting and co-cultivation were the same between treatment and control groups. Detailed media compositions including different selective agents is described in Table 1 and Table S1. For Phi selection media preparation, basal salts of N6 and MS with no P were specially formulated by PhytoTechnology Laboratories (Overland Park, KS). Monopotassium phosphite (KH2PO3, CAS No. 13977-65-6) was purchased from Wanjie International (ZheJiang, China) while monopotassium phosphate (KH2PO4) was obtained from Sigma-Aldrich (St.Louis, MO). Regeneration media for somatic embryo maturation and germination for Hi II and B104 were prepared using MS salts (Murashige and Skoog 1962).
Maize transformation
Agrobacterium-mediated maize transformation was conducted following protocols described by Frame et al. (2011). Immature zygotic embryos of Hi II and B104 were infected with A. tumefaciens EHA101 (Hood et al. 1986) harboring the A522 plasmid expressing the ptxD gene, then transferred to cocultivation media, and kept in the dark at 20 °C for 3 days. Immature embryos were subcultured onto resting media and kept in dark at 28 °C for 7 days before being transferred onto selection media. Unless otherwise stated, all tissue culture plates were maintained in dark at 28 °C from this step forward. Embryos were transferred onto different selection media containing Phi or bialaphos as summarized in Table 1. The difference in the selection between Hi II and B104 is described as follows.
Selection of transgenic events for Hi II transformation
Responding embryos were cultured on Selection 1 (S1) media (Table S1) in the dark (25 °C) for 2 weeks. The media used for Phi selection treatment groups were depleted with P as described above. After 2 weeks, calli proliferating on the S1 media were subcultured onto Selection 2 (S2) media (Table S1). Subculture duration for S2 stage was 3 weeks for Phi and 2 weeks for the bialaphos selection groups, respectively. Putative transgenic events forming Type II calli that survived the S2 selection stage were subcultured for another round of S2 before being transferred onto pre-regeneration media for 2 weeks. Embryogenic calli were transferred onto Regeneration 1 (R1) media (Table S1) for 2 weeks for somatic embryo maturation and then subcultured to Regeneration 2 (R2) media (Table S1) and germinated in the light (25 °C, 80–100 µE/m2/s, 16:8 photoperiod) for 7–10 days. Transgenic Hi II plantlets were advanced in the greenhouse to obtain flowering T0 plants.
Selection of transgenic events for B104 transformation
B104 embryos were cultured on B104 S1 media in the dark (25 °C) for 2 weeks, and then subcultured onto S2 media (Table S1). As for Hi II transformation, the media used for Phi selection were depleted with P. Duration for the S2 stage were 4 weeks on Phi selection (freshly subcultured into new media every 2 weeks) and 2 weeks on bialaphos selection group. Putative transgenic Type I calli were subcultured on S2 media for two more rounds before being transferred onto R1 media for 2 weeks (Table S1). Mature somatic embryos were then subcultured to R2 media (Table S1) in the light (25 °C, 80–100 µE/m2/s, 16:8 photoperiod) for 7–10 days for germination.
Molecular analyses
Putative transgenic calli were subjected to molecular analysis to confirm the presence of the ptxD gene. Polymerase Chain Reaction (PCR) was used for DNA analysis. DNA from calli were extracted using cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle 1987) and utilized as a PCR template to amplify the ptxD gene (±1 Kb) using primer forward 5′-GGTGTTACTTCTGCAGATGC-3′ and reverse 5-GAGAGATAGATTTGTAGAGA-3′. The PCR product was analyzed with agarose gel electrophoresis.
Quantitative real time PCR (qPCR) was used to estimate copy number of ptxD in T0 Hi II plants. Template genomic DNA (20 ng) was added in a reaction containing QuantiTect SYBR Green PCR master mix (Qiagen) following the manufacturer’s instruction. Each sample was analyzed in duplicate. Copy number estimation was determined against a known concentration of the plasmid A522 DNA control (Gadaleta et al. 2011; Song et al. 2002).
