Abstract
Key message
The zeaxanthin epoxidase gene ( MsZEP ) was cloned and characterized from alfalfa and validated for its function of tolerance toward drought and salt stresses by heterologous expression in Nicotiana tabacum.
Abstract
Zeaxanthin epoxidase (ZEP) plays important roles in plant response to various environment stresses due to its functions in ABA biosynthetic and the xanthophyll cycle. To understand the expression characteristics and the biological functions of ZEP in alfalfa (Medicago sativa), a novel gene, designated as MsZEP (KM044311), was cloned, characterized and overexpressed in Nicotiana tabacum. The open reading frame of MsZEP contains 1992 bp nucleotides and encodes a 663-amino acid polypeptide. Amino acid sequence alignment indicated that deduced MsZEP protein was highly homologous to other plant ZEP sequences. Phylogenetic analysis showed that MsZEP was grouped into a branch with other legume plants. Real-time quantitative PCR revealed that MsZEP gene expression was clearly tissue-specific, and the expression levels were higher in green tissues (leaves and stems) than in roots. MsZEP expression decreased in shoots under drought, cold, heat and ABA treatment, while the expression levels in roots showed different trends. Besides, the results showed that nodules could up-regulate the MsZEP expression under non-stressful conditions and in the earlier stage of different abiotic stress. Heterologous expression of the MsZEP gene in N. tabacum could confer tolerance to drought and salt stress by affecting various physiological pathways, ABA levels and stress-responsive genes expression. Taken together, these results suggested that the MsZEP gene may be involved in alfalfa responses to different abiotic stresses and nodules, and could enhance drought and salt tolerance of transgenic tobacco by heterologous expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abscisic acid is an important phytohormone in plants and participates in regulation of plant growth and development. It is a regulator of seed dormancy and germination (Siriwardana et al. 2014), cell division and elongation (Zeevaart and Creelman 1988), stomata closure (Lind et al. 2015) and nodule development (Ding et al. 2008). Furthermore, ABA is very important in plant responses to various environmental stresses such as drought (Schroeder et al. 2001; Sreenivasulu et al. 2012), salt (Hou et al. 2013), cold and heat (Baron et al. 2012). These diverse functions require a tight control of synthesis, signal perception and transduction. ABA biosynthesis is regulated by plant development (Xiong and Zhu 2003) and environmental signals such as drought, salt, temperature, and light (Seiler et al. 2011; Seung et al. 2012).
ZEP catalyzes the conversion of zeaxanthin (Zx) into violaxanthin (Vx) in plastids (Marin et al. 1996). This reaction contributes to not only ABA biosynthesis but also the xanthophyll cycle and carotenoid biosynthesis (Nambara and Marion-Poll 2005; DellaPenna and Pogson 2006). Therefore, ZEP plays central roles in plant response to various environmental stresses. The expression and regulation of ZEP gene have been investigated in many plant species. Mutants impaired in ZEP have been isolated in several species, including Arabidopsis (Xiong et al. 2002; Barrero et al. 2005), Nicotiana plumbaginifolia (Marin et al. 1996; Xiong et al. 2002), and rice (Agrawal et al. 2001). The ZEP transcript levels were different among species and tissues under various environment stresses. It was reported that the ZEP transcript levels increased in roots of both tobacco and tomato during rapid or progressive drought stress, while showing little changes in leaves (Thompson et al. 2000; Audran et al. 2001). Besides, Audran et al. (1998) found ZEP expression decreased in the leaves of N. plumbaginifolia. An early study reported that the ZEP gene involved in the regulation of ABA biosynthesis in roots and contributed to the plant response to drought (Audran et al. 2001). The functions of ZEP gene of plant tolerance to abiotic stress such as drought, salt and chilling stress in tomato (Wang et al. 2008) and Arabidopsis (Park et al. 2008) have been studied by transgenic technology.
Alfalfa (Medicago sativa), an important leguminous herbages, can fix nitrogen by interaction symbiotically with rhizobia in root nodules (Perret et al. 2000; Hou et al. 2013). Both the growth of legume plants and the nodules functions are limited by drought (Clement et al. 2008), salt (Delgado et al. 1994), high temperature (Hungria and Kaschuk 2014) and abscisic acid (ABA) (Ding et al. 2008). It was shown that overexpression of a bacteria feature gene can enhance the legume plants tolerance to abiotic stresses including drought (Suárez et al. 2008) and salt (Bianco and Defez 2009), and rhizobia homospermidine metabolism acts as a stress tolerance strategy under salt stress (Fujihara 2008). Egamberdieva et al. (2013) reported that the increase of ABA in root nodules under salt stress can alleviate the damages in legumes. Many genes associated with ABA have been studied in alfalfa so far (Luo et al. 1992; Kovács et al. 1998; Jin et al. 2010).
Our previous study demonstrated that active nodules can enhance the alfalfa tolerance to drought stress (Yang et al. 2013) and salt stress (unpublished). Moreover, using DNA microarray technology, differentially expressed genes have been identified between alfalfa inoculated rhizobium (NA) and alfalfa not-inoculated rhizobium (NN) under drought stress. However, little is known about these candidate genes. Here, MsZEP, one of the candidate genes, was isolated and analyzed from alfalfa. To understand the expression pattern of MsZEP to abiotic stresses and nodules in alfalfa, alfalfa inoculated rhizobium (NA) and alfalfa not-inoculated rhizobium (NN) were treated with drought, cold, heat and ABA, and then the transcripts levels were detected using qRT-PCR. Moreover, we also investigated the biological functions of MsZEP in response to drought and salt stresses by heterologous expression the gene in Nicotiana tabacum.
Materials and methods
Plant materials
Seeds of alfalfa (M. sativa L. cv. baoding) were surfaced sterilized with 70 % ethanol for 30 s, then immersed in 0.5 % sodium hypochloride solution for 10 min, and rinsed 4–5 times with sterile water. Sterilized seeds were germinated on wet filter paper in Petri dishes at 24 °C under a 16-h photoperiod. Five-day-old seedlings were transplanted to plastic pots (6 × 11 cm) filled with sterilized sandy soil in the greenhouse (1 plant/pot). One set of plants (NA) was inoculated at transplanting with Rhizobium meliloti strain Dormal. This set received a N-free nutrient solution every day. The other set (NN) was watered with 1/4 strength Hoagland (Hoagland and Arnon 1950) nutrient solution. Plants were cultured in the greenhouse with the average temperature of 30 ± 5 and 20 ± 3 °C, and the relative humidity of 55 ± 5 and 70 ± 5 % during day and night.
