Abstract
Key message
TaWD40D that encodes a member of WD40 family proteins is a novel gene involved in the wheat response to abiotic stress. TaWD40D functions as a positive regulator of plant responses to salt stress and osmotic stress in plant.
Abstract
Abiotic stresses can severely affect plant growth and crop productivity. WD40 repeat-containing proteins play a key role in protein–protein or protein–DNA interactions by acting as scaffolding molecules and promoting protein activity. In this study, a stress-inducible gene, TaWD40D, was identified from Chinese spring wheat (Triticum aestivum L.). TaWD40D encodes a protein containing seven WD40 domains. Subcellular localization in Nicotiana benthamiana mesophyll cells and Arabidopsis root cells showed the presence of TaWD40D in the cytoplasm and nucleus. Heterologous overexpression of TaWD40D in Arabidopsis greatly increased plant tolerance to abscisic acid (ABA), salt stress, and osmotic stress during seed germination and seedling development. The expression patterns of two genes from the SOS pathway (SOS2 and SOS3) and three ABA genes (ABI2, RAB18 and DREB2A) functioning in ABA-dependent and ABA-independent pathways were altered in the transgenic lines overexpressing TaWD40D under the treatments. Notably, the basal level of the ABI2 expression was substantially increased in the TaWD40D overexpression lines. The down-regulation of TaWD40D in wheat by virus-induced gene silencing resulted in a decreased relative water content and less vigorous growth compared to non-silenced lines. Our results suggest that TaWD40D functions as a positive regulator of plant responses to salt stress and osmotic stress that could be utilized for the genetic improvement of stress tolerance in crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses, such as drought and salinity, limit the areas where crops can be produced and adversely affect crop quality and productivity worldwide. To overcome these limitations, plants have evolved various mechanisms to respond to and cope with environmental stresses at both the physiological and biochemical levels (Byrt and Munns 2008; Munns and Tester 2008; Zhu 2002). Both high salinity and drought can cause hyperosmotic stress that can injure plant cells (Adamec 1984), thus, osmotic adjustment is considered to be a common adaptive strategy for plant survival under conditions of drought and high salinity. In addition to osmotic stress, high salinity can cause cellular damage by disrupting ion homeostasis, resulting in subsequent cellular malfunction and even cell death (Shabala 2009; Zhu 2003). Therefore, the maintenance of cellular ionic equilibrium is critical for plant adaptation to salt stress conditions.
During the past two decades, significant progress has been made in understanding the molecular mechanisms underlying plant physiological and biochemical responses to osmotic stress and high salinity using Arabidopsis as a model organism. Indeed, many genes involved in osmosensing (Tran et al. 2007; Wohlbach et al. 2008), abscisic acid (ABA) biosynthesis (Bray 1997; Iuchi et al. 2001; Seo et al. 2004), osmolyte biosynthesis and metabolism (Kishor et al. 2005), and stress signal transduction pathways (Knight and Knight 2001) have been identified. Notably, several signaling pathways that regulate plant responses to salt or osmotic stress have been defined, including the salt overly sensitive (SOS) pathway, which mediates plant-specific responses to ionic stress, and ABA-dependent and dehydration-responsive element-binding protein (DREB) mediated (ABA-independent) pathways, which modulate plant adaptation to drought (Budak et al. 2013; Ergen et al. 2009; Mahajan and Tuteja 2005; Raghavendra et al. 2010; Vinocur and Altman 2005). Stress tolerance is a complex trait involving many genes and multiple pathways, however, accumulating evidence from the past two decades has revealed multifaceted cross-talk between ABA-dependent and -independent pathways, as well as the SOS signaling pathway. It has been shown that a range of regulatory factors is involved in both ABA-dependent and -independent signaling (Chinnusamy et al. 2004; Haake et al. 2002; Narusaka et al. 2003). For example, SOS2, a protein kinase and a key component of the SOS pathway (Qiu et al. 2002), is able to physically interact with ABA-insensitive 2 (ABI2), a protein phosphatase 2C family protein that is a key negative regulator of the ABA signal transduction pathway (Ohta et al. 2003; Rodriguez et al. 1998). Thus, the SOS signaling pathway is directly connected to the ABA signaling pathways, so that plants are able to cope with ionic and osmotic stresses caused by high salinity.
Despite significant progress in our understanding of the mechanisms involved in plant stress responses and adaptation, a coherent picture of how these components and signaling pathways act in concert to regulate the homeostatic responses of plant cells to salt and water stress has not yet emerged. Scaffold proteins are key regulators of many signaling pathways that interact and/or bind with multiple components of a signaling pathway or the components of different pathways, tethering them into complexes that switch signaling on/off or cross-link pathways together (Ananieva et al. 2008; Hall and Lefkowitz 2002; Heinrich et al. 2002). WD40 repeat-containing proteins are a group of proteins that serve as platforms for the assembly of protein complexes or mediate transient interplay among other proteins. They are highly conserved and are extremely abundant in eukaryotic organisms (Stirnimann et al. 2010). A common feature of WD40 repeat-containing proteins is a short core region of approximately 40 amino acids that contains a glycine–histidine (GH) dipeptide at the N terminus and a C-terminal tryptophan-aspartate (WD) dipeptide separated by a linker of variable length (Smith et al. 1999). Studies have shown that WD40 proteins are part of a broad spectrum of components in the cytoplasm and nucleoplasm (Ouyang et al. 2012) that may play key roles in a number of important biological processes, including signal transduction, protein trafficking, chromatin modification, and transcription (Smith 2008; van Nocker and Ludwig 2003). In Arabidopsis, several WD40 repeat-containing proteins have been shown to mediate plant growth and development (McNellis et al. 1994; Ramsay and Glover 2005). A recent genome-wide analysis of stress-responsive genes in rice revealed five genes encoding Salt Responsive WD40 proteins (SRWD1-5) that are responsive to salt stress, and the expression of SRWD1 may be correlated with the sensitivity of rice cultivars to salt stress (Huang et al. 2008). Another gene encoding a putative WD40 protein, Setaria italica WD40 (SiWD40), in foxtail millet was also highly induced by various abiotic stresses and may be regulated by dehydration-responsive elements during environmental stress (Mishra et al. 2012b). Most recently, Arabidopsis GIGANTUS1 (GTS1), a WD40 repeat-containing protein, was also shown to mediate seed germination and biomass accumulation through a protein interaction in Arabidopsis (Gachomo et al. 2014). Those results highlight the importance of WD40 repeat-containing proteins in plant stress responses and the urgent need for functional analyses of WD40 proteins.
