Abstract
Key message
Cowpea cultivars differing in salt tolerance reveal differences in protein profiles and adopt different strategies to overcome salt stress. Salt-tolerant cultivar shows induction of proteins related to photosynthesis and energy metabolism.
Abstract
Salinity is a major abiotic stress affecting plant cultivation and productivity. The objective of this study was to examine differential proteomic responses to salt stress in leaves of the cowpea cultivars Pitiúba (salt tolerant) and TVu 2331 (salt sensitive). Plants of both cultivars were subjected to salt stress (75 mM NaCl) followed by a recovery period of 5 days. Proteins extracted from leaves of both cultivars were analyzed by two-dimensional electrophoresis (2-DE) under salt stress and after recovery. In total, 22 proteins differentially regulated by both salt and recovery were identified by LC–ESI–MS/MS. Our current proteome data revealed that cowpea cultivars adopted different strategies to overcome salt stress. For the salt-tolerant cultivar (Pitiúba), increase in abundance of proteins involved in photosynthesis and energy metabolism, such as rubisco activase, ribulose-5-phosphate kinase (Ru5PK) (EC 2.7.1.19), glycine decarboxylase (EC 1.4.4.2) and oxygen-evolving enhancer (OEE) protein 2, was observed. However, these vital metabolic processes were more profoundly affected in salt-sensitive cultivar (TVu), as indicated by the down-regulation of OEE protein 1, Mn-stabilizing protein-II, carbonic anhydrase (EC 4.2.1.1) and Rubisco (EC 4.1.1.39), leading to energy reduction and a decline in plant growth. Other proteins differentially regulated in both cultivars corresponded to different physiological responses. Overall, our results provide information that could lead to a better understanding of the molecular basis of salt tolerance and sensitivity in cowpea plants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Salinity is one of the major abiotic stresses limiting plant growth and yield in many areas of the world, particularly on irrigated land. NaCl is the predominant salt in most saline environments (Türkan and Demiral 2009). Many crop species are sensitive to high concentrations of salt, with negative impacts on agricultural production. As a result, the development of improved levels of tolerance to salt stress has become an urgent priority for many crop breeding programs (Vinocur and Altman 2005), and much research effort has been devoted to gain a better understanding of the mechanisms adopted by plants to combat salt stress.
Salt stress causes water deficit, ion toxicity, nutrient imbalance, and oxidative stress and leads to cellular damage, growth reduction and even plant death (Tester and Davenport 2003; Munns 2005; Miller et al. 2010). Under natural conditions, plants have evolved several mechanisms to cope with the presence of salt. Physiological studies have demonstrated that changes in water relation, nutrient uptake, hormonal metabolism, carbon metabolism and antioxidant metabolism play an important role in salt tolerance. In addition, dramatic alterations in gene expression, which lead to changes in the protein profile, have been found to be closely related to these processes (Du et al. 2010; Sobhanian et al. 2011). Therefore, a clear understanding of stress-response proteins is important for further understanding the molecular mechanisms controlling plant salt tolerance.
Proteomics, the study of global changes in proteins, including information on protein abundances, their variations and modifications, has proven to be a powerful approach for the identification of proteins and mechanisms involved in salt response and tolerance (Veeranagamallaiah et al. 2008; Aghaei et al. 2009; Pang et al. 2010; Liu et al. 2012). Recently, Du et al. (2010), while studying the effects of salinity on the proteome of cucumber (Cucumis sativus) roots, identified 29 protein spots with functions related to metabolism, energy and transport that were involved in regulating reactions and defending against stress. According to Kosová et al. (2011), information on proteome changes in response to stress has been fragmentary. Typically, studies have only compared non-stressed (control) and stressed plants. Moreover, few studies in this area have taken into account the dynamic processes of the plant response to stress that depend on stress intensity and duration. In this context, an even less studied process is that of recovery following the cessation of stress treatment. In this respect, the great majority of these studies were performed on plant subjected to drought stress (Salekdeh et al. 2002; Gazanchian et al. 2007; Ingle et al. 2007; Echevarría-Zomeño et al. 2009). However, very limited information has been available concerning protein changes in plants during recovery from salt stress. Chourey et al. (2003) reported that several salinity-stress-induced LEA proteins were accumulated during the salt-stress-triggered growth arrest of young rice (Oryza sativa L.) seedlings and that the recovery from salt stress was accompanied by the degradation of these proteins.