Stress treatments
Sixty days after inoculation, the plants were subject to different stress treatments. For drought treatment, alfalfa roots without sand were immediately wrapped in a wet tissue paper (18 cm × 18 cm), and the plants were then placed on a rack for dehydration stress. Shoots and roots were harvested after 0, 4, 8, 12 h dehydration stress and re-watered for 12 h (Yang et al. 2013). For cold or heat treatment, plants were performed at 4 or 42 °C with regard to the control at 25 °C. For ABA treatment, alfalfa roots wrapped in a wet tissue paper were put into nutrient solution supplemented with 10 μM ABA, whereas the control was treated with nutrient solution. For all the cold, heat and ABA stress as well as for the control conditions, plant shoots and roots were sampled after 2, 4, 8, 12 and 24 h. The tissues were immediately frozen in liquid nitrogen and stored at −80 °C until use. The experiment was repeated three times.
Isolation of full-length MsZEP cDNA
Total RNA was extracted from alfalfa leaves with the RNeasy extraction Kit (Invitrogen, Carlsbad, CA, USA). First strand complementary DNA (cDNA) was generated using Superscript III reverse transcriptase (Invitrogen, USA). Based on the sequence of Medicago truncatula ZEP gene, two primers (ZEP-F/ZEP-R, Table 1) were used to amplify alfalfa partial cDNA sequence. Products were cut from the gel and TA cloned using the pMD-18T vector kit (Takara, Japan), and sequenced by Invitrogen Co. (Shanghai, China). To obtain the full-length MsZEP cDNA, a SMART™ RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions was used. RACE primers were designed using Primer Premier 5 software. The primers of 5′ GSP and 5′ NGSP (Table 1) were used for 5′ RACE PCR, and 3′ GSP (Table 1) was used for 3′ RACE PCR. PCR was performed using a touch down cycling profile starting with five cycles of amplification at 94 °C/30 s, 72 °C/3 min, followed by another five cycles at 94 °C/30 s, 70 °C/30 s, 72 °C/3 min, then 25 cycles at 94 °C/30 s, 68 °C/30 s, 72 °C/4 min, and finally an extension at 72 °C for 10 min. PCR products were electrophoresed on 1.0 % agarose gel, gel purified and cloned into pMD18-T (TaKaRa, Japan) plasmid vector, then sequenced by Invitrogen Co. (Shanghai, China). The full-length cDNA sequences were created by assembly of the obtained partial cDNA, 3′ and 5′ fragments. Subsequently, primers (LZEP-F and LZEP-R, Table 1) were designed to obtain the full-length cDNA sequence.
Sequence analysis
The nucleotide sequence, deduced amino acid sequence and open reading frame (ORF) of MsZEP were analyzed using bioinformatic tools at the websites (http://www.expasy.org/ and http://www.ncbi.nlm.nih.gov/). The deduced amino acid sequences alignments were performed and a phylogenetic tree was constructed using neighbor-joining method with DNAMAN version 5.0 (Lynnon Biosoft, Vaudreuil, QC, Canada).
Gene expression analysis
Total RNA was extracted from each tissue with different stress treatments and first strand cDNA was synthesized with a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s instructions. qRT-PCR was performed with Roche FastStart Universal SYBR Green Master on the Roche 480 II real-time PCR detection system (Roche Diagnostics), using alfalfa β-actin gene as a reference gene (Long et al. 2014). Three independent biological replicates and three technical replicates for each sample were used for the qRT-PCR. A melt curve was performed at the end of each reaction to verify PCR product specificity. The qRT-PCR gene expression was quantified from three technical replicates using the 2−ΔΔCT comparative methods as described by Livak and Schmittgen (2001) and calibrated by amplification efficiency, where ΔΔCT = (CT, Target − CT, Actin) Time x − (CT, Target − CT, GAPDH) Time0. The primer sequences for transcript analyses (qZEP-F/qZEP-R and Actin-F/Actin-R) are shown in Table 1.
Nicotiana tabacum transformation
An overexpression vector was constructed based on the pCAMBIA1300-35S-sGFP vector. The ORF of the MsZEP gene was amplified with specific primers modified by SmaI and XbaI restriction sites, MsZEP-SmaI and MsZEP-XbaI. The amplification products were inserted into a pCAMBIA1300-35S-sGFP vector downstream from a CaMV 35S promoter. The MsZEP binary vector was transformed into Agrobacterium tumefaciens GV3101. Nicotiana tabacum was transformed by the leaf disc co-cultivation method (Horsch et al. 1985; Wu et al. 2014). Transgenic plants were selected on MS solid medium containing 30 mg/L hygromycin B and confirmed by semi-quantitative RT-PCR analysis using specific primers (MsZEP-F/MsZEP-R, Table 1) for the MsZEP gene. Five-week-old wild type tobacco plants (WT) and two selected T1 transgenic lines (Lines 3 and 6) were used for salt and drought stress tolerance analysis.
Analysis of drought and salt tolerance in MsZEP-overexpression tobacco
Five-week-old wild type tobacco and two T1 transgenic lines were subjected to drought and salt stress treatments. For drought treatment, well-watered plants were treated by withholding irrigation for 0, 3, 7, and 14 days. For salt treatment, plants from the two transgenic lines and the wild type were watered every day with 1/2 Hoagland nutrient solution supplemented with 200 mM NaCl for 2 weeks, whereas the control was treated with nutrient solution. Chlorophyll fluorescence was measured using pulse-amplitude-modulation (PAM) chlorophyll fluorometer (Heinz-Walz-GmbH, Effeltrich, Germany). Fv/Fm ratio was recorded during a saturating photon pulse (4000 μmol m−2 s−1) using a whole plant (Yang et al. 2014). For imaging stomatal aperture, leaves of plants were fixed as described (Park et al. 2008). The third leaves from wild-type and transgenic plants were harvested in darkness. Epidermal strips prepared from the underside of leaves were placed in centrifugal tube containing 2 mL opening buffer (10 mM Mes/KOH, 50 mM KCl, pH 6.0). Epidermal strips were kept in the opening buffer for 2 h in darkness at 24 °C to standardize the initial state, and then incubated for 3 h in the light (200 μmol m−2 s−1). To investigate the effect of drought on stomatal aperture, epidermal strips were kept in the opening buffer supplemented with 150 mM d-mannitol for 3 h in the light. Stomatal apertures were observed under a light microscope. More than 50 guard cells from each sample were used to image stomatal aperture. Stomatal conductance (Gs) of transgenic and WT plants was determined using a Li-COR 6400 system (Lincoln, NE, US) (Elhaddad et al. 2014). Measurements were taken from three leaves from three separate plants of each line.