Wheat (Triticum aestivum L.) is one of the most important crops in the world. Drought and salt are the most common environmental stresses affecting wheat growth and grain yields (Morran et al. 2011; Rhoades and Loveday 1990). Although significant progress has been made in understanding the physiological mechanisms underlying wheat responses to drought and high salinity, the molecular mechanisms of stress tolerance to drought and salt in wheat remain largely unknown. Previously, we identified a wheat gene encoding a WD40 repeat-containing protein (TaWD40D) that was highly induced by polyethylene glycol (PEG)-simulated water stress based on microarray data (unpublished data). In this study, we found that TaWD40D is responsive to both drought and salt, and that the expression of TaWD40D is also regulated by ABA. TaWD40D encodes a functionally unknown protein containing seven WD40 repeats that is expressed in both the cytoplasm and nucleus. Furthermore, we showed that overexpression of TaWD40D in Arabidopsis significantly reduced the sensitivity of the transgenic plants to osmotic stress and ABA. Virus-induced gene silencing (VIGS) of TaWD40D resulted in reduced drought tolerance in wheat seedlings. Our results indicate that TaWD40D is a positive regulator of stress tolerance in plants.
Materials and methods
Plant materials and abiotic stress experiments
Hexaploid wheat (T. aestivum L. Cv. Chinese spring) plants were used in this study. Following surface sterilization with 50 % ethanol for 2 min and washing twice with sterilized water, wheat seeds were germinated and cultured with double-distilled water in a growth chamber under a 16-/8-h photoperiod at 23 °C. Wheat seedlings at the two-leaf stage were subjected to stress by replacing the hydroponic medium with 300 mM NaCl, 20 % (w/v) PEG 6000, and 100 µM ABA. To investigate the expression of the target gene at different developmental stages, wheat seedling leaves and roots, spindle leaves at booting, and spikes at the heading stage were sampled. Seedlings were grown in a greenhouse without environmental stress. To further identify the genomic location of the target gene, 21 nulli-tetrasomic (NT) lines of Chinese spring were used for chromosome location (Sears 1966).
All stocks of Arabidopsis were in the same genetic background (Columbia [Col-0] ecotype) and were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Plants were grown either in soil or on half-strength Murashige and Skoog (MS; pH 5.7) medium (Murashige and Skoog 1962) containing 2 % w/v sucrose at 22 °C in a growth room under a 16-/8-h light/dark photoperiod. Abiotic stress treatments were carried out by adding 200 mM NaCl, 300 mM mannitol, and 0.8 µM ABA to MS medium, respectively.
Cloning and sequencing of TaWD40D cDNA
A dramatically upregulated gene (TaWD40D) represented by TaAffx.51040.1.S1_at which derived from a wheat EST (Gene Bank CA731812.1) was selected from Microarray data (6-day-old wheat plants were treated with 20 % PEG6000 solution for 0, 8 and 72 h. Labeled cRNA sample was hybridized to Affymetric GeneChip® containing 55 K wheat transcripts). A wheat cDNA library of Chinese Spring (Professor Jizeng Jia, Chinese Academy of Agricultural Sciences) was screened for cloning of the putative full-length TaWD40D cDNA. Primers were listed in Table S1. The homologues of TaWD40D were searched from European Nucleotide Archive of EMBL-EBI (http://www.ebi.ac.uk/ena), the DFCI wheat gene index database (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gireport.pl?gudb=wheat) and wheat genome database (http://www.wheatgenome.org/). Full-length cDNA sequences and deduced amino acid sequences were produced using NCBI/GenBank/Blast and the CerealsDB (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/Documents/). The detection of conserved domains was done using the SMART motif search program (http://smart.embl-heidelberg.de/).
Phylogenetic tree construction
TaWD40D protein homologs in other plant species were retrieved from GenBank using the BLASTP program. The sequences were aligned by CLUSTALW (2.0), and a phylogenetic tree was constructed using MEGA 5.2 software by the neighbor-joining method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1,000 replicates). The numbers at the nodes indicate the bootstrap values in percentages.
Subcellular localization assay
To create a cauliflower mosaic virus (CaMV) 35S::GFP-TaWD40D fusion, the full-length cDNA sequences were amplified by PCR and cloned in-frame into the binary vector pEGAD between the EcoRI and BamHI sites. Positive recombination was confirmed by sequencing. The plasmids were introduced into Col-0 plants and Nicotiana benthamiana leaves through Agrobacterium tumefaciens-mediated (strain EHA101) transformation. The epidermal cell layers of Arabidopsis roots and N. benthamiana leaves expressing the GFP-TaWD40D fusion were assayed for fluorescence using a confocal microscope at an excitation wavelength of 488 nm (Leica SP8; Leica Microsystems, Wetzlar, Germany).
Gene expression analysis
Total RNA extraction and reverse transcriptase reaction were mentioned above. For reverse transcription RT-PCR analysis, PCR reactions were conducted as follows: 94 °C for 3 min followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Wheat Tubulin (AF326781) and Arabidopsis Actin2 (At3g18780) were used as loading controls for expression analysis in wheat and Arabidopsis, respectively. Real-time quantitative RT-PCR (qRT-PCR) was performed in an ABI PRISM 7500 system and detected using a SYBR Green PCR master mix kit (Applied Biosystems, Carlsbad, CA, USA). Relative gene expression levels were detected using the 2−ΔΔCT method (Livak and Schmittgen 2001). Three biological replicates were performed, and for each biological replicate, at least three technical replicates were performed. Values of expression level are mean ± SE. Statistical significance was reported at P < 0.05 with t test. The sequences of the relevant primers are given in Table S1.
Generation of transgenic Arabidopsis plants
The 35S::GFP-TaWD40D expression cassette was introduced into wild-type plants through A. tumefaciens-mediated transformation using the floral dip method (Clough and Bent 1998). Positive transgenic plants were screened on half-strength MS agar medium supplemented with the commercial herbicide Basta and then confirmed by examining TaWD40D expression using RT-PCR. Southern blotting was performed as described previously (Zhang et al. 2011) to confirm the presence and copy number of TaWD40D. Genomic DNA from the transgenic lines and wild-type plants was isolated and then digested overnight with BamHI without cutting the TaWD40D sequence. The signals on the hybridized blot were captured using a Typhoon Variable Mode Imager (Amersham Biosciences Ltd., Amersham, UK).