The ptxD transcription level was analyzed using reverse-transcriptase PCR (RT-PCR) method. Total RNA was extracted from T0 Hi II plants using Qiagen RNeasy Plant Mini kit (Qiagen Inc, Valencia, CA) following the supplier’s instructions. Complementary DNA (cDNA) was synthesized from 1 µg of the total RNA using SuperSCript™ III reverse transcriptase (Invitrogen, Grand Island, NY) in a total volume of 20 µl. An aliquot of 2 µl cDNA (100 ng) was used as template for PCR to amplify approximately 300 bp fragment of ptxD using primer pair forward (5-TGAGCTGCTCGCACTCGTTA-3′) and reverse (5′-AGTGCTCGCCGGGGCGAGAC-3). A housekeeping gene, actin, was used here as an internal control. An approximately 500 bp actin fragment was amplified using the following primers, forward (5′-ATTCAGGTGATGGTGTGAGCCACAC-3′) and reverse (5′-GCCACCGATCCAGACACTGTACTTCC-3′). Equal amounts of PCR product were analyzed by agarose gel electrophoresis and transcription level was determined semi-quantitatively. Quantitative real time PCR (qPCR) was used to analyze the relative gene expression level of ptxD in T0 Hi II plants. Template cDNA was added in a reaction containing QuantiTect SYBR Green PCR master mix (Qiagen) following the manufacturer’s instructions. Each sample was analyzed in duplicate. Relative ptxD gene expression was determined using the ΔΔct method and normalized against the actin housekeeping gene.
Results and discussion
Phosphite selection kill curve establishment
In previous work on tobacco transformation using the ptxD/Phi system as selection, it was established that 1 mM of Phi in regeneration media was adequate to select for transgenic events (López-Arredondo and Herrera-Estrella 2013). To determine the Phi selection level for maize transformation, we first evaluated a number of Phi concentrations (kill curve) for their effect on embryogenic callus induction for both maize Hi II and B104 genotypes. Maize immature zygotic embryos (IZEs) were aseptically dissected from ears of Hi II and B104. Approximately 25 IZEs/plate were cultured in Pi-free media but containing different concentrations of Phi (from 0 to 3.5 mM). One control plate (standard N6S for Hi II and MSS for B104) was included. Due to limited explant sources, the evaluation was repeated only twice.
Visual examination suggested that all Phi concentrations including 0 mM Phi suppressed callus formation and development regardless of callus types (Type II callus for Hi II and Type I callus for B104, Figure S1). It is likely that the lack of Pi in the callus induction media severely affected the callus formation. Based on these initials observations, we designed treatment groups as shown in Table 1. We used 0.5 and 1.25 mM Phi for S1 and S2, respectively, in Hi II transformation (groups B and C); and 1.25 mM Phi for S1 and either 1.25 or 2 mM Phi for S2 in B104 transformation (groups E–H). Because of the poor callus induction responses in the absence of Pi, we also included treatment groups (groups C, F and H) in which a low concentration of Pi (50 µM KH2PO4) was included in the S1 media to favor callus initiation and proliferation.
Hi II transformation with phosphite selection
In total, over 1600 Hi II maize IZEs were infected with A. tumefasciens EHA101 carrying the A522 plasmid expressing both bar and ptxD genes in three independent experiments (Table 2). As shown in Fig. 1, this construct carries the bar gene under a constitutive nos promoter and the ptxD gene under another constitutive and strong maize ubiquitin gene promoter. After co-cultivation, the infected embryos were divided into two or three selection groups for either bialaphos or Phi selection. In the first Hi II transformation experiment maize embryos selected with Phi were grown on S1 media containing 0.5 mM Phi for 2 weeks then transferred onto S2 containing 1.25 mM Phi for 3 weeks (Table 1). Control embryos were grown on S1 containing 1.5 mg/L bialaphos for 2 weeks then transferred to S2 containing 3 mg/L bialaphos for another 2 weeks (Frame et al. 2011). Callus initiation from IZEs cultured on Phi-containing media was considerably slower than for the control IZEs on bialaphos selection. Embryogenic Type II callus emerged approximately 3 weeks after IZEs were transferred onto S2 media under Phi selection, while callus from the control group emerged as early as 2 weeks after being sub-cultured on S2. This delayed callus response on Phi-containing media may be attributed to the fact that under Phi selection, plants have first to convert Phi into Pi before it can be metabolized.