Relative water content (RWC) was determined according to Yang et al. (2013). Proline content was determined spectrophotometrically following the method of Bates et al. (1973). Malonyldialdehyde (MDA) content was measured using a modified thiobarbituric acid (TBA) method (Puckette et al. 2007; Yang et al. 2013). The soluble sugar content was determined following the method described by Dreywood (1946). The activity of superoxide dismutase (SOD) was measured by nitroblue tetrazolium (NBT) method (Giannopolitis and Ries 1977) with minor modification. Three plants from each line were used in each experiment. The experiment was repeated three times.
ABA levels in wild-type and transgenic plants were determined following the method described by Pēna-Cortés et al. (1989) and Park et al. (2008) with minor modification. In brief, about 1 g plant material was frozen in liquid nitrogen and grounded to a powder, and then samples were kept in 5 mL 80 % (v/v) methanol overnight in dark at 4 °C. The extract was centrifuged at 2000g for 10 min at 4 °C, and the resulting pellet was re-extracted with another 5 mL 80 % (v/v) methanol, as described above. The extract was purified through Sep-Pak C18 cartridges (Waters, Milford, MA, USA). The supernatants were taken to dryness under a stream of nitrogen and re-dissolved in 1 mL methanol, then filtered through a 0.45 μm syringe filter. ABA levels were quantified using the Phytodetek-ABA kit (made by China Agricultural University), following the manufacturer’s protocol. A standard curve was established on each plate. Raw values for ABA levels were standardized by plant mass and extraction volume.
Besides, the expression levels of two stress-responsive genes from tobacco, NtDREB4 and NtP5CS,were also analyzed (as described above). Tobacco actin gene (NtActin) was used as a reference gene. The primer sequences for transcript analyses (NtDREB4-F/NtDREB4-R; NtP5CS-F/NtP5CS-R and NtActin-F/NtActin-R) were shown in Table 1.
Statistical analysis
All data are presented as the mean ± standard error (SE) from three biological replicates of each experiment. Statistical significance was calculated by analysis of Student’s T test. The significant difference among various treatment groups are represented by ‘*’ at p ≤ 0.05 and ‘**’ at p ≤ 0.01. Analyses were performed with IBM SPSS Statistics 18.0 software. Figures were created using SigmaPlot 10.0 (Systat Software, Inc., Germany).
Results
Isolation and characterization of the MsZEP gene
The full-length ORF of MsZEP contains 1992 bp nucleotides and encodes a protein of 663 amino acid residues (Fig. 1) with a predicted relative molecular mass of 72.97 kD and a theoretical pI of 8.63. Amino acid sequence alignment indicated that the deduced amino acid sequence of MsZEP was highly homologous to other plant ZEP sequences, and it contained four conserved motifs of the plant ZEP proteins: a long monooxygenase domain (236–541), two lipocalin conserved motifs (158–175; 266–296) and a Forkhead associated domain (573–640). According to the alignment, MsZEP shares 99, 80, 74, 73, 71, 71, 70, 69 and 68 % amino acid sequence identities with M. truncatula ZEP (XP_003638746), Glycine max ZEP (NP_001276261), Prunus mume ZEP (XP_008241462), Camellia sinensis ZEP (AJB84624), Morus alba var. multicaulis ZEP (AIU94746), Cucumis melo ZEP (XP_008457782), N. plumbaginifolia ZEP (Q40412.1), Solanum tuberosum ZEP (ADF28630), Arabidopsis thaliana ZEP (NP_851285), respectively (Fig. 2). Phylogenetic analysis showed that MsZEP was grouped into a branch with other legume plants, such as M. truncatula, Cicer arietinum, G. max, Lupinus luteus, Glycine soja, Vigna unguiculata and Phaseolus vulgaris (Fig. 3).
Amino acid sequence alignment of MsZEP from alfalfa with ZEP from other selected plant species. Conserved residues are shaded in black. Gray shading indicate similar residues in more than 80 % of all the sequences. The residues highlighted with boxes indicate two lipocalin conserved motifs, a long monooxygenase domain is shown with a rough solid underline and a Forkhead associated domain is shown with a dash underline. The species and corresponding GenBank accession number are as follows: M. truncatula (XP_003638746); G. max (NP_001276261); P. mume (XP_008241462); C. sinensis (AJB84624); Morus alba var. multicaulis (AIU94746); C. melo (XP_008457782); N. plumbaginifolia (Q40412.1); S. tuberosum (ADF28630); AtZEP, A. thaliana (NP_851285)
Phylogenetic tree of MsZEP and other plant ZEP proteins was constructed by neighbor-joining method with 1000 bootstrap replication using DNAMAN software. GenBank accession number for amino acid sequences: C. melo (XP_008457782); Morus. alba var. multicaulis (AIU94746); Medicago sativa (KM044311); M. truncatula (XP_003638746); C. arietinum (XP_004509142); G. max (NP_001276261); G. soja (KHN26473); P. vulgaris (XP_007155924); V. unguiculata (BAB11934); L. luteus (AHI87686); Malus domestica (XP_008340140); Pyrus × bretschneideri (XP_009343160); P. mume (XP_008241462); A. thaliana (NP_851285); Ricinus communis (XP_002523587); C. sinensis (AJB84624); N. plumbaginifolia (Q40412.1); S. tuberosum (ADF28630); Vitis vinifera (AAR11195)
Tissue-specific expression of MsZEP
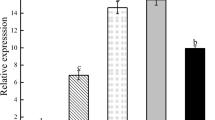
As shown in Fig. 4, although the MsZEP was expressed in all tissues, the expression levels were significantly different (p ≤ 0.01). The transcript levels of MsZEP in leaves were the highest, and that of in stems and roots was about 0.24- and 0.03-fold, respectively.