Cowpea is widely grown in arid and semiarid regions of the world (Ehlers and Hall 1997), and it is one of the most important crops for people living in developing countries. However, cowpea is subject to salt stress that reduces its yield, like many other crops. Studies of the salt tolerance of cowpea using physiological and biochemical approaches have produced a large body of data. Most of these studies have focused on biomass production, photosynthesis, compatible solute accumulation and antioxidant enzyme activities (Silva et al. 2003; Murillo-Amador et al. 2006; Cavalcanti et al. 2007). Earlier studies conducted in our laboratory have indicated varying behaviors of key enzymes of several metabolic pathways, varying levels of metabolites and altered photosynthetic and N assimilation efficiency in plants of two cowpea cultivars. Our data showed that Pitiúba is a salt-tolerant cultivar, while TVu 2331 is a salt-sensitive cultivar (Costa et al. 2003; Praxedes et al. 2010). However, the protein dynamics of salt tolerance has not been investigated. In the present study, two-dimensional electrophoresis (2-DE) in conjunction with mass spectrometry (MS) analysis was used to investigate leaf proteome-level differences between cultivars of cowpea exhibiting differential salinity tolerance. The main goal of this study was to characterize the differential abundance of proteins in leaves during salt stress and recovery and to discuss their roles in mediating the response of cowpea to salt stress.
Materials and methods
Plant material and growth conditions
Seeds of cowpea [Vigna unguiculata (L.) Walp], cultivars Pitiúba (salt tolerant) and TVu 2331 (salt sensitive) were surface sterilized in 1 % (v/v) sodium hypochlorite for 5 min and then rinsed three times in distilled water. Seeds of each cultivar were sown in vermiculite moistened with half-strength Hoagland’s nutrient solution in the absence (control) or presence of 75 mM NaCl (salt stress). Five days after sowing, seedlings of each cultivar were transferred to 6-L pots with full-strength Hoagland’s nutrient solution remaining a set of plants exposed to control conditions (Hoagland solution without NaCl) and another set of plants exposed to salt stress (Hoagland solution with 75 mM NaCl). Seventeen days after sowing, a set of salt-stressed plants was transferred to pots containing Hoagland’s nutrient solution without NaCl (recovery treatment). All nutrient solutions were replaced every 5 days and routinely aerated. Water loss by evapotranspiration was estimated by the pot weight loss and replaced daily with distilled water. Twenty-two days after sowing, leaves (second and third fully expanded leaves from the apex) of all treatments (control, salt stress and recovery) were flash frozen in liquid nitrogen, lyophilized and stored at −80 °C for proteomic analysis. Physiological parameters such as leaf area and shoot dry mass were also evaluated. Leaf area was determined with a Li-Cor area meter (LI-3100, Li-Cor Inc., Lincoln, NE, USA). Plant shoots were separated, dried for 72 h at 60 °C and weighed for determination of dry mass. The experiment was conducted in Fortaleza (3.74°S, 38.58°W), northeastern Brazil, under greenhouse conditions with mean midday photosynthetically active radiation of approximately 1,500 μmol m−2 s−1, a temperature of 29.7 ± 0.6 °C, and a relative humidity of 70 ± 5 %.