Virus-induced gene silencing of TaWD40D in wheat
TaWD40D gene silencing in wheat was performed by the VIGS-Barley Stripe Mosaic Virus (BSMV) method as previously described (Yuan et al. 2011). A 296 bp TaWD40D-specific fragment was amplified and then inserted into the vector pCa-γbLIC (Yuan et al. 2011) using a primer (Table S1) containing an LIC site. For agroinfiltration, BSMVα, BSMVβ, and BSMVγbLIC were mixed equally and infiltrated immediately into N. benthamiana leaves. At 10 days post-infiltration, the infiltrated leaves were harvested and ground in inoculated buffer. Next, leaves from Chinese Spring plants at the two-leaf stage were inoculated into the grinding solution. The original BSMV (BSMV0) was used as a viral control, infection buffer without BSMV transcripts was used as a mock control. BSMVPDS obtained from Professor Dawei Li lab (State Key Laboratory of Agro-Biotechnology, China Agricultural University, Beijing, China) was used as a positive control for silencing of the phytoene desaturase gene (TaPDS) (Yuan et al. 2011), and BSMV WD40D was used for TaWD40D silencing. The transcript levels of TaPDS and TaWD40D were analyzed by RT-PCR at 12 days after infection, and the wheat plants with reduced expression of TaPDS and TaWD40D and the control plants were then used for phenotypic analysis.
Drought treatment and water status determination of the VIGS-TaWD40D wheat plants
For drought treatment, we divided the infected wheat and mock control into two subsets. One set of plants was maintained at 100 % field capacity and stress was imposed on the other set of plants by withholding watering when photobleaching of the BSMVPDS plants appeared at 12 days post-inoculation. Each of the infection groups consisted of 5 plants and individual leaves from third to seventh per plant (total 25 leaves) were evaluated for wilting, symptoms and expression analysis of TaWD40D gene at 20 days after drought treatment based on consistent bleaching phenotype of these leaves in the BSMVPDS positive control. To quantify the wilting level of the leaves, the pooled samples with five leaves per plant were used for measurement of the leaf relative water content (RWC). The leaf relative water content of the wheat plants was estimated according to the method as previously described (Ekanayake et al. 1993). Leaves from stressed and non-stressed plants were weighed to determine the fresh weight (FW). The leaf segments were then floated on deionized water for 24 h to determine their turgid weight (TW). The leaves were then oven-dried for 24 h at 80 °C to a constant dry weight (DW). The RWC was calculated using the formula RWC (%) = [(FW − DW)/(TW − DW)] × 100. Three biological repeats were performed for the above experiment. Values are mean ± SE (n = 5). Statistical significance was reported at P < 0.05 with t test.
Results
Cloning and sequence analysis of TaWD40D
Based on the Microarray data (unpublished data), a dramatically upregulated gene (TaWD40D) represented by TaAffx.51040.1.S1_at which derived from a wheat EST (Gene Bank CA731812.1) was selected and cloned from a wheat cDNA library of Chinese spring. The TaWD40D cDNA is 1,986 bp in length and consists of 174 bp of the 5′-untranslated region (UTR), 1428 bp of the open reading frame (ORF), and 384 bp of the 3′-UTR (Fig. 1a). Blasting European Nucleotide Archive of EMBL-EBI, TaWD40D is located on Scaffold IWGSC_CSS_4DS_scaff_1637308: 1,515–2,129 region which match to 1–613 bp of TaWD40D cDNA and Scaffold IWGSC_CSS_4DS_scaff_2291115: 16,609-15,167 region which match to 544–1,986 bp of TaWD40D cDNA, and there are 70 bp (544–613 bp of TaWD40D) overlapping sequence between these two Scaffold (scaff_1637308: 2,059–2,129 region and scaff_2291115: 16,609–16,539 region). So TaWD40D is located on chromosome 4D which consistent with the data by PCR-based analysis of genomic DNA of Chinese spring nulli-tetrasomic lines using specific primer (Fig. S1), and contains only one exon which confirmed by analyzing the genomic sequence of TaWD40D via PCR amplification (Fig. 1a). The sequence of TaWD40D has been deposited in the GenBank database and the accession number has been signed as KJ511492.
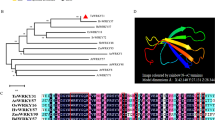
Structure and phylogeny analysis of TaWD40D. a The structure of TaWD40D gene. Exon shown as black box, and the 5′-UTR and 3′-UTR as black bars. b Amino acid sequence of TaWD40D. The WD40 domains are underlined. c Structural analysis of TaWD40D. The two boxes show low complexity regions, while triangles show the WD40 domain. The scale bar indicates the position of the WD40 domain. d Phylogenetic tree of TaWD40D and its homologs from various organisms. Bootstrap values based on 1,000 replications are represented beside the branches. The scale bar indicates the relative amount of change along the branches
Further analysis showed that the TaWD40D ORF encodes a protein of 476 amino acid residues with a predicted molecular mass of 50.16 kDa and a theoretical pI of 6.55 (Fig. 1b). A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and conserved domain analysis based on the predicted peptide sequence of the coding sequence revealed that TaWD40D protein contains seven WD40 domains and belongs to the WD40 protein family (Fig. 1c). A seven-bladed β-propeller structure is the most stable β-sheet geometry and a major factor in WD40 protein identification (Murzin 1992). To see whether the TaWD40D protein has the similar structure, we predicted the three-dimensional structure of TaWD40D. The result revealed that the TaWD40D protein also has a typical seven-bladed β-propeller architecture, with each blade consisting of a four-stranded anti-parallel β sheet (Fig. S2).
Using a BLASTP search of the NCBI database, we found that the deduced amino acid sequence of TaWD40D showed strong similarity to a predicted protein in Hordeum vulgare (GenBank: BAJ99241.1), with 85 % identity and 87 % positives (E value = 0). In addition, TaWD40D was found to be closely related to several uncharacterized proteins from different plants, including an Oryza sativa WD40 domain protein (GenBank: AAR01655.1; 73 % identity; E value = 0), Zea mays nucleotide-binding protein (GenBank: DAA50409.1; 65 % identity; E value = 2e-168), and a Ricinus communis F-box and WD40 domain protein (GenBank: XP_002510317; 51 % identity; E value = 6e-148). To study the evolutionary relationships between TaWD40D and its homologs, we performed a phylogenetic analysis and found that TaWD40D had higher homology with the proteins from monocots (e.g., O. sativa and Z. mays), and lower homology with the proteins from dicots (e.g., Vitis vinifera and Arabidopsis) (Fig. 1d).