Relative expression analysis of MsZEP in different tissues of alfalfa. The relative level of mRNA was normalized to that of the Medicago Actin gene. Transcript levels are expressed relative to Actin. Bars represent the mean ± SE (n = 3). Bars with different letters indicate significant difference (p ≤ 0.01)
Expression of MsZEP in response to various stresses and nodules
Using qRT-PCR, the expression of MsZEP in response to drought, cold (4 °C), heat (42 °C) and ABA in either shoots or roots were analyzed. In response to all these treatments, the relative expression levels of MsZEP showed similarly patterns in shoots but varied in roots (Fig. 5). Under drought treatment, the expression levels of MsZEP in the shoots of both NA and NN gradually decreased (Fig. 5a). In contrast to shoots, a significant up-regulation of the MsZEP expression in roots of both NA and NN was observed at 4 h, by up to 3.54- and 2.11-fold of the control, then down to 0.21- and 0.33-fold at 8 h, respectively (Fig. 5b). When seedlings were exposed to 4 °C, MsZEP transcription levels fell sharply in both shoots and roots of NA and NN (Fig. 5c, d). Under the 42 °C heat simulation conditions, the expression patterns of MsZEP in shoots were similar to that at 4 °C (Fig. 5e). The expression levels in roots of NA clearly declined to about half of the control except at 4 h, while the expression of NN fluctuated slowly, and increased at 4 and 8 h (Fig. 5f). For ABA treatment, the transcription levels in shoots decreased in both NA and NN (Fig. 5g). The MsZEP expression in the roots of NA showed no significant changes, except a decrease at 4 and 12 h. In contrast to NA, transcripts of MsZEP in NN increased to 1.8- and 1.2-fold of the control at 2 and 4 h, respectively. When seedlings were subjected to ABA up to 8 h, the MsZEP expression declined and maintained 0.5- to 0.6-fold of the control (Fig. 5h).
Relative expression analysis of MsZEP induced by abiotic stress and ABA in alfalfa inoculated rhizobium (NA) or alfalfa not-inoculated rhizobium (NN). a Expression levels in shoots induced by drought. Rewater, plants were subject to drought stress for 12 h and then put into nutrient solution for 12 h; b expression levels in roots induced by drought. Rewater, plants were subject to drought stress for 12 h and then put into nutrient solution for 12 h; c expression levels in shoots induced by cold; d expression levels in roots induced by cold; e expression levels in shoots induced by heat; f expression levels in roots induced by heat; g expression levels in shoots induced by 10 μM ABA; h expression levels in roots induced by 10 μM ABA. The relative level of mRNA was normalized to that of the Medicago Actin gene. Transcript levels are expressed relative to Actin. Bars represent the mean ± SE of three biological replicates, Asterisk and double asterisk indicate p ≤ 0.05 and p ≤ 0.01, respectively
In addition, the results showed that the transcription of MsZEP in NA was higher (p ≤ 0.01 or p ≤ 0.05) than that of NN in both roots and shoots before stress treatments (Fig. 5). When seedlings were treated with abiotic stress including drought, cold or heat, the expression of the MsZEP of NA in both roots and shoots was significant higher (p ≤ 0.01 or p ≤ 0.05) than that of NN in the earlier stage of these treatments (Fig. 5a–f). For ABA treatment, the expression levels in shoots of NA were also higher (p ≤ 0.01 or p ≤ 0.05) than that of NN (Fig. 5g). However, the transcripts levels in roots of NN were higher (p ≤ 0.01) than that of NA in the earlier stage before 4 h, and then the expression levels in NA were higher than NN in the later stage after 8 h (Fig. 5h).
Function validated of MsZEP-overexpression tobacco toward drought and salt stresses
To investigate whether heterologous expression of MsZEP-affected plant drought and salt stress response, the MsZEP gene was introduced into tobacco. The MsZEP expression levels of three individual T1 tobacco lines were determined using semi-quantitative RT-PCR. The results showed that the expression of MsZEP of transgenic lines is much higher (p ≤ 0.01 or p ≤ 0.05) than that of WT plants (Fig. 6a). Lines 3 and 6 were selected for further analysis.
Reverse transcription PCR amplification of MsZEP transgenic plant lines and effect of drought stress on resistant phenotype of non-transgenic and MsZEP-overexpression tobacco. a Reverse transcription PCR amplification of MsZEP in non-transgenic and MsZEP-overexpression tobacco. WT wild type tobacco, 3, 6, 9, MsZEP-overexpression tobacco T1 plant lines; tobacco actin, loading control; b Wild type and MsZEP transgenic tobacco withholding irrigation for 0 (control), 7, and 14 days. WT wild type tobacco; Lines 3 and 6, MsZEP-overexpression tobacco T1 plant lines
Five-week-old transgenic and WT plants were treated by withholding irrigation. It was found that WT plants were more wilted than transgenic lines under drought stress for 7 and 14 days (Fig. 6b). To examine the physiological changes induced by the overexpression of the MsZEP gene, RWC, proline content, soluble sugar content, MDA content, SOD activity, the maximum photochemical efficiency (Fv/Fm), stomatal aperture and stomatal conductance were measured in the leaves under 7 days drought treatment and 200 mM NaCl treatment. Results showed that the RWC of MsZEP-overexpression lines was significantly higher (p ≤ 0.01 or p ≤ 0.05) than that of WT after 7 days drought treatment (Fig. 8b). No significant difference (p ≤ 0.05) was observed in proline and MDA contents between MsZEP transgenic and WT plants before both the drought and salt stress. Both the proline and MDA contents increased after drought and salt stress in both MsZEP transgenic plants and WT plants, but the proline content in MsZEP transgenic lines was significant higher (p ≤ 0.01 or p ≤ 0.05), and accumulated more quickly compared with WT (Fig. 7a, b). In contrast, the MDA content in MsZEP transgenic lines was significant lower (p ≤ 0.01 or p ≤ 0.05), and accumulated more slowly than that of WT plants (Fig. 7e, f). Similar to proline, soluble sugar content and SOD activity in both MsZEP transgenic and WT plants increased after drought and salt stress, but the contents in MsZEP transgenic lines accumulated significantly higher and more quickly than that of WT (p ≤ 0.01 or p ≤ 0.05) (Fig. 7c, d, g, h).