In the subsequent Hi II transformation experiments (experiment 2 and 3), an additional Phi selection group (group C) was included whereby the S1 media was supplemented with 50 µM Pi in addition to the 0.5 mM Phi selective agent. The small amount of Pi supplementation was intended to support immature embryo response in the early stage of development and improve the recovery of transformed cells. Interestingly, there was no noticeable difference in terms of callus growth and transgenic events recovery between group B and C (Table 2). Slower callus growth was also observed in the group C.
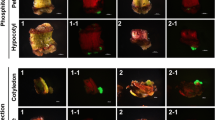
To increase the likelihood that emerging calli were indeed transgenic events, they were transferred to S2 media for another round of selection before regeneration. All Hi II putative events identified from either bialaphos or Phi selection were regenerable (Fig. 2). No morphological differences were observed from T0 plants regenerated from either selection system.
B104 transformation under phosphite selection
The B104 maize genotype is a better suited inbred for functional genomic studies compared to the hybrid genotype Hi II. It is one of the few inbred lines that are amenable for Agrobacterium-mediated method using the bar/bialaphos selection system, and as such it was also tested in this study. Four independent experiments were conducted using maize B104 IZEs for transformation with the A522 construct. Based on observations from the kill curve experiments, the nature of B104 callus (Type I), and our experience in Hi II and B104 transformation using bialaphos as a selective agent (Frame et al. 2011), we used 1.25 mM Phi in the S1 stage for B104 transformation. This was higher than the Phi concentration (0.5 mM) that was used for Hi II selection (Type II callus) at the same stage (Table 1). As we did for Hi II transformation, supplementation with 50 µM Pi (group F) or without Pi (group E) in Phi selection were compared. Similar to what was observed in Hi II transformation, slower callus initiation was observed in both B104 Phi selection treatments (groups E and F) when compared to control embryos on the bialaphos selection media. In fact, callus initiation for B104 was even slower than that for Hi II. Type I embryogenic calli formation was apparent 4 weeks after sub-cultured onto Phi-containing S2 media, while Type I callus initiation for the bialaphos selection control group (group D) was observed 2 weeks after being subcultured onto S2 media.
Unlike Type II callus of Hi II genotype that is friable and relatively homogenous, Type I callus of B104 is often compact and heterogeneous. Resistant calli generated from the infected embryos may contain a mixture of transformed and non-transformed cells. Therefore, resistant callus of B104 were further selected with another 2 rounds on S2 media. Each subculture of B104 was performed by cutting callus clump into small pieces and placing them onto fresh S2 media containing 2 mM Phi (groups E and F, Table 2). However, no putative embryogenic callus events survived after 2 weeks on S2 Phi selection media in the experiments 1 and 2. In contrast, a total of 13 bialaphos resistant putative callus events (6 and 7 events from experiment 1 and 2, respectively) were recovered from the bialaphos selection control group (Table 2).
A slight modification in the Phi selection media was made for B104 transformation experiments 3 and 4 (Table 2) whereby the concentration of Phi in S2 media was at 1.25 mM (group G and H) throughout selection. Slow emergence of Type I callus was also observed in groups G and H. However, after another two rounds in 1.25 mM Phi selection on S2, some putative transgenic calli could be identified. From these two independent experiments, a total of six putative events from Phi selection (two events in group G, four events in group H) and 12 putative events from the bialaphos selection (group D) were recovered (Table 2).
Transformation frequencies
A summary of transformation frequencies (TF) for Hi II and B104 with the treatment regimens described in Table 2 is presented in Fig. 3. Due to limited resources including plant materials and person power, only three independent experiments for Hi II and four independent experiments for B104 were performed. Nevertheless, in each experiment, similar numbers of embryos were treated to evaluate the Phi selection.