Protein extraction
Total soluble protein was extracted in five replicates from leaves of each treatment according to the method of Shen et al. (2002) with some modifications. The samples were ground to powder in liquid nitrogen with a pre-cooled mortar and pestle. Approximately, 200 mg of leaf samples was mixed with 200 mg of polyvinylpolypyrrolidone (PVPP) and homogenized in 5 mL of extraction buffer containing 40 mM Tris–HCl (pH 7.5), 250 mM sucrose, 10 mM ethylenediamine tetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT) and 1 % (v/v) Triton X-100. The mixture was vortexed, shaken at 4 °C for 1–2 h, and centrifuged at 15,000×g for 15 min at 4 °C. The supernatant was precipitated with 10 mL 10 % (w/v) trichloroacetic acid (TCA) in cold acetone for 12 h at −20 °C. After centrifugation at 15,000×g for 15 min at 4 °C, the supernatants were discarded, and the pellets were washed four times with ice-cold acetone. The resulting pellet was vacuum dried and dissolved in 300 μL lysis buffer (7 M urea, 2 M thiourea, 2 % (w/v) 3-[(3-Cholamidopropyl) dimethylammonio] propanesulfonic acid (CHAPS), 2 % (v/v) immobilized pH gradient (IPG) buffer 3–10 and 1 % (w/v) DTT) (GE Healthcare: Life sciences, USA) at room temperature for 1 h. The suspension was centrifuged at 12,000×g for 10 min at 25 °C. The supernatant was collected as the protein extract and used for 2-DE analysis. Protein concentrations were determined in duplicate according to Bradford (1976) using bovine serum albumin (Sigma-Aldrich, USA) as the standard.
2-DE separation and image analysis
The proteins extracted were diluted with rehydration buffer containing 8 M urea, 2 % (w/v) CHAPS, 2 % (v/v) IPG buffer (pH 3–10), 0.3 % (w/v) DTT and 0.002 % (w/v) bromophenol blue (GE Healthcare: Life sciences, USA). Approximately 400 μg of protein was loaded onto a 13-cm, pH 4–7 linear gradient IPG strip (GE Healthcare, USA). Isoelectric focusing (IEF) was performed using a Multiphor™ II apparatus (Amersham Pharmacia Biotech) according to the manufacturer’s instructions. Briefly, focusing was performed at 20 °C with the following steps: gradient at 300 V for 5 min, gradient at 3,500 V for 1:30 h, and 3,500 V for 4:10 h. After IEF, the protein in the strips was denatured for 15 min with equilibration buffer [50 mM Tris–HCl (pH 8.8), 6 M urea, 30 % (v/v) glycerol, 2 % (w/v) sodium dodecyl sulfate (SDS), 0.002 % (w/v) bromophenol blue and 1 % (w/v) DTT] and then incubated with the same buffer containing 2.5 % (w/v) iodoacetamide instead of DTT for 15 min at room temperature. The second dimension of electrophoresis (SDS-PAGE) was performed on a 12.5 % (v/v) gel using a Hoefer SE 600 Ruby electrophoresis unit (GE Healthcare: Life sciences, USA). Strips were embedded on the top of SDS-PAGE gels and then sealed using 0.5 % (v/v) molten agarose solution. Electrophoresis was run at 15 mA per gel for 30 min, followed by 25 mA per gel until the bromophenol blue tracking dye ran off the gel. After SDS-PAGE, gels were fixed overnight with a fixing solution containing 10 % (v/v) acetic acid and 40 % (v/v) ethanol and stained with colloidal Coomassie Blue G-250 (Neuhoff et al. 1988).
2-DE gels were scanned at 300 dpi using an Image Scanner (GE Healthcare: Life sciences, USA). Image analysis was performed using ImageMaster™ 2-D Platinum software (version 6.0, GE Healthcare, USA). At least three gels from each treatment (control, salt stress and recovery) from three independent experiments were used for the analysis. Background subtraction, landmark identification, the aligning/matching of spots within gels, the quantification of matched spots and spot analysis were performed according to the software guide. Before spot matching, one of the gel images was selected as the reference gel. To correct the variability due to staining, the abundance of each protein spot was estimated by the percentage intensity (intensity %), i.e., the spot intensity of one spot in relation to the sum of all detected spots on the gel. The normalized percentage intensity of protein spots from triplicate biological samples was subjected to statistical analysis by ImageMaster 2-D Platinum software. Quantitative image analysis was conducted to reveal protein spots with reproducible differences in relative abundance (intensity % >2.0-fold and p value <0.05). The experimental MW (kDa) of each protein was estimated using 2-DE markers (Sigma-Aldrich, USA), and the experimental pI was determined by the migration of the markers on the IPG strip.