Effect of drought and salt stress on the physiology of wild type (WT) and MsZEP transgenic tobacco (Lines 3, 6). a Content of free proline in transgenic lines and wild-type tobacco under drought stress for 7 days; b Content of free proline in transgenic lines and wild-type tobacco under salt stress (200 mM NaCl) for 7 days; c Content of the soluble sugar in transgenic lines and wild-type tobacco under drought stress for 7 days; d Content of free the soluble sugar in transgenic lines and wild-type tobacco under salt stress (200 mM NaCl) for 7 days; e Content of the MDA in transgenic lines and wild-type tobacco under drought stress for 7 days; f Content of free MDA in transgenic lines and wild-type tobacco under salt stress (200 mM NaCl) for 7 days; g Activity of SOD in transgenic lines and wild-type tobacco under drought stress for 7 days; h Activity of SOD in transgenic lines and wild-type tobacco under salt stress (200 mM NaCl) for 7 days. Bars represent the mean ± SE (n = 3), asterisk and double asterisk indicate p ≤ 0.05 and p ≤ 0.01, respectively
As shown in Fig. 9a, b, the maximum photochemical efficiency of PS II (Fv/Fm) in MsZEP transgenic lines was significantly higher (p ≤ 0.01 or p ≤ 0.05) than that of WT before and after drought and salt stress, and the stomatal conductance (Gs) was significantly higher (p ≤ 0.01 or p ≤ 0.05) in MsZEP transgenic plants than that of WT line under drought and salt stress. Besides, the stomatal aperture of MsZEP transgenic plants was smaller than that of WT under light and drought treatment (Fig. 8a).
Stomatal aperture and RWC in wild type and MsZEP transgenic tobacco during drought stress. a Stomatal aperture of wild type (WT) and MsZEP transgenic tobacco (Lines 3, 6) under dark, light and 150 mM D-mannitol treatment, bars 100 μm; b RWC of wild type (WT) and MsZEP transgenic tobacco (Lines 3, 6) under drought stress for 0, 3, 7 and 14 days. Bars represent the mean ± SE (n = 3), double asterisk indicates p ≤ 0.01
ABA levels either at drought and salt stress conditions or in transgenic plants were measured using an immunoassay. As shown in Fig. 10, the ABA content in MsZEP-overexpression tobacco was significantly higher (p ≤ 0.01) than that of WT before and after drought and salt stress. In addition, our results demonstrated that the expression levels of two endogenous genes, DREB and P5CS, were significantly higher (p ≤ 0.01 or p ≤ 0.05) in MsZEP transgenic plants than WT plants, under both drought and salt stress (Fig. 9c, d).
Effect of drought and salt stress on Fv/Fm, Gs and two stress-responsive genes in MsZEP transgenic lines (Lines 3, 6) and wild-type (WT) tobacco. a Fv/Fm ratio in MsZEP transgenic lines (Lines 3, 6) and wild-type (WT) tobacco under drought stress (7 days) and salt stress (200 mM NaCl). b Gs in MsZEP transgenic lines (Lines 3, 6) and wild-type (WT) tobacco under drought stress (7 days) and salt stress (200 mM NaCl). c Expression level of DREB in MsZEP transgenic lines (Lines 3, 6) and wild-type (WT) tobacco induced by drought stress (7 days) and salt stress (200 mM NaCl). d Expression level of P5CS in MsZEP transgenic lines (Lines 3, 6) and wild-type (WT) tobacco induced by drought stress (7 days) and salt stress (200 mM NaCl). Bars represent the mean ± SE (n = 3), asterisk and double asterisk indicate p ≤ 0.05 and p ≤ 0.01, respectively
Discussion
It is demonstrated that the enzyme ZEP plays an important role in the ABA biosynthesis and the photoprotective xanthophyll cycle. The ZEP genes have been isolated in many plants, such as N. plumbaginifolia (Audran et al. 1998), Arabidopsis (North et al. 2005), tomato (Thompson et al. 2000) and Gentiana lutea (Zhu et al. 2002). Further studies have shown that the regulation of ZEP gene is reflected at the transcript levels (Thompson et al. 2000; Borel et al. 2001; Schwarz et al. 2014). Here, we have cloned a novel ZEP gene from alfalfa, named MsZEP. The MsZEP shared a high degree of identity with ZEP proteins from other plants (Fig. 2), especially with M. truncatula, indicating that the MsZEP protein is a typical ZEP protein. Besides, the MsZEP protein was the nearest to ZEP from M. truncatula, and clustered together with other legume plants in the phylogenetic tree (Fig. 3). These results implicated the evolution of the MsZEP and the orthologous and paralogous relationships of ZEP protein within the legumes.
Transcripts of the ZEP gene were found in all tissues of different plants, including flowers, seeds, roots, leaves and stems (Audran et al. 1998; Thompson et al. 2000; Ruiz-Sola et al. 2014). In agreement with previous findings (Audran et al. 1998), the MsZEP mRNA was expressed in all alfalfa tissues, but transcript levels were found to be higher in green tissues (stems and leaves) than in roots (Fig. 4). Recently, Schwarz et al. (2014) reported that the ZEP was a tissue-specific accumulation protein, and it was mostly localized in leaf chloroplasts and root plastids. Within chloroplasts, ZEP was mainly deposited in the thylakoid membrane and stroma.