Comparison of Hi II and B104 transformation frequencies (TF) under different selection systems. The bars and error bars represent the mean and standard deviation of TF for each treatment groups from three Hi II experiments and four B104 experiments presented in Table 2. Infected embryos were cultured on three different selection media (bialaphos, Phi, or Phi + Pi). Hi II transformation experiments were divided into three groups (A–C). B104 transformation experiments were divided into five groups (D–H). See Table 1 for group description
In general, Phi selection resulted in lower TF compared than bialaphos selection in both Hi II and B104. Based on 3 independent experiments in Hi II, the TF with ptxD/Phi selection (group B) ranging from 1.3 to 2 % (Table 2) with an average of 1.5 ± 0.4 % (Fig. 3). The inclusion of Pi in S1 media (group C) did not enhance TF (ranging from 1.3 to 1.7 % with an average of 1.5 ± 0.3 %). On the other hand, in the control group (group A) selected with bialaphos, TF ranged between 2.4 and 4.3 % with an average of 3.7 ± 1.1 %. It is worth noting that the TF of Hi II using the bar/bialaphos selection was below the TF previously reported using this protocol (Frame et al. 2011). This could be attributed to a number of reasons including sub-optimal ear quality and less experienced researchers.
TF for B104 using ptxD/Phi selection was lower than for Hi II. The high Phi concentration (2 mM) in S2 media groups E and F did not support callus growth. Reduction of the Phi concentration to 1.25 mM resulted in a TF of 0.4 ± 0 % (group G) while Pi supplementation (group H) resulted in TF between 0.4 and 1.1 % with an average of 0.75 ± 0.5 % (Fig. 3). Similar to Hi II transformation, higher TF was achieved for B104 using the bar/bialaphos for selection (4.7 ± 0.4 %). However, it must be stated that due to limited resources, the small number of experiments did not allow the optimization of ptxD/Phi transformation and most likely this could be significantly improved with further experiments.
Plantlets were readily produced from putative positive callus events obtained from both Phi and bialaphos selection regimens with no noticeable differences in growth patterns or morphology. In this report, none of the T0 plants were grown to maturity for seeds due to very limited resources. However, in separate experiments we had generated large numbers of transgenic maize plants with the same construct using the bar/bialaphos system for selection. All ptxD expressing plants grew well in the greenhouse condition and produced seeds under both phosphite and phosphate fertilization regime. The progeny of these transgenics also performed well with no noticeable alteration in morphology comparing to non-transgenic null segregants (D. Lopez-Arredondo, in preparation).
Effect of phosphite selection on callus growth
In order to use ptxD/Phi as a selection system, plant materials have to be cultured on growth media that lacks Pi. If the ptxD gene is transformed and integrated into the genome and expressed, the transformed cells will be able to convert the non-metabolizable Phi into metabolizable Pi and thus survive and thrive on the media minus Pi. Lack of Pi in plant growth media severely inhibits maize callus growth as shown in the Phi kill curve experiments (Figure S1).
In embryos infected with the ptxD carrying construct and selected on Phi, slower emergence of embryogenic callus was observed both in Hi II and B104 under Phi selection as compared to the control groups that were selected on bialaphos. Phi selection also affected embryogenic callus end-point size wherein smaller callus were observed for Hi II (Figure S2) and B104 (Figure S3) when compared with eventual callus size for the bialaphos selected explants. In the case of B104, further delay in the development of Type I callus compared to the Type II callus of Hi II in the experiments was observed. This may be due to the fact that a higher concentration of Phi (1.25 mM) was used in S1 for B104 compared to 0.5 mM Phi used for Hi II. To enhance callus initiation at the early embryo development stage, low concentration of metabolizable Pi was supplemented in the media to overcome the total lack of P at the S1 stage. However, this attempt did not improve callus growth or transgenic event recovery for Hi II or B104. Although Phi selection resulted in slower and smaller callus growth in Hi II and B104, callus morphology and characteristics were similar to the bialaphos selection group.
The concentration of Phi in S1, S2 and regeneration media should be optimized for future Hi II and B104 transformation. In addition, supplementation of Pi in the early callus initiation stage can be fine-tuned to further improve the overall transformation frequencies.