Expression patterns of TaWD40D in wheat
To investigate whether TaWD40D has a role in wheat development and the response to abiotic stimuli, we first analyzed the gene expression pattern of TaWD40D using RT-PCR and qRT-PCR assays. As shown in Fig. 2a, TaWD40D was expressed in multiple tissues, including young seedlings, booting spindles, and heading spikes, when Chinese Spring wheat plants were grown under normal conditions. Notably, the transcript level of TaWD40D in the leaves was higher than that in the roots of 2-week-old seedlings (Fig. 2a). When the 2-week-old seedlings were subjected to abiotic stress, the transcript levels of TaWD40D in the leaves were dramatically altered. In the presence of 20 % (w/v) PEG 6000, TaWD40D was upregulated and reached its peak at 3 h of treatment, after which the expression level declined sharply (Fig. 2b). In plants exposed to 300 mM NaCl, TaWD40D expression in the leaves reached its highest level at 1 h after treatment and the transcript level then gradually decreased during prolonged treatment (Fig. 2c). These results suggest that TaWD40D functions in the plant response to osmotic stress and salt stress. To further test whether TaWD40D is regulated by ABA, we treated young wheat seedlings with 100 µM ABA. As shown in Fig. 2d, the expression of TaWD40D was also responsive to ABA, suggesting a role for TaWD40D in ABA-mediated stress responses.
qRT-PCR and RT-PCR analysis of TaWD40D expression patterns in wheat. a Expression patterns of TaWD40D in wheat tissues at different developmental stages. SL seedling leaf, SR seedling root, BS booting spindle, HS heading spike. b Expression patterns of TaWD40D in response to 20 % (w/v) PEG 6000, c 300 mM NaCl, and d 100 µM abscisic acid (ABA). The vertical coordinates are fold changes, while the horizontal ordinates are the treatment time (0, 1, 3, 6, 12, and 24 h). The 2−ΔΔCT method was used to measure the relative expression level of the target gene. The wheat Tubulin gene was used as an internal control. The expression of TaWD40D in seedling leaves (a) or at 0 h (b–d) was regarded as standard, and all other values were compared with it. Error bars represent the standard deviation among three biological replicates. Gels: upper, TaWD40D-specific fragments amplified by RT-PCR. Lower, wheat Tubulin gene fragments amplified by RT-PCR as an internal control
Subcellular localization of TaWD40D
WD40 proteins are localized in a variety of organelles, including the nucleus (Zhang et al. 2006), prevacuolar compartment, and Golgi complex (Lee et al. 2006). To determine the subcellular localization of the TaWD40D protein, we made a construct harboring a GFP-TaWD40D fusion protein under the control of the constitutive CaMV 35S promoter. The expression and distribution of TaWD40D was first examined by the transient transformation of N. benthamiana mesophyll cells. As shown in Fig. 3a, GFP fluorescence in the N. benthamiana mesophyll cells expressing GFP-TaWD40D was observed in the peripheral membrane, cytoplasm, and nucleus. To further validate the subcellular localization of TaWD40D, we generated transgenic Arabidopsis plants expressing GFP-TaWD40D. A confocal microscopic analysis of root cells from the transgenic plants revealed the same subcellular localization of GFP-TaWD40D as in N. benthamiana mesophyll cells (Fig. 3b), suggesting TaWD40D is distributed in multiple cellular locations.
Subcellular localization of TaWD40D. a GFP and a GFP-TaWD40D fusion protein were transiently expressed in N. benthamiana mesophyll cells. Left images, darkfield microscopy. Middle images, brightfield microscopy. Right images, the left and middle images combined. Scale bar 50 µm. b Expression of GFP and GFP-TaWD40D in Arabidopsis root cells
TaWD40D overexpression confers tolerance to salt, mannitol, and ABA during seed germination in Arabidopsis
To further investigate whether TaWD40D mediates stress responses in plants, we generated transgenic Arabidopsis plants overexpressing TaWD40D driven by the CaMV 35S promoter (Fig. 4a), and two T3 homozygotes containing a single copy of TaWD40D (Fig. 4b) were selected for phenotypic analysis. Under normal conditions, there was no obvious difference between the TaWD40D overexpression lines and the wild-type control during the whole growth cycle (Fig. S3). The germination rates of the TaWD40D overexpression lines were also comparable to that of the wild type when germinated on MS medium (Fig. 4c). Intriguingly, when sown on MS medium containing 300 mM mannitol or 200 mM NaCl, the germination rates of the TaWD40D overexpression lines were much higher than those of wild type and plants transformed with empty vector. For example, in the presence of 200 mM NaCl, approximately 70 % of the transgenic seeds germinated at 5th day after stratification, whereas only about 40 % of the wild-type seeds germinated (Fig. 4c). Notably, nearly all of the germinating seedlings of the TaWD40D overexpression lines remained green at 12th day after stratification, by contrast, many germinating seedlings of wild-type and empty vector lines became bleached and died in the presence of 200 mM NaCl (Fig. 4d). These results indicate that ectopic expression of TaWD40D greatly enhances plant tolerance to both osmotic stress and ionic stress in Arabidopsis.
Transgene identification and stress response analysis in wild-type and transgenic plants. a RT-PCR profiles of TaWD40D in wild-type and transgenic plants. The Arabidopsis Actin2 gene was used as an internal control. b Southern blot analysis of wild-type and transgenic Arabidopsis. Genomic DNA from the transgenic lines and wild-type plants was digested with BamHI. A TaWD40D-specific probe was used, and the plasmid DNA carried TaWD40D as a positive control (+). c Effect of 200 mM NaCl, 300 mM mannitol, and 0.8 µM ABA on the germination of seeds. 50 Arabidopsis seeds for each line were planted in MS medium, then the rate of germination was calculated at the 5th day after cold stratification. Radicle extends fully is the criterion of germination. Error bars represent the standard deviation among three biological replicates. Significant differences (t test, *P < 0.05) are marked with asterisks. d Both wild-type and transgenic seeds were germinated in MS medium as a control, while MS medium supplemented with 200 mM NaCl, 300 mM mannitol, and 0.8 µM ABA was used to induce abiotic stress. The day indicates the time when the photos were taken after stratification. EV empty vector transgenic line. OE1 and OE2, TaWD40D overexpression transgenic lines. WT wild type
To examine whether TaWD40D mediates the ABA response, we also analyzed the response of the TaWD40D overexpression lines to ABA. When germinated on MS medium containing 0.8 µM ABA, the transgenic lines overexpressing TaWD40D exhibited obviously higher seed germination rates than wild-type and vector control plants (Fig. 4c). In addition, the transgenic plants overexpressing TaWD40D showed improved growth at 12th day after stratification compared with controls, suggesting that TaWD40D overexpression causes reduced sensitivity of Arabidopsis plants to the ABA-mediated inhibition of postgerminative development (Fig. 4d). Taken together, these results demonstrate that TaWD40D plays an important role in plant responses to abiotic stress.