Zeaxanthin epoxidase (ZEP) plays important roles in responding to various environmental stresses due to its functions in the ABA biosynthesis and xanthophyll cycle. It is widely reported that ABA content increases upon abiotic stress, such as drought, salt, cold and heat. However, the transcript levels of ZEP vary among species and tissues under different environmental stresses. In Arabidopsis, N. plumbaginifolia and tomato, the expression levels of ZEP under drought stress are up-regulated in roots, but down-regulated (Audran et al. 1998; Thompson et al. 2000; North et al. 2005) or unchanged (Ruiz-Sola et al. 2014) in leaves; the transcripts of ZEP in both roots and leaves of cowpea are not changed under drought stress (Iuchi et al. 2000). In the present study, expression of MsZEP was down-regulated significantly in shoots of both NA and NN under drought, cold, heat and ABA treatments, while the expression levels in roots shows different trends under different stresses (Fig. 5). In contrast to the previous studies (Audran et al. 1998; Thompson et al. 2000), MsZEP transcripts in roots of both NA and NN were up-regulated at 4 h after drought stress, but then down-regulated at 8 h (Fig. 5b). These results suggested that the expression of alfalfa ZEP gene (MsZEP) in roots is up-regulated by rapid drought stress and down-regulated by progressive drought stress. The expression of ZEP gene in tomato was not induced by temperature, but overexpression of ZEP gene can enhance the sensitivity to chilling stress (Wang et al. 2008). Similar to the expression patterns in shoots, the MsZEP gene in roots also decreased clearly under cold stress conditions (Fig. 5d). For heat stress, the transcript levels of MsZEP fluctuated in root of both NA and NN, but the fluctuating range was larger in NA (Fig. 5f). However, the transcript levels of ABA1/ZEP in imbibed seeds of Arabidopsis were up-regulated at high temperature (Toh et al. 2008). ABA feedback stimulates the expression of the biosynthetic genes, which is also likely through a Ca2+ dependent phosphoprotein cascade (Xiong and Zhu 2003). Exogenous ABA enhanced the expression of ZEP in Arabidopsis (Xiong et al. 2002). However, the present study showed that exogenous ABA down-regulated the expression of MsZEP in shoots of both NA and NN, while the expression in roots of NN was up-regulated in the earlier stage of treatment and then decreased, and no significant changes were found in NA except a decrease at 4 and 12 h. Moreover, we found that the expression of the MsZEP in both roots and shoots of NA was higher than that of NN under non-stressful conditions and in the earlier stage of different abiotic stress treatments (Fig. 5). These results indicated that nodules can up-regulated the expression of ZEP gene in both roots and shoots of alfalfa under non-stressful conditions and rapid abiotic stress such as drought, cold and heat.
Abiotic stresses such as drought and salt can cause membrane injury, decrease leaf relative water content, reduce hydrolytic enzyme activity, increase lipid peroxidation level, induce stomatal closure to prevent from water loss and reduce plants’ photosynthetic capabilities (Medrano et al. 2002; Demiral and Türkan 2005; Tarchoune et al. 2010). These adaptable strategies are necessary for plants to combat against abiotic stresses. MDA, as an end product of lipid peroxidation, has been used extensively as an indicator for membrane injury under various abiotic stress conditions (Yang et al. 2014). Proline and soluble sugar are generally assumed to serve as a physiologically compatible solute that increases as needed to maintain a favorable osmotic potential between the cell and its surroundings (Chaves et al. 2003). SOD is one of the important antioxidant enzymes to scavenge reactive oxygen species (ROS) serving a protective function during oxidative stress when plants are subjected to drought and salt (Kim et al. 2010). Under drought and salt stress, the increase of SOD enzyme activity has been reported in different plants such as maize, tomato and Arabidopsis (Hernandez et al. 2000; Tarchoune et al. 2010). An early study reported that the ZEP gene involved in the regulation of ABA biosynthesis in roots and contributed to the plant response to drought (Audran et al. 2001). It was also shown that overexpression of a ZEP gene could increase the sensitivity to high light and chilling stress in tomato (Wang et al. 2008), and enhance salt and drought tolerance in Arabidopsis (Park et al. 2008). In the present study, the MsZEP-overexpression plants showed higher proline content, higher soluble sugar content, higher SOD activity and lower MDA content compared to WT plants, under both drought and salt stress (Fig. 7). These results suggested that MsZEP-overexpression plants could enhance drought and salt tolerance by regulating their physiological and biochemical processes.
Stoma regulates the gas exchange and water status of leaves, and protects the plants from extensive water loss during drought stress condition (Saliendra et al. 1995; García-Mata and Lamattina 2001). Here, the stomatal aperture of MsZEP-overexpression plants under light and drought was smaller than that of WT (Fig. 8a), suggesting that these plants would be expected to lose less water. RWC is used as the most meaningful index of water stress tolerance including drought and salt stress (Nayyar and Gupta 2006). In this study, the results showed that the RWC of MsZEP-overexpression plants was higher than that of WT plants under drought stress (Fig. 8b). These results suggested that MsZEP-overexpression tobacco enhanced drought tolerance by the closure of stomatal aperture to reduce water loss. Recently, Park et al. (2008) reported the similar function in Arabidopsis under drought conditions.
The leaf maximal photochemical efficiency (Fv/Fm) represents the photosynthetic capability of plants. It is acknowledged that the abiotic stresses such as drought and salt adversely affect plant photosynthesis (Negi et al. 2015). Here, we found that MsZEP-overexpressing tobacco maintained higher Fv/Fm under both drought and salt condition in comparison to WT plants (Fig. 9a). Stomatal conductance (Gs) was regarded as a reference parameter in plant photosynthesis regulation (Medrano et al. 2002). In the present study, stomatal conductance of MsZEP-overexpression plants was higher compared to WT plants under both drought and salt stress (Fig. 9b). These results demonstrated that MsZEP-overexpression plants showed stronger photosynthetic capability.
ABA has been postulated as the main regulator in mediating stomatal responses to environmental stimuli (Dodd 2003). A recently study reported that overexpression of NCED gene could enhance drought resistance in petunia by increasing transcripts of NCED mRNA, elevating leaf ABA and proline contents (Estrada-Melo et al. 2015). In this study, MsZEP-overexpression tobacco showed higher ABA content under both normal and stress condition (Fig. 10). These results suggested that MsZEP-overexpression tobacco enhanced drought and salt tolerance by elevating endogenous ABA levels, and the increase of ABA levels may be due to the expression of MsZEP in transgenic plants.
Dansana et al. (2014) reported that OsiSAP1 overexpression enhanced drought stress tolerance in transgenic rice by affecting expression of endogenous stress-responsive genes. The expression levels of stress-responsive genes including RD29A, RD29B, RD19, RD22 and P5CS in Arabidopsis LOS6/ABA1 mutants, deficient in AtZEP, all decreased under osmotic stress (Xiong et al. 2002). In our study, the results showed that the expression of endogenous DREB gene and P5CS was much higher in MsZEP-overexpressing plants than in WT plants, under both drought and salt stress (Fig. 9c, d), suggesting that MsZEP-overexpressing plants confer drought and salt tolerance by affecting expression of endogenous stress-responsive genes.