Transgene co-presence and copy number analyses
Resistant events generated in the transformation experiments were subjected to molecular analyses to confirm the presence of the ptxD gene. In total, 32 Hi II and 31 B104 putative events were analyzed using the polymerase chain reaction (PCR) to detect the ptxD gene. Analysis results are summarized in Table 2 and representative events are presented in Figure S4. For Hi II, 14 events were generated from group A, 10 events from group B and 8 events from group C. All events, regardless whether they were selected from the bialaphos system (group A) or the Phi system (groups B and C), contained an intact copy of the ptxD and bar gene fragments (Figure S4A). For B104, all events generated from Phi selection (2 from group G and 4 from group H) were positive for the ptxD and bar genes (Figure S4B). These results indicate that both selection systems can also deliver an accompanying gene of interest for both maize Hi II and B104 transformed in this study.
To evaluate whether different selection systems have any effect on copy number of the ptxD gene, we performed quantitative PCR (qPCR) on T0 plants of Hi II events (Fig. 4a). Plasmid A522 was utilized as a control to establish a standard curve for copy number estimation (Figure S5). Analysis of transgene copy number using qPCR has been reported in numerous studies as a reliable and fast method that is comparable to Southern blot hybridization (Gadaleta et al. 2011; Ingham et al. 2001; Song et al. 2002), especially in laboratories with resource constraints. Due to large genome size of maize, Southern blot hybridization can present technical difficulty for laboratories that do not perform this technique regularly.
Copy number and gene expression analyses of the ptxD gene in T0 Hi II transgenic plants. a Copy number estimation of the ptxD gene determined by quantitative PCR. Error bars represent standard deviation of mean from two technical duplicates. b relative ptxD gene expression normalized to actin housekeeping gene. Data from 22 representative transgenic callus events generated from different selection systems: bialaphos (group A), Phi (group B), and Phi + Pi (group C)
Both single and multiple-copy insertions are observed in the events analyzed (Fig. 4a). Seven out of 22 events carry a single copy of ptxD. Six of the eight events tested were from the bialaphos selection (group A) and the remaining one event was from the Phi selection (group B). Group A (bialaphos selection) generated 67 % (6/9) single copy events, 11 % (1/9) low copy (2–4 copies) events, and 22 % (2/9) high copy (>5 copies) events. Group B (Phi selection) generated 14 % (1/7) single copy events, 86 % (6/7) low copy events and no high copy event. Group C (Phi + Pi selection) generated no single copy event, 67 % low copy events and 33 % high copy events.
The Agrobacterium-mediated maize transformation method is reported to generate transgenic events with single and low copy insertions (Ishida et al. 1996). Our work here, using either bialaphos or Phi selection, is in agreement with previous observations. It appears that a higher percentage of single copy events was obtained from the bialaphos selection group. However, the number of events obtainesd from each treatment groups are too low to be conclusive.
The reason why some constructs or transformation procedures generate a higher percentage of single or multiple copy insertions remains a topic of discussion. Expression strength of a selectable marker gene expression may attribute to the copy number of an inserted marker gene (Gendloff et al. 1990; Hobbs et al. 1993). In this study, a single construct A522 carrying both selectable marker gene, the bar and ptxD genes; the bar gene was driven by nos promoter and the ptxD was driven by a maize ubiquitin promoter. Agrobacterium-A522 strain was used for maize embryo infection and cocultivation. The infected embryos were divided into 2 or 3 groups, either plated on media for bialaphos or for Phi selection. For future Phi selection optimization, different promoters could be evaluated to determine whether copy number insertions could be reduced to improve transgenic event quality.
Transgene expression and phosphite tolerance tests
The ptxD gene expression analysis was performed using reverse transcript (RT-PCR). Semi quantitative RT-PCR analysis detected the ptxD transcript in all T0 Hi II plants (Figure S6). Quantitative PCR analysis further verified relative expression level (low to high) of ptxD gene in T0 Hi plants (Fig. 4b). Not surprisingly, medium to high ptxD gene expression was generally observed in transgenic events selected on Phi (group B and C). It is worth noting that almost all high expressers were the events that also carried high copy number inserts (Fig. 4a vs b). For example, the highest expression event C-1 carried 9 copies of the insert from the Phi selection group C. All single copy events, whether selected on bialaphos media (group A) or Phi media (group B), displayed low ptxD gene expression.