Heterologous expression of TaWD40D altered the expression of some abiotic stress-related genes in Arabidopsis
To explore how the overexpression of TaWD40D alters the responses of the transgenic plants to abiotic stress, we performed qRT-PCR to analyze the expression profiles of some abiotic stress-responsive genes. These include dehydration inducible genes (DREB1A, DREB2A, RD29A), the genes in ABA biosynthesis and ABA signaling pathway (RD29B, RAB18, ABI1, ABI2, and ABA1), and the salt stress-related genes (SOS1, SOS2, SOS3, NHX1, and HKT1). qRT-PCR analysis results showed that overexpression of TaWD40D affects the expression patterns of some tested genes. As shown in Fig. 5 and Fig. S4, the transcripts of nearly all the tested genes were not changed when the TaWD40D overexpression seedlings were grown under normal growth conditions. However, ABI2 was the exception and its expression was markedly upregulated in the TaWD40D transgenic lines grown under normal conditions (Fig. 5a, f, k). When the transgenic plants were subjected to NaCl, mannitol and ABA treatments, we found that expression patterns of eight tested genes, such as ABI1, DREB1A, RD29A and NHX1, showed similar trends as in the wild-type and vector control in the TaWD40D transgenic lines, although some minor changes between the transgenic lines and the controls were observed (Fig. S4). Some differences in the levels of expression of tested genes were also observed among two individual transgenic lines. However, the expression patterns of some genes were substantially altered by overexpression of TaWD40D. For example, the upward trend in ABI2 gene expression remained the same in the transgenic plants overexpressing TaWD40D under NaCl, mannitol and ABA treatments, but their transcript levels of ABI2 were significantly higher than that in the control plants (Fig. 5a, f, k). SOS2 and SOS3 in the TaWD40D transgenic lines were dramatically upregulated, about eight- and sixfold, respectively, compared with WT under salt stress (Fig. 5b, c), although the transcript levels of SOS2 and SOS3 in the transgenic plants were somewhat different from that in controls under mannitol and ABA treatments (Fig. 5g–i, m). Interestingly, DREB2A and RAB18 in the TaWD40D transgenic plants showed different patterns in response to NaCl and mannitol/ABA treatments. In the presence of NaCl, DREB2A and RAB18 were greatly downregulated and not altered, respectively (Fig. 5d, e), whereas when treated with mannitol-simulated osmotic stress and ABA, the transcript levels of DREB2A and RAB18 were significantly higher in the TaWD40D overexpression lines than in the WT (Fig. 5i, j, n, o). Together, these results indicate that overexpression of TaWD40D greatly affects the expression patterns of the abiotic stress-related genes and ABA-inducible genes.
The expression of stress-responsive genes in the Arabidopsis plants overexpressing TaWD40D. Six-day-old seedlings were exposed to either 200 mM NaCl (a–e), 300 mM mannitol (f–j), or 100 µM ABA (k–o) for 3 h. Gene-specific primers were used to detect the relative transcript levels of stress-responsive genes. The Arabidopsis Actin2 gene was used as an internal control. Under normal conditions (Control), the gene expression in WT was regarded as standard, and EV, OE1, and OE2 values were compared with it. Under the stress conditions, the gene expression in WT was regarded as standard, and EV, OE1, and OE2 values were compared with it. The gene expression of each genotype under normal conditions was compared with the genotype under the stress condition, respectively. Error bars represent the standard deviation among three biological replicates. Significant differences (t test, *P < 0.05) are marked with asterisks. EV empty vector transgenic line. OE1 and OE2, TaWD40D overexpression transgenic lines. WT wild type
TaWD40D gene silencing reduces drought tolerance in wheat
Barley stripe mosaic virus (BSMV) is a positive sense RNA virus and its tripartite genome is composed of RNAα, RNAβ and RNAγ (Yuan et al. 2011). The γb ORF in the RNAγ cDNA clone can be manipulated to accommodate the transcription of non-viral sequences in infected barley or wheat plants (Holzberg et al. 2002; Scofield et al. 2005). Recently, BSMV-VIGS has been successfully used for silencing of wheat genes. To further characterize the biological function of TaWD40D in wheat, we adopted BSMV-VIGS-mediated tool to downregulate TaWD40D expression in wheat seedlings. It has been shown that TaPDS gene (wheat phytoene desaturase) causes photobleaching phenotype through BSMV-VIGS (Holzberg et al. 2002; Scofield et al. 2005). To confirm the BSMV-VIGS system is effective in silencing wheat gene, we used a BSMV-mediated silencing of TaPDS as a positive control. As shown in Fig. 6a, when inoculated with BSMV harboring the TaPDS, BSMVPDS-infected wheat seedling leaves especially the leaves from 3rd to 7th showed a clear and consistent photobleaching phenotype at about 12 days post-inoculation, and the gene expression of TaPDS was markedly downregulated accordingly (Fig. 6c). These results indicate that the VIGS system can efficiently silence the target gene in wheat, which is consistent with the previous results (Holzberg et al. 2002; Scofield et al. 2005).
Virus-induced gene silencing of TaWD40D in wheat and phenotypic analysis of the wheat plants. a The phenotype of leaves subjected to TaPDS suppression and drought stress. b Comparison of drought tolerance between TaWD40D-silenced and non-silenced plants. c RT-PCR profiles of TaWD40D in mock and transgenic lines. The wheat Tubulin gene was used as an internal control. d Relative water contents between silenced and non-silenced plants under drought stress. Error bars represent the standard deviation among three biological replicates. Significant differences (t test, *P < 0.05) are marked with asterisks. Mock, infection buffer without BSMV. BSMV0, plants inoculated with empty BSMV viral vector, all other plants were inoculated with BSMV targeted to specific genes. The plants were well-watered to maintain 100 % field capacity. Photobleaching of the BSMVPDS plants appeared at 12 days post-inoculation, drought stress was then implemented by withholding watering and the photos were taken at 32 days post-inoculation
Next, we analyzed the wheat seedlings inoculated with BSMV WD40D . We found that nearly all the BSMV WD40D .inoculated leaves and the newly emerged leaves including the leaves from 3rd to 7th showed the typical mosaic symptoms as BSMV0 (Fig. 6a). Further RT-PCR analysis result revealed that the transcript levels of TaWD40D were also greatly reduced in the inoculated leaves compared with the controls at 12 days post-inoculation (Fig. 6c). These BSMV-VIGS-silenced plants and viral-uninoculated plants were subjected to water stress treatment by withholding water, and the well-watered plants were used as control. Under well-watered conditions, slight leaf chlorosis was observed in all of the silenced and viral control plants after inoculation with the BSMV constructs because of the response to infection, this is in agreement with previous studies in wheat (Scofield et al. 2005; Van Eck et al. 2010). Under drought conditions, typical wilting symptoms and poor vigor were observed in all plants (Fig. 6a, b). However, the BSMV WD40D plants showed more severe wilting symptoms compared with the mock, including the BSMV PDS plants. Notably, all the tested leaves of the BSMV WD40D plants were severely wilted and some of them even dried out. These data indicate that TaWD40D gene silencing may reduce the ability of plants to retain leaf cellular water.