In summary, a novel ZEP gene, MsZEP, was cloned and characterized in alfalfa. The MsZEP may be involvement of alfalfa responses to drought, cold and heat, demonstrating that the MsZEP gene is under stress regulation. Besides, nodules can upregulate the MsZEP expression levels in alfalfa. Heterologous expression of MsZEP conferred drought and salt stress tolerance in transgenic tobacco. MsZEP-overexpression plants showed higher RWC, higher proline and soluble sugar contents, higher SOD activity and lower MDA content under drought and salt stress. Moreover, smaller stomatal aperture, higher stomatal conductance, higher Fv/Fm ratio and higher ABA levels were also detected in MsZEP-overexpression plants. We also found that the expression levels of stress-responsive genes such as DREB and P5CS were significantly up-regulated in MsZEP-overexpression plants. Taken together, these results suggested that the MsZEP gene may be involved in alfalfa responses to different abiotic stresses and nodules, and could affect various physiological pathways, ABA levels, stomatal aperture and stress-responsive genes to enhance the salt and drought tolerance in transgenic plants. Furthermore, this study would provide valuable insights into the role of nodules in regulating MsZEP gene expression patterns under abiotic stresses and a candidate gene for enhancing plant salt and tolerance.
Abbreviations
- ABA:
-
Abscisic acid
- Fv/Fm:
-
Maximum photochemical efficiency
- GFP:
-
Green fluorescent protein
- Gs:
-
Stomatal conductance
- MDA:
-
Malonyldialdehyde
- NA:
-
Alfalfa inoculated rhizobium
- NN:
-
Alfalfa not-inoculated rhizobium
- ORF:
-
Open reading frame
- qRT-PCR:
-
Quantitative real-time PCR
- RACE:
-
Rapid amplification of cDNA ends
- RWC:
-
Relative water content
- SOD:
-
Superoxide dismutase
- Vx:
-
Violaxanthin
- ZEP:
-
Zeaxanthin epoxidase
- Zx:
-
Zeaxanthin
References
Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol 125:1248–1257
Audran C, Borel C, Frey A, Sotta B, Meyer C, Simonneau T, Marion-Poll A (1998) Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol 118:1021–1028
Audran C, Liotenberg S, Gonneau M, North H, Frey A, Tap-Waksman K, Vartanian N, Marion-Poll A (2001) Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Funct Plant Biol 28:1161–1173
Baron KN, Schroeder DF, Stasolla C (2012) Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci 188:48–59
Barrero JM, Piqueras P, Gonzalez-Guzman M, Serrano R, Rodriguez PL, Ponce MR, Micol JL (2005) A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot 56:2071–2083
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Bianco C, Defez R (2009) Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J Exp Bot 60:3097–3107
Borel C, Audran C, Frey A, Marion-Poll A, Tardieu F, Simonneau T (2001) N. plumbaginifolia zeaxanthin epoxidase transgenic lines have unaltered baseline ABA accumulations in roots and xylem sap, but contrasting sensitivities of ABA accumulation to water deficit. J Exp Bot 52:427–434
Chaves MM, Maroco JP, Pereira JS (2003) Understanding plant responses to drought—from genes to the whole plant. Funct Plant Biol 30:239–264
Clement M, Lambert A, Herouart D, Boncompagni E (2008) Identification of new up-regulated genes under drought stress in soybean nodules. Gene 426:15–22
Dansana PK, Kothari KS, Vij S, Tyagi AK (2014) OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep 33:1425–1440
Delgado M, Ligero F, Lluch C (1994) Effects of salt stress on growth and nitrogen fixation by pea, faba-bean, common bean and soybean plants. Soil Biol Biochem 26:371–376
DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57:711–738
Demiral T, Türkan I (2005) Comparative lipid peroxidation, antioxidant defense systems and proline content in roots of two rice cultivars differing in salt tolerance. Environ Exp Bot 53:247–257
Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GE (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20:2681–2695
Dodd IC (2003) Hormonal interactions and stomatal responses. J Plant Growth Regul 22:32–46
Dreywood R (1946) Qualitative test for carbohydrate material. Ind Eng Chem Anal Ed 18:499
Egamberdieva D, Jabborova D, Wirth S (2013) Alleviation of salt stress in legumes by co-inoculation with Pseudomonas and Rhizobium. In: Arora NK (ed) Plant microbe symbiosis: fundamentals and advances, 11th edn. Springer, New York, pp 292–299
Elhaddad NS, Hunt L, Sloan J, Gray JE (2014) Light-induced stomatal opening is affected by the guard cell protein kinase APK1b. PLoS One 9(5):e97161
Estrada-Melo AC, Reid MS, Jiang C-Z (2015) Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Hortic Res 2:15013
Fujihara S (2008) Biogenic amines in rhizobia and legume root nodules. Microbes Environ 24:1–13
García-Mata C, Lamattina L (2001) Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol 126:1196–1204
Giannopolitis CN, Ries SK (1977) Superoxide dismutases I. Occurrence in higher plants. Plant Physiol 59:309–314
Hernandez J, Jimenez A, Mullineaux P, Sevilia F (2000) Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ 23:853–862
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circ Calif Agric Exp Stn 47:32
Horsch R, Fry J, Hoffmann N, Eichholtz D, Sa Rogers, Fraley R (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Hou X, Liang Y, He X, Shen Y, Huang Z (2013) A novel ABA-responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in Arabidopsis thaliana. Plant Mol Biol Rep 31:791–801
Hungria M, Kaschuk G (2014) Regulation of N2 fixation and NO3−/NH4+ assimilation in nodulated and N-fertilized Phaseolus vulgaris L. exposed to high temperature stress. Environ Exp Bot 98:32–39
Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123:553–562
Jin T, Chang Q, Li W, Yin D, Li Z, Wang D, Liu B, Liu L (2010) Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Tissue Org 100:219–227
Kim MD, Kim YH, Kwon SY, Yun DJ, Kwak SS (2010) Enhanced tolerance to methyl viologen-induced oxidative stress and high temperature in transgenic potato plants overexpressing the CuZnSOD, APX and NDPK2 genes. Physiol Plant 140:153–162
Kovács I, Ayaydin F, Oberschall A, Ipacs I, Bottka S, Pongor S, Dudits D, Tóth ÉC (1998) Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. Plant J 15:185–197