The ability of Hi II transgenic maize expressing the ptxD gene to utilize Phi as a P source and tolerate Phi was confirmed by performing a Phi tolerance test. Two transgenic callus events each from group A (A-1 and A-6), group B (B-1 and B-4), and group C (C-4 and C-6) were grown on media containing three different levels of Phi (Fig. 5). Their growth was compared with non-transgenic Hi II callus. Figure 5a compares the growth of one Phi-positive event (B-1) and non-transgenic Hi II callus on three different media. On regular media (N6S) without any selective agents, both the transgenic and non-transgenic calli grow equally well. On N6S media containing 1.5 mg/L bialaphos only transgenic callus thrived while the non-transgenic callus growth was inhibited. This result also indicates that transgenic events generated in Phi selection were also resistant to bialaphos. Callus growth for both transgenic and non-transgenic events was severely inhibited when cultured on media containing no Phi and Pi (the bottom panel in Fig. 5a). This growth inhibition is due to the complete lack of a P source necessary for cell growth in the cell media.
Phosphite tolerance test of transgenic Hi II callus. a Comparison of event B-1 callus of generated from phosphite (Phi) selection and non-transgenic Hi II callus on media (N6S) without selective agents, N6S containing 1.5 mg/L bialaphos, or N6S without Phi and phosphate (Pi). The ability of transgenic Hi II callus to utilize and tolerate Phi in the media containing different concentration of Phi b 1.25, c 2.5 and d 5 mM Phi were tested. Two transgenic Hi II events generated from bialaphos (A-1 and A-6), Phi (B-1 and B-4), and Phi + Pi (C-4 and C-6) selection groups were compared to non-transgenic (NT) Hi II callus
Figure 5b–d illustrates the growth of the six transgenic callus lines subcultured on media containing 1.25–5 mM of Phi. Event A-1 was a single copy but low expression ptxD event (Fig. 4). This event could only grow on media containing 1.25 mM Phi but not on the media containing 2.5 and 5 mM Phi. Events B-1 and C6 both had 2-copies of the ptxD transgene and were medium expressors (Fig. 4). These two events were able to grow on both 1.25 and 2.5 mM Phi-containing media, while their growth was slightly suppressed on 5 mM Phi-containing media. Events A-6, B-4 and C-4 were multiple copy number events (>4) and were high expressors (Fig. 4). Events B-4 and C-4 were able to grow on all media including 5 mM Phi; however, event A-6 grew well on the two low Phi media but had slight growth inhibition when cultured in 5 mM Phi media.
The phenotypic performance observed from the selected transgenic callus lines correlates positively with their genotypic data, that is, the higher the ptxD gene expression, the better the callus grows on increasing concentration of Phi in the culture media. These results suggested that transgenic callus could metabolize Phi as P source and the ability of transgenic events to metabolize Phi depend on the level of gene expression. Although the high expressors identified in this work all possessed higher transgene copy number, it is worth noting that these tests were performed on T0 transgenic callus lines due resource constraints. Multiple-copy transgene insertions often cause more genome disruption than that of single-copy insertions. Multiple-copy insertion events also have high probability for transgene instability and reduced transgene expression in subsequent generations (Shou et al. 2004; Zhong et al. 1999).
Conclusion
We have demonstrated that a bacterial gene encoding a phosphite oxidoreductase, PTXD, can be used as a selectable marker gene for Agrobacterium-mediated maize transformation. This system allows transformed cells expressing the ptxD gene to grow in the presence of Phi as a sole P source while non-transformed cells cannot. Since ptxD/Phi is not an antibiotic or herbicide resistance based selection system, it is less likely to cause resistance that commonly raise concern in the selectable system. Furthermore, Phi has been used as a biopesticide that does not pose harm to human health or the environment based on US environmental Protection Agency classification.