In-gel digestion and MS analysis
Selected protein spots were manually excised from the gel and digested with sequencing-grade trypsin as described by Shevchenko et al. (2006). Briefly, gel pieces were destained by washing three times with 25 mM ammonium bicarbonate in 50 % (v/v) acetonitrile (ACN). After removing the destaining buffer, the gel pieces were dehydrated twice with 100 % ACN and lyophilized before digestion at 37 °C overnight (16 h) with 20 μL of 40 mM ammonium bicarbonate in 10 % (v/v) ACN containing 20 ng μL−1 trypsin (Promega, Madison, WI, USA). The peptides were extracted three times with 5 % (v/v) formic acid in 50 % (v/v) ACN using an ultrasonicator (UltraCleaner 1400A, Unique, SP, Brazil). The resulting peptide extracts were pooled, lyophilized in a vacuum centrifuge (SpeedVac SVC100H, Savant, USA) and stored at −80 °C until identification by mass spectrometry (MS) analysis.
The extracted tryptic fragments were analyzed by capillary liquid chromatography/nanoelectrospray ionization tandem mass spectrometry (LC–ESI–MS/MS), using a Q-ToF mass spectrometer (Waters Corp., Milford, MA, USA) coupled with a Waters Nano high-performance liquid chromatography (UPLC) unit. The product ion spectra collected were processed using Protein Lynx Global Server 2.0 (Waters Corp., MA, USA) and were converted to peak-list text files for database searching.
Protein identification
To identify proteins, MS/MS ion searches were performed on the processed spectra in the NCBI non-redundant database using MASCOT Daemon (Matrix Science, London, UK), under the taxon Viridiplantae (Green Plants). All peptides were assumed to be monoisotopic and [M+H]+ (protonated molecular ions). The other search parameters were the following: a mass accuracy of ±1.2 Da, one missed cleavage, complete carbamidomethylation of cysteine residues and partial oxidation of methionine residues. To identify a protein by LC–ESI–MS/MS, the following criteria were used: a minimum of three independent matching peptides, significant protein scores given by MASCOT (p < 0.05), and a minimum of 10 % coverage of protein by matching peptides. The identified proteins had to be in the top hit. Additional searches were also performed against the cowpea EST database to achieve or confirm the protein identification. In addition, the theoretical pI and MW of identified proteins were compared with the experimental pI and MW. The sub-cellular location of identified proteins was estimated using the software TargetP (http://www.cbs.dtu.dk/services/TargetP/). The server predicted whether the sequences contained chloroplast transit peptides (cTP), a mitochondrial targeting peptide (mTP), or a signal peptide (SP) for secretion. In addition, Uniprot (http://www.uniprot.org/) was used to search for the pathways and biological reactions associated with the identified proteins.
Experimental design and statistical analysis
The plants were distributed over a completely randomized two-plant plot design, with six treatment combinations (two cultivars and three salt treatments) and five replicates. The results of physiological parameters were analyzed using two-way ANOVA test performed with Assistat software, version 7.6 beta (Silva and Azevedo 2002). Differences were analyzed using the Tukey test at a p < 0.05 level. Results were reported as the mean ± SD of the replicates. Spot intensities of differential proteins in a 2D gel were calculated from three spots in three replicated gels. Significance of abundance protein spot differences was assessed by Student’s t test using the ImageMaster™ 2-D Platinum software. The protein spots showing a significant difference (p < 0.05) of abundance change (up or down) were considered as “proteins differentially regulated”.