Lind C, Dreyer I, López-Sanjurjo EJ, von Meyer K, Ishizaki K, Kohchi T, Lang D, Zhao Y, Kreuzer I, Al-Rasheid KA (2015) Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr Biol 25:928–935
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Long R, Wang H, Shen Y, Kang J, Zhang T, Sun Y, Zhang Y, Li M, Yang Q (2014) Molecular cloning and functional analysis of a salt-induced gene encoding an RNA-binding protein in alfalfa. Mol Breed 34:1465–1473
Luo M, Liu J, Mohapatra S, Hill R, Mohapatra S (1992) Characterization of a gene family encoding abscisic acid-and environmental stress-inducible proteins of alfalfa. J Biol Chem 267:15367–15374
Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15:2331
Medrano H, Escalona JM, Bota J, Gulías J, Flexas J (2002) Regulation of photosynthesis of C3 plants in response to progressive drought: stomatal conductance as a reference parameter. Ann Bot-Lond 89:895–905
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Nayyar H, Gupta D (2006) Differential sensitivity of C3 and C4 plants to water deficit stress: association with oxidative stress and antioxidants. Environ Exp Bot 58:106–113
Negi NP, Shrivastava DC, Sharma V, Sarin NB (2015) Overexpression of CuZnSOD from Arachis hypogaea alleviates salt and drought stress in tobacco. Plant Cell Rep 34:1109–1126
North HM, Frey A, Boutin JP, Sotta B, Marion-Poll A (2005) Analysis of xanthophyll cycle gene expression during the adaptation of Arabidopsis to excess light and drought stress: changes in RNA steady-state levels do not contribute to short-term responses. Plant Sci 169:115–124
Park HY, Seok HY, Park BK, Kim SH, Goh CH, Lee Bh, Lee CH, Moon YH (2008) Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochem Biophys Res Commun 375:80–85
Pēna-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S (1989) Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci 86:9851–9855
Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol R 64:180–201
Puckette MC, Weng H, Mahalingam R (2007) Physiological and biochemical responses to acute ozone-induced oxidative stress in Medicago truncatula. Plant Physiol Biochem 45:70–79
Ruiz-Sola MÁ, Arbona V, Gómez-Cadenas A, Rodríguez-Concepción M, Rodríguez-Villalón A (2014) A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS One 9:e90765
Saliendra NZ, Sperry JS, Comstock JP (1995) Influence of leaf water status on stomatal response to humidity, hydraulic conductance, and soil drought in Betula occidentalis. Planta 196:357–366
Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410:327–330
Schwarz N, Armbruster U, Iven T, Brückle L, Melzer M, Feussner I, Jahns P (2014) Tissue-specific accumulation and regulation of zeaxanthin epoxidase in Arabidopsis reflect the multiple functions of the enzyme in plastids. Plant Cell Physiol 56:346–357
Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62:2615–2632
Seung D, Risopatron JPM, Jones BJ, Marc J (2012) Circadian clock-dependent gating in ABA signalling networks. Protoplasma 249:445–457
Siriwardana CL, Kumimoto RW, Jones DS, Holt BF III (2014) Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol Biol Rep 32:971–986
Sreenivasulu N, Harshavardhan VT, Govind G, Seiler C, Kohli A (2012) Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506:265–273
Suárez R, Wong A, Ramírez M, Barraza A, Orozco MdC, Cevallos MA, Lara M, Hernández G, Iturriaga G (2008) Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol Plant Microbe Interact 21:958–966
Tarchoune I, Sgherri C, Izzo R, Lachaal M, Ouerghi Z, Navari-Izzo F (2010) Antioxidative responses of Ocimum basilicum to sodium chloride or sodium sulphate salinization. Plant Physiol Biochem 48:772–777
Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42:833–845
Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146:1368–1385
Wang N, Fang W, Han H, Sui N, Li B, Meng QW (2008) Overexpression of zeaxanthin epoxidase gene enhances the sensitivity of tomato PSII photoinhibition to high light and chilling stress. Physiol Plant 132:384–396
Wu D, Ji J, Wang G, Guan C, Jin C (2014) LchERF, a novel ethylene-responsive transcription factor from Lycium chinense, confers salt tolerance in transgenic tobacco. Plant Cell Rep 33:2033–2045
Xiong L, Zhu J-K (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133:29–36
Xiong L, Lee H, Ishitani M, Zhu J-K (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277:8588–8596
Yang P, Zhang P, Li B, Hu T (2013) Effect of nodules on dehydration response in alfalfa (Medicago sativa L.). Environ Exp Bot 86:29–34
Yang Y, Sun X, Yang S, Li X, Yang Y (2014) Molecular cloning and characterization of a novel SK3-type dehydrin gene from Stipa purpurea. Biochem Biophys Res Commun 448:145–150
Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Ann Rev Plant Physiol Plant Mol Biol 39:11439–11473
Zhu C, Yamamura S, Koiwa H, Nishihara M, Sandmann G (2002) cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol Biol 48:277–285
Acknowledgments
This work was supported by the Project of National Natural Science Foundation of China (31372357, 31272490), the National Key Technology R&D Program in the 12th 5-year plan of China (2011BAD17B05) and the major project for Tibetan forage industry (Z2014C02N02).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Author contribution statement
Z.Q. Zhang performed the experiment, analyzed data and wrote the manuscript; Y.F. Wang cloned and analyzed the gene; L.Q. Chang, T. Zhang and J. An sampled the material and determined physiological indexes; Y.S. Liu and Y.M. Cao performed transgenic tobacco experiment; X. Zhao and X.Y. Sha performed qRT-PCR experiment; P.Z. Yang and T.M. Hu provided ideas, designed the research, and edited the manuscript; all authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by L. Peña.
Rights and permissions
About this article
Cite this article
Zhang, Z., Wang, Y., Chang, L. et al. MsZEP, a novel zeaxanthin epoxidase gene from alfalfa (Medicago sativa), confers drought and salt tolerance in transgenic tobacco. Plant Cell Rep 35, 439–453 (2016). https://doi.org/10.1007/s00299-015-1895-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1895-5