It has been reported that the ptxD/Phi selection system can be used for tobacco and Arabidopsis transformation (López-Arredondo and Herrera-Estrella 2013). Here we confirm that it can be used effectively as a positive selectable marker system for the transformation of a major crop, maize. Despite the apparently lower transformation frequencies and the lack of progeny test due to limited resources in this assessment study, the fact that Phi resistant transgenic lines were recovered from two maize genotypes, could be readily generated by researchers with little transformation skills is evidence of the robustness of this selection system. Further improvements, including optimization of Phi concentrations in selection media will be needed to enhance the quantity and quality of transgenic events. It is also important to mention that the cost of Phi is significantly lower and more stable than herbicides or antibiotics, making it more amenable for the development of transformation systems of other crop plants.
Author contribution statement
KW and LHE conceptualized the research, XX designed and performed kill curve experiment, HN designed and performed all experiments. LHE and DLA provided materials and expertise. HN and KW analyzed data and wrote manuscript. All the authors read and approved the manuscript.
References
Armstrong CL, Green CE, Phillips RL (1991) Development and availability of germplasm with high type II culture formation response. Maize Genet Cooperative Newsl 65:92–93
Bevan M, Barnes WM, Chilton MD (1983) Structure and transcription of the nopaline synthase gene region of T-DNA. Nucl Acids Res 11(2):369–385
Block MD, Botterman J, Vandewiele M, Dockx J, Thoen C, Gosselé V, Movva NR, Thompson C, Montagu MV, Leemans J (1987) Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J 6(9):2513–2518
Breyer D, Kopertekh L, Reheul D (2014) Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants—scientific developments, current use, operational access and biosafety considerations. Crit Rev Plant Sci 33(4):286–330
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18(4):675–689
Chu CC (1975) Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Sci Sin 18:659–668
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Frame B, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger T, Pegg SE, Li B, Nettleton D, Pei D, Wang K (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129(1):13–22
Frame B, McMurray J, Fonger T, Main M, Taylor K, Torney F, Paz M, Wang K (2006) Improved Agrobacterium-mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep 25(10):1024–1034
Frame B, Main M, Schick R, Wang K (2011) Genetic transformation using maize immature zygotic embryos. In: Yeung E, Thorpe TA (eds) Plant embryo culture: methods and protocols. Springer Science and Business Media, LLC, New York, pp 327–341
Gadaleta A, Giancaspro A, Cardone MF, Blanco A (2011) Real-time PCR for the detection of precise transgene copy number in durum wheat. Cell Mol Biol Lett 16(4):652–668
Gendloff EH, Bowen B, Buchholz WG (1990) Quantitation of chloramphenicol acetyl transferase in transgenic tobacco plants by ELISA and correlation with gene copy number. Plant Mol Biol 14(4):575–583
Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O’Brien JV, Chambers SA, Adams WR, Willetts NG, Rice TB, Mackey CJ, Krueger RW, Kausch AP, Lemaux PG (1990) Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2(7):603–618
Hallauer AR, Lamkey KR, White PR (1997) Registration of five inbred lines of maize: B102, B103, B104, B105, and B106. Crop Sci, pp 1405–1406
Hobbs SLA, Warkentin TD, DeLong CMO (1993) Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol 21(1):17–26
Hood EE, Helmer GL, Fraley RT, Chilton MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168(3):1291–1301
Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. BioTechniques 31(1):132–134, 136–140
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996) High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14(6):745–750
Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2(7):1614–1621
Joersbo M, Okkels FT (1996) A novel principle for selection of transgenic plant cells: positive selection. Plant Cell Rep 16(3–4):219–221
Kanda K, Ishida T, Hirota R, Ono S, Motomura K, Ikeda T, Kitamura K, Kuroda A (2014) Application of a phosphite dehydrogenase gene as a novel dominant selection marker for yeasts. J Biotechnol 182–183:68–73
Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7(5):193–195
Knapp JE, Kausch AP, Chandlee JM (2000) Transformation of three genera of orchid using the bar gene as a selectable marker. Plant Cell Rep 19(9):893–898
Lopez-Arredondo DL, Herrera-Estrella L (2012) Engineering phosphorus metabolism in plants to produce a dual fertilization and weed control system. Nat Biotechnol 30(9):889–893
López-Arredondo DL, Herrera-Estrella L (2013) A novel dominant selectable system for the selection of transgenic plants under in vitro and greenhouse conditions based on phosphite metabolism. Plant Biotechnol J 11(4):516–525
Lucca P, Ye X, Potrykus I (2001) Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed 7(1):43–49
McDonald AE, Grant BR, Plaxton WC (2001) Phosphite (phosphorous acid): its relevance in the environment and agriculture and influence on plant phosphate starvation response. J Plant Nutr 24(10):1505–1519
Miki B, McHugh S (2004) Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J Biotechnol 107(3):193–232
Min BW, Cho YN, Song MJ, Noh TK, Kim BK, Chae WK, Park YS, Choi YD, Harn CH (2007) Successful genetic transformation of Chinese cabbage using phosphomannose isomerase as a selection marker. Plant Cell Rep 26(3):337–344
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147(3):969–977
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Negrotto D, Jolley M, Beer S, Wenck AR, Hansen G (2000) The use of phosphomannose-isomerase as a selectable marker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep 19(8):798–803
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313(6005):810–812
Ramessar K, Peremarti A, Gomes-Galera S, Naqvi S, Moralejo M, Munoz P, Capell T, Christou P (2007) Biosafety and risk assessment framework for selectable marker genes in transgenic crop plants: a case of the science not supporting the politics. Transgenic Res 16(3):261–280
Reed J, Privalle L, Powell ML, Meghji M, Dawson J, Dunder E, Sutthe J, Wenck A, Launis K, Kramer C, Chang YF, Hansen G, Wright M (2001) Phosphomannose isomerase: an efficient selectable marker for plant transformation. InVitro Cell Dev Biol Plant 37(2):127–132
Shou H, Frame B, Whitham S, Wang K (2004) Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breed 13(2):201–208
Song P, Cai C, Skokut M, Kosegi B, Petolino J (2002) Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERS™-derived transgenic maize. Plant Cell Rep 20(10):948–954
van den Elzen PJM, Townsend J, Lee KY, Bedbrook JR (1985) A chimaeric hygromycin resistance gene as a selectable marker in plant cells. Plant Mol Biol 5(5):299–302
White J, Chang SY, Bibb MJ, Bibb MJ (1990) A cassette containing the bar gene of Streptomyces hygroscopicus: a selectable marker for plant transformation. Nucl Acids Research 18(4):1062
Zhao ZY, Gu W, Cai T, Tagliani L, Hondred D, Bond D, Schroeder S, Rudert M, Pierce D (2002) High throughput genetic transformation mediated by Agrobacterium tumefaciens in maize. Mol Breed 8(4):323–333
Zhong GY, Peterson D, Delaney DE, Bailey M, Witcher DR, Register JC III, Diane B, Li CP, Marshall L, Kulisek E, Ritland D, Meyer T, Hood EE, Howard JA (1999) Commercial production of aprotinin in transgenic maize seeds. Mol Breed 5(4):345–356
Acknowledgments
HN and KW thank Bronwyn Frame for her expertise in maize transformation, scientific discussion and critical review of the manuscript. This work was supported in part by the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch Project No. IOW05162 to KW), the Iowa State University Crop Bioengineering Consortium (KW), the Howard Hughes Medical Institute, Grant 55005946 (LHE) and Charoen Pokphand Indonesia (HN).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
HN, XX, LHE and KW declare that they have no conflict of interest. DLA is an employee and shareholder and LHE is a shareholder of StelaGenomics México, which provided materials and expertise for the work. However, this does not alter the authors’ adherence to all the Plant Cell Reports policies on sharing data and materials.
Additional information
Communicated by P. Lakshmanan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nahampun, H.N., López-Arredondo, D., Xu, X. et al. Assessment of ptxD gene as an alternative selectable marker for Agrobacterium-mediated maize transformation. Plant Cell Rep 35, 1121–1132 (2016). https://doi.org/10.1007/s00299-016-1942-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-1942-x