Results
Plant growth response to salt stress and recovery
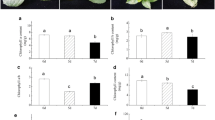
Salt stress with 75 mM NaCl caused a significant (p < 0.05) reduction in evaluated growth parameters, especially in the salt-sensitive cultivar (Fig. 1). Salinity reduced the leaf area by 48 % in Pitiúba and 64 % in TVu as compared to plants under control conditions (Fig. 1a). Shoot dry mass was also reduced in both cultivars, but the reduction was more pronounced in TVu (54 %) than in Pitiúba (38 %) as compared to their respective control values (Fig. 1b). After recovery treatment (complete NaCl withdrawal from the root medium), leaf area and shoot dry mass were significantly (p < 0.05) higher than in plants under salt stress.
Leaf area (a) and shoot dry mass (b) of Vigna unguiculata, cv. Pitiúba (open square) and cv. TVu (filled square), when subjected to control or salt-stress (75 mM NaCl) conditions and after recovery for 5 days. Vertical bars indicated the mean value ±SD of five replicates. Within each treatment, small letters indicate significant differences with p < 0.05 (Tukey test)
2-DE protein profile in response to salt stress and recovery
The effects of salt stress and recovery on the leaf protein profiles of cowpea cultivars were analyzed with 2-DE. In the preliminary experiments, major differences in protein profiles were observed at 75 mM NaCl (data not shown) which corresponded to the same concentration used previously by Costa et al. (2003) and Praxedes et al. (2010) in V. unguiculata cultivars. This salt concentration was selected as the reference for all subsequent differential 2-DE analyses. These experiments revealed that many of the proteins resolved were in the pH range of 4–7 with a molecular mass of 10.8–73.4 kDa. Accordingly, IEF was carried out within 4–7 pH range, and the second dimension of SDS-PAGE was performed with 12.5 % polyacrylamide.
To detect differences in abundance of proteins from cowpea leaves, 2-DE maps obtained from control and salt-treated plants from both stress and recovery treatments were compared with ImageMaster™ 2-D Platinum software. At least three 2-DE gels obtained from five biological replicates of the control and treated leaf samples were used for image analysis. Representative images from 2D gels of the Pitiúba and TVu cultivars are shown in Figs. 2 and 3, respectively. Quantitative image analysis revealed the protein spots that significantly changed their relative abundance (int. %) by 2.0-fold at p < 0.05. Changes in relative abundance of differentially regulated proteins during salt stress and recovery are available in Online Resource 1. In our research, 10 spots (spots 95, 100, 112, 118, 120, 134, 185, 212, 230 and 233) exhibited increases in abundance (up-regulation), and 13 spots (spots 93, 101, 105, 119, 121, 140, 149, 157, 179, 181, 184, 189 and 207) exhibited a decrease in abundance (down-regulation) in Pitiúba plants under salt stress (Figs. 2, 4; Online Resource 1). In TVu, 13 spots (spots 57, 58, 60, 79, 80, 111, 124, 143, 172, 178, 187, 232 and 236) were found to be up-regulated and 14 spots (spots 38, 82, 139, 146, 159, 163, 190, 195, 199, 200, 229, 233, 262 and 265) were down-regulated under salt-stress conditions (Figs. 3, 4; Online Resource 1).