To validate this prediction, we measured the RWC of silenced and non-silenced plants under drought stress. As shown in Fig. 6d, the control plants had approximately a 15–30 % reduction in RWC under water stress, by contrast, a more significant reduction in RWC (about 60 %) was observed in the plants infected with BSMV WD40D (t test, *P < 0.05). This physiological measurement matched the wilting phenotype of the BSMV WD40D plants under drought stress. Together, these data indicate that TaWD40D is required for maintenance of leaf vigor and acts as a positive regulator of drought tolerance in wheat.
Discussion
TaWD40D protein possesses the typical features of WD40 family proteins
The WD40 repeat-containing proteins comprise a diverse superfamily of regulator proteins, and play indispensable roles in the development and environmental adaptation of eukaryotes (Mishra et al. 2012a). Structurally, seven or more WD40 repeats, each containing 44–60 residues, are the principal characteristics of a WD40 domain (Neer et al. 1994). The WD40 repeats are highly conserved and these repeating units can form a highly stable β-propeller structure that serves as a rigid scaffold for protein interactions. The current evidence has shown that WD40 repeats within a protein provide binding sites for proteins and foster protein–protein interactions; they can also act as a modular interaction domain of large proteins. Based on these features, the WD40 repeat-containing proteins may function as an integral component of protein complexes (Xu and Min 2011). One well-studied example is the G protein complex. The crystal structure of the G-βγ complex shows that the β subunit of the G protein adopts a seven-bladed β propeller and an N-terminal amphipathic α helix to interact with the G-γ subunit (Lambright et al. 1996). Many WD40 repeat-containing proteins have been reported to serve as adaptors or presentation molecules in various cellular processes in budding yeast (Saccharomyces cerevisiae), fruit fly (Drosophila melanogaster) and humans (Homo sapiens) (Ruthenburg et al. 2007).
In plants, the roles of WD40 repeat-containing proteins have drawn attention. Genome-based analysis revealed that most Arabidopsis WD40 repeat-containing proteins are strongly conserved in their structural features across eukaryotes, including those that modulate plant-specific processes (Van Nocker and Ludwig 2003). In crop plants, very few WD40 repeat-containing proteins have been reported. However, the WD40 repeat proteins are increasingly recognized as key regulators of specific developmental events or responses to environmental stresses in plants (Mishra et al. 2012a, b). In this study, we cloned a wheat TaWD40D gene that encodes a member of WD40 family proteins. TaWD40D protein contains seven conserved WD40 domains (Fig. 2b, c). Phylogenetic analysis showed that TaWD40D protein shares high protein sequence identity to WD40 proteins in plants (Fig. 2d). Taking into account the strong similarity between the predicted three-dimensional structure of TaWD40D (Fig. S2) and the G protein β subunit, these results confirm that TaWD40D belongs to the WD40 protein family.
TaWD40D is a novel gene involved in the wheat response to abiotic stress
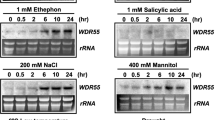
Despite the critical function of WD40 repeat-containing proteins, their functions in plants remain largely unknown. Recent studies of WD40 repeat proteins in Arabidopsis, rice, and foxtail millet have highlighted the diverse roles of WD40 repeat proteins in plant development and responses to the environment (Mishra et al. 2014; Ouyang et al. 2012; Van Nocker and Ludwig 2003). The large WD40 protein family includes many members that are involved in plant responses to abiotic stress. Pleiotropic regulatory locus 1 (PRL1) is a WD40 protein with multiple roles in the regulation of plant development and stress responses in Arabidopsis. It has been shown that prl1 mutant plants are hypersensitive to abiotic stress, and the loss of function of PRL1 in Arabidopsis causes increased sensitivity to several growth hormones, including cytokinin, ethylene, ABA, and auxin (Németh et al. 1998). PRL1 may integrate these hormonal signaling pathways to control the developmental plasticity of plants in response to abiotic stresses. In Arabidopsis, several other WD40 proteins that modulate plant stress responses have been identified, including high expression of osmotic stress-responsive genes 15 (HOS15) (Zhu et al. 2008), DWA1, and DWA2 (Lee et al. 2010a). Functional analyses of these WD40 proteins have suggested multiple regulatory roles in histone modification and protein degradation. Over the past several years, the roles of WD40 proteins in crop plants have also drawn attention. In Brassica napus, BnSWD1 has been shown to be upregulated under salt stress and various hormonal treatments such as ABA, salicylic acid, and methyl jasmonate (Lee et al. 2010b). Recently, the identification of stress-responsive genes such as SRWD1 in rice and SiWD40 in foxtail millet has indicated critical roles for WD40 proteins in modulating responses to abiotic stress in monocots (Mishra et al. 2012b; Ouyang et al. 2012).
Here, we reported that TaWD40D is a ubiquitously expressed gene in various wheat tissues and that it is highly induced by NaCl and PEG 6000-simulated water stress (Fig. 1b, c). More importantly, TaWD40D is a functional gene that mediates plant stress tolerance. For example, overexpression in Arabidopsis conferred enhanced stress tolerance to salt and osmotic stress during seed germination and postgerminative development (Fig. 4c, d). Moreover, TaWD40D gene silencing induced by the VIGS method resulted in substantially reduced tolerance of wheat plants to water stress (Fig. 6b). The reduced drought tolerance caused by TaWD40D gene silencing may be due to increased water loss because those plants with a low transcript level of TaWD40D had a low RWC (Fig. 6d). Thus, we have identified a wheat gene encoding a WD40 protein that plays an important role in plant tolerance to salt stress and water stress. Since the transcript level of TaWD40D was also responsive to ABA (Fig. 1d) and Arabidopsis transgenic plants overexpressing TaWD40D were insensitive to ABA during early development (Fig. 4c, d), we speculate that TaWD40D modulates in part stress responses in an ABA-dependent manner. The low RWC detected in plants with reduced TaWD40D expression may be caused by reduced sensitivity of the stomata to ABA.