Coomassie-stained two-dimensional gels of proteins extracted from Vigna unguiculata cv. Pitiúba leaves grown under salt-stress (75 mM NaCl) and control conditions and after recovery treatment. Proteins (400 μg) were separated in 13-cm IPG strips (pH 4–7 linear) in the first dimension and by SDS-PAGE on 12.5 % polyacrylamide gels in the second. Differentially regulated protein spots (intensity % ≥2.0-fold) are numbered. Spot no. 77 was detected only under the condition of stress. The nine framed zones are enlarged in Fig. 5
Coomassie-stained two-dimensional gels of proteins extracted from Vigna unguiculata cv. TVu 2331 leaves grown under salt-stress (75 mM NaCl) and control conditions and after recovery treatment. Proteins (400 μg) were separated in 13-cm IPG strips (pH 4–7 linear) in the first dimension and by SDS-PAGE on 12.5 % polyacrylamide gels in the second. Differentially regulated protein spots (intensity % ≥2.0-fold) are numbered. Spot no. 263 was detected only under the condition of stress. The nine framed zones are enlarged in Fig. 6
The effects of recovery from salt stress on the protein spot relative abundance in cowpea leaf were also studied (Figs. 2, 3). After recovery, the abundance of some responsive protein spots returned to the control levels for both cultivars. For example, spots 95, 100 and 185 increased (3.7-fold, 2.3-fold and 2.6-fold, respectively) in Pitiúba leaves under salt stress and then decreased to control levels after the end of stress treatment (Fig. 2; Online Resource 1). A similar result was observed in TVu plants, where some protein spots increased in size under salt stress and then shrank to control levels after recovery (for example, spots 79, 80 and 143) (Fig. 3; Online Resource 1). In contrast, spots 181 and 184 were strongly down-regulated by 3.1- and 2.3-fold, respectively, in Pitiúba leaves during salt stress compared to the control, and then increased after recovery (Fig. 2; Online Resource 1). Similarly, spots 139, 195 and 199 in TVu leaves during salt stress were down-regulated by 2.0-, 4.3- and 2.7-fold, respectively, but were subsequently restored in recovered plants (Fig. 3; Online Resource 1). In addition, salinity caused the appearance of two new protein spots (spot 77 in Pitiúba and spot 263 in TVu) that exhibited significant reductions in their intensity levels after recovery from salt stress (Figs. 2, 3; Online Resource 1). In summary, for Pitiúba, the comparison between stress and recovery revealed nine spots that increased (spots 101, 105, 106, 140, 181, 182, 184, 222 and 253) and eight spots that decreased (spots 77, 94, 95, 100, 112, 156, 185 and 230) in abundance in the recovered plants. For TVu, seven spots augmented (spots 139, 163, 190, 199, 200, 201 and 233) and 17 spots decreased (spots 33, 38, 58, 60, 85, 79, 80, 123, 124, 143, 149, 157, 161, 172, 187, 193 and 263) in abundance after recovery from salt stress (Fig. 4). Some of the changes in protein abundance under salt and recovery treatment are enlarged in Figs. 5 and 6.
Enlargement of the nine zones (I–IX) within the 2-DE gels of Vigna unguiculata cv. Pitiúba (Fig. 2) showing significant protein spot intensity changes during salt stress (75 mM NaCl) and after recovery. The arrows indicate the spot positions on the gel. Some of these spots were identified by LC–ESI–MS/MS (see Table 1). Vertical bars represent the relative abundance of each spot (Mean ± SD of three independent gels). Difference between the treatments was significant at *p < 0.05; **p < 0.01; ***p < 0.001 according to Student’s t test
Enlargement of the nine zones (I–IX) within the 2-DE gels of Vigna unguiculata cv. TVu 2331 (Fig. 3) showing significant protein spot intensity changes during salt stress (75 mM NaCl) and after recovery. The arrows indicate the spot positions on the gel. Some of these spots were identified by LC–ESI–MS/MS (see Table 1). Vertical bars represent the relative abundance of each spot (Mean ± SD of three independent gels). Difference between the treatments was significant at * p < 0.05; ** p < 0.01; *** p < 0.001 according to Student’s t test
Analysis of the 2-DE gels for control and recovered plants revealed variations in the abundance of some spots in both cultivars. Among the differentially regulated proteins in Pitiúba, nine spots showed an increased accumulation (spots 112, 117, 118, 134, 140, 182, 184, 233, 247) and eight protein spots showed a decreased accumulation (spots 119, 121, 157, 174, 181, 189, 207, 295) in the recovered plants when compared with controls (Figs. 2, 4; Online Resource 1). In TVu, five spots were up-regulated (spots 35, 139, 201, 229, 236) and four were down-regulated (spots 85, 146, 195, 265) during the recovery period relative to the control (Figs. 3, 4; Online Resource 1).