Despite the important roles of TaWD40D in Arabidopsis and wheat using ectopic overexpression and VIGS-mediated gene silencing approach, there is still a need for further confirmation of the role of TaWD40D in plant tolerance to abiotic stress. In particular, the previous studies have shown that there are some limitations in VIGS-mediated gene silencing in crop plants including incomplete silencing of the target gene, negative effects of viral infections on phenotypic analysis of the gene function, inhibition of viral spread by the target gene, etc. (Yuan et al. 2011). In this study, we found that viral inoculation indeed caused some infection symptom in wheat leaves, but the symptoms did not affect plant growth vigor (Fig. 6a). Therefore, our result provides support for use of VIGS-mediated gene silencing technology for rapid functional analysis of TaWD40D gene in plant drought tolerance. Importantly, the BSMV TaWD40D did not affect viral spread in wheat plants because inoculated plants showed typical viral infection symptoms and the TaWD40D expression was successfully silenced especially in the leaves from 3rd to 7th of the inoculated plant (Fig. 6c). Functional analysis of TaWD40D using stable transgenic wheat plants overexpression and knockdown/knockout of TaWD40D gene will help us for eventual elucidation of the role of the gene in wheat response to abiotic stresses.
Possible mechanisms of the TaWD40D-mediated plant response to abiotic stress
Although our understanding of the specific mechanism of TaWD40D-mediated stress tolerance is limited, we have gained some insights into the molecular mechanism by which TaWD40D regulates stress tolerance in Arabidopsis overexpressing TaWD40D. A gene expression analysis of transgenic Arabidopsis plants overexpressing TaWD40D revealed that TaWD40D may play crucial role in regulating ABA signaling pathway because the transcript level of ABI2, the well-known negative regulator of the ABA signaling pathway, was markedly upregulated regardless of ABA treatment (Fig. 5k). The fact that RAB18 was also greatly upregulated in TaWD40D-overexpressing plants under ABA stress (Fig. 5o) supports the role of TaWD40D in the ABA signaling pathway. ABA is a phytohormone controlling ABA-induced seed germination inhibition and the inhibition of postgerminative growth (Himmelbach et al. 1998). Indeed, compelling evidence indicates that the malfunction of any of the key regulators of ABA signaling can alter ABA-mediated responses to salinity or osmotic stress (Li et al. 2010). RAB18 is a regulatory component in ABA signaling (Jeannette et al. 1999). ABI2 is a key negative regulator involved in ABA signaling and upregulation of ABI2 reduces ABA sensitivity during seed germination (Finkelstein and Somerville 1990; Meyer et al. 1994). The evidence that overexpression of TaWD40D results in decreased sensitivity of germinating seedlings to ABA (Fig. 4d) favors the notion and, therefore, it is likely that TaWD40D modulates plant responses to stress in an ABA-dependent manner.
TaWD40D may also regulate plant stress responses in an ABA-independent manner because the overexpression of TaWD40D resulted in substantially increased expression of DREB2A (Fig. 5i, n), a well-known component of the ABA-independent pathway. It has been shown that the upregulation of DREB2A increased the expression of downstream drought stress-responsive genes and enhanced plant tolerance to drought (Liu et al. 1998). Thus, it is likely that activation of DREB2A in the TaWD40D overexpression lines may also contribute to their increased tolerance to osmotic stress. DREB1A/CBF3 transcription factor can also specifically bind to the DRE/CRT cis-element and regulate cold stress-responsive gene expression and cold tolerance (Sakuma, et al. 2002). In our study, expression of Arabidopsis DREB1A was not affected by overexpression of TaWD40D under normal and stress conditions (Fig. S4). These results suggest that TaWD40D may be involved in drought tolerance, but not cold response. RD29A and RD29B are quickly and strongly induced by salt stress and drought (Yamaguchi-Shinozaki and Shinozaki 1993). However, RD29A and RD29B are induced through ABA-independent and ABA-dependent pathways, respectively (Fujita et al. 2005; Narusaka et al. 2003). Recent results showed that knockout of RD29A and RD29B results in increased salt tolerance, although the functions of two proteins remain unknown (Msanne, et al. 2011). In our work, we found that expression levels of both RD29A and RD29B was not altered in the TaWD40D overexpression lines under normal or stress treatments (Fig. S4). Our results indicate that wheat TaWD40D may regulate plant stress response in a manner independent of RD29A and RD29B.
Furthermore, TaWD40D has a role in the maintenance of ion homeostasis in plant cells. The SOS signaling pathway, consisting of the key components SOS1, SOS2, and SOS3, is the only well-defined pathway regulating the response to ion disequilibrium in Arabidopsis (Zhu 2000). Overexpression of a single SOS gene, and especially two or three genes, resulted in enhanced salt tolerance in Arabidopsis plants (Yang et al. 2009). TaWD40D overexpression in Arabidopsis dramatically increased the expression levels of SOS2 and SOS3 in response to salt stress (Fig. 5b, c). Thus, TaWD40D positively regulates SOS signaling and plant responses to salt stress. Unexpectedly, the transcription abundance of Arabidopsis NHX1 and HKT1 was not affected by ectopic expression of TaWD40D (Fig. S4). NHX1 encodes a vacuolar sodium/proton antiporter and HKT1 encodes a high-affinity K+ transporter, and both genes play key role in ion excretion and compartmentation (Darley et al. 2000; Uozumi et al. 2000). The result suggests that TaWD40D may not regulate transcription of these two genes, but we do not exclude the possibility that TaWD40D modulate their function at protein level.
Plant stress tolerance is a complex trait that is regulated by a complicated signaling network intertwined with various cellular signaling pathways. Considering that TaWD40D is a WD40 repeat-containing protein, it is conceivable that TaWD40D may be a key component that dynamically integrates multiple regulatory pathways mediating plant tolerance to abiotic stress. Genome-wide transcriptome analysis of the transgenic Arabidopsis plants overexpressing TaWD40D will help us reveal genes and pathways that are predicted to be mediated by TaWD40D in plant stress response. Despite of lack of a definitive role of TaWD40D and the TaWD40D-mediated molecular mechanism in plant stress response, our result provides a starting point for further elucidation of the function of the TaWD40D and genetic engineering of crops for enhanced tolerance to abiotic stress.
Author contribution statement
Conceived and designed the experiments: XL DK. Performed the experiments: DK ML. Analyzed the data: DK ZD. Contributed reagents/materials/analysis tools: ML HJ. Wrote the paper: DK XL.