Identification of differentially regulated proteins
We have selected the spots that presented the highest differences in response to salt stress and recovery. For each cultivar, a total of 11 differential protein spots were subjected to tryptic digestion, and the resulting peptides were analyzed by LC–ESI–MS/MS. Nineteen proteins identified through tandem mass spectra and non-redundant NCBI database searches using the MASCOT tool (http://www.matrixscience.com/) are listed in Table 1. The majority of these protein spots corresponded to proteins involved in photosynthesis and energy metabolism, e.g., three rubisco activase proteins (spots 77, 93 and 95 in Pitiúba), two ribulose-5-phosphate kinase proteins (spot 100 in Pitiúba and spot 60 in TVu), two ATP synthase beta subunit proteins (spot 184 in Pitiúba and spot 172 in TVu), one oxygen-evolving enhancer (OEE) protein 2 (spot 185 in Pitiúba), one OEE protein 1 (spot 139 in TVu), one carbonic anhydrase protein (spot 163 in TVu), and one ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) large subunit protein (spot 195 in TVu). Other proteins in Pitiúba included one vicilin (spot 140), one lectin precursor (spot 149), and two proteins with unknown function (spots 157 and 212). In TVu, we also identified proteins involved in folding and stability, such as photosystem II (PSII) stability/assembly factor HCF136 (spot 80) and manganese-stabilizing protein-II (spot 199) and proteins involved in carbohydrate metabolism and signal transduction, such as sedoheptulose-1,7-bisphosphatase (SBP) isoform 1 (spot 79) and calreticulin (spot 262).
Spots 212 and 230 in Pitiúba and spots 143 and 263 in TVu were identified only after additional searches using the MASCOT tool and the plant EST database, which revealed the highest score for ESTs from cowpea. Protein spots 143, 212, 230 and 263 generated the best scores (1,142; 351; 160; 156) with the cowpea ESTs FC460400, FF382103, FC459361 and DR068329, respectively. Further, BLASTn searches comparing these ESTs with the NCBI nr database revealed that spots 143, 212, 230 and 263 were highly similar to Arabidopsis FAD/NAD(P)-binding oxidoreductase (NM_101382) [73 % of identity (coverage 60 %)]; soybean peptidyl-prolyl cis–trans isomerase (XM_003556536) [84 % of identity (coverage 99 %)]; soybean glycine decarboxylase complex H-protein (XM_003539121) [86 % of identity (coverage 91 %)]; and Medicago truncatula plastocyanin (XM_003603795) [72 % of identity (coverage 75 %)], respectively.
The differential pattern of main regulated proteins in leaves of V. unguiculata cultivars during salt-stress and recovery is showed in a radar plot (Fig. 7).
Discussion
Salinity is a major abiotic stress that affects plant growth and development. To cope with salt stress, plants have developed many strategies mediated by changes in gene expression, which result from protein changes altering certain metabolic and signal pathways (Vinocur and Altman 2005). Previously, our research group has identified two V. unguiculata cultivars revealing different levels of salt-stress tolerance: Pitiúba (salt tolerant) and TVu 2331 (salt sensitive) (Costa et al. 2003; Praxedes et al. 2010). In the present study, salt stress caused physiological changes in cowpea plants, as shown by a significant reduction in leaf area and in shoot dry mass, especially in sensitive cultivar (Fig. 1). In this context, we conducted proteomic analyses in leaves of Pitiúba and TVu cultivars to gain insights into the regulatory mechanisms involved in salt tolerance of V. unguiculata plants. Although a high similarity of protein profiles was observed in both cultivars, there were significant differences between them in abundance of some salt responsive proteins. The present study indicated that the variation in salt tolerance and sensitivity in cowpea plants might be associated with the differential accumulation of certain stress-responsive proteins. The main differentially regulated proteins from leaves of both cultivars during salt stress and recovery are discussed below.
Stress-responsive proteins in Pitiúba
Two spots corresponding to