Abbreviations
- ABA:
-
Abscisic acid
- BSMV:
-
Barley stripe mosaic virus
- CaMV:
-
Cauliflower mosaic virus
- GFP:
-
Green fluorescent protein
- MS:
-
Murashige and Skoog
- ORF:
-
Open reading frame
- PEG:
-
Polyethylene glycol
- qRT-PCR:
-
Quantitative RT-PCR
- RT-PCR:
-
Reverse transcription-PCR
- UTR:
-
Untranslated region
- VIGS:
-
Virus-induced gene silencing
References
Adamec L (1984) The effect of plasmolysis and deplasmolysis on the permeability of plant membranes. Biol Plantarum 26:128–131
Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL (2008) Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol 148:1868–1882
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Budak H, Kantar M, Kurtoglu KY (2013) Drought tolerance in modern and wild wheat. Sci World J 2013:548246
Byrt CS, Munns R (2008) Living with salinity. New Phytol 179:903–905
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Darley CP, van Wuytswinkel OC, van der Woude K, Mager WH, de Boer AH (2000) Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J 351:241–249
Ekanayake IJ, Dedatta SK, Steponkus PL (1993) Effect of water-deficit stress on diffusive resistance, transpiration, and spikelet desiccation of rice (Oryza-Sativa L). Ann Bot 72:73–80
Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9:377–396
Finkelstein RR, Somerville CR (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94:1172–1179
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488
Gachomo EW, Jimenez-Lopez JC, Baptiste LJ, Kotchoni SO (2014) GIGANTUS1 (GTS1), a member of Transducin/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol 14:37
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
Hall RA, Lefkowitz RJ (2002) Regulation of G protein-coupled receptor signaling by scaffold proteins. Circ Res 91:672–680
Heinrich R, Neel BG, Rapoport TA (2002) Mathematical models of protein kinase signal transduction. Mol Cell 9:957–970
Himmelbach A, Iten M, Grill E (1998) Signalling of abscisic acid to regulate plant growth. Philos Trans R Soc Lond B Biol Sci 353:1439–1444
Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30:315–327
Huang J, Wang MM, Bao YM, Sun SJ, Pan LJ, Zhang HS (2008) SRWD: a novel WD40 protein subfamily regulated by salt stress in rice (Oryza sativa L.). Gene 424:71–79
Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333
Jeannette E, Rona JP, Bardat F, Cornel D, Sotta B, Miginiac E (1999) Induction of RAB18 gene expression and activation of K+ outward rectifying channels depend on an extracellular perception of ABA in Arabidopsis thaliana suspension cells. Plant J 18:13–22
Kishor PBK, Sangam S, Amrutha RN, Laxmi PS, Naidu KR, Rao KRSS, Rao S, Reddy KJ, Theriappan P, Sreenivasulu N (2005) Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci 88:424–438
Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci 6:262–267
Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379:311–319
Lee MH, Lee SH, Kim H, Jin JB, Kim DH, Hwang I (2006) A WD40 repeat protein, Arabidopsis Sec13 homolog 1, may play a role in vacuolar trafficking by controlling the membrane association of AtDRP2A. Mol Cells 22:210
Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai MQ, Li JG, Byun MO, Deng XW (2010a) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-Based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22:1716–1732
Lee S, Lee J, Paek K-H, Kwon S-Y, Cho HS, Kim SJ, Park JM (2010b) A novel WD40 protein, BnSWD1, is involved in salt stress in Brassica napus. Plant Biotechnol Rep 4:165–172
Li S, Xu C, Yang Y, Xia G (2010) Functional analysis of TaDi19A, a salt-responsive gene in wheat. Plant Cell Environ 33:117–129
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
McNellis TW, von Arnim AG, Deng XW (1994) Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6:1391–1400
Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264:1452–1455
Mishra AK, Puranik S, Prasad M (2012a) Structure and regulatory networks of WD40 protein in plants. J Plant Biochem Biotechnol 21:32–39
Mishra AK, Puranik S, Bahadur RP, Prasad M (2012b) The DNA-binding activity of an AP2 protein is involved in transcriptional regulation of a stress-responsive gene, SiWD40, in foxtail millet. Genomics 100:252–263
Mishra AK, Muthamilarasan M, Khan Y, Parida SK, Prasad M (2014) Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS One 9:e86852
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249
Msanne J, Lin J, Stone JM, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Murzin AG (1992) Structural principles for the propeller assembly of beta-sheets: the preference for seven-fold symmetry. Proteins 14:191–201
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148
Neer EJ, Schmidt CJ, Nambudripad R, Smith TF (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature 371:297–300
Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12:3059–3073
Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100:11771–11776
Ouyang Y, Huang X, Lu Z, Yao J (2012) Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom 13:100
Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Rhoades J, Loveday J (1990) Salinity in irrigated agriculture. Agronomy, pp 1089–1142
Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421:185–190
Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25:15–30
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138:2165–2173
Sears E (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. Chromosome manipulations and plant genetics. Springer, pp 29–45
Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45:1694–1703
Shabala S (2009) Salinity and programmed cell death: unravelling mechanisms for ion specific signalling. J Exp Bot 60:709–712
Smith TF (2008) Diversity of WD-repeat proteins. The coronin family of proteins. Springer, pp 20–30
Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24:181–185
Stirnimann CU, Petsalaki E, Russell RB, Muller CW (2010) WD40 proteins propel cellular networks. Trends Biochem Sci 35:565–574
Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104:20623–20628
Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122:1249–1259
Van Eck L, Schultz T, Leach JE, Scofield SR, Peairs FB, Botha AM, Lapitan NL (2010) Virus-induced gene silencing of WRKY53 and an inducible phenylalanine ammonia-lyase in wheat reduces aphid resistance. Plant Biotechnol J 8:1023–1032
Van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genom 4:50
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Wohlbach DJ, Quirino BF, Sussman MR (2008) Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20:1101–1117
Xu C, Min J (2011) Structure and function of WD40 domain proteins. Protein Cell 2:202–214
Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340
Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z (2009) Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31
Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6:e26468
Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M (2006) Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci USA 103:1422–1427
Zhang H, Mao X, Jing R, Chang X, Xie H (2011) Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot 62:975–988
Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124:941–948
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, Bressan RA (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA 105:4945–4950
Acknowledgments
We are grateful to Professor Jizeng Jia (Key Laboratory of Crop Germplasm and Biotechnology, Ministry of Agriculture, Institute of Crop Germplasm Resources, Chinese Academy of Agricultural Sciences, 100081, Beijing, China) and Professor Dawei Li (State Key Laboratory of Agro-Biotechnology, China Agricultural University, Beijing, China) for providing the wheat cDNA library of Chinese Spring and the VIGS vectors, respectively. This research was financially supported by National Program on Key Basic Research Project (2012CB114300) and the National Transgenic Key Project of the Ministry of Agriculture of China (2011ZX08009-003-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Wendy Harwood.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2014_1717_MOESM1_ESM.doc
Fig. S1. Chromosomal location of TaWD40D. Fig. S2 The three-dimensional structure of TaWD40D protein. Fig. S3 TaWD40D overexpression lines are comparable to the wild type at different developmental stages. Fig. S4 Expression of stress-responsive genes in the wild-type and TaWD40D overexpression lines under normal or stress conditions. Table S1 Primers used in this study. (DOC 1976 kb)
Rights and permissions
About this article
Cite this article
Kong, D., Li, M., Dong, Z. et al. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep 34, 395–410 (2015). https://doi.org/10.1007/s00299-014-1717-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1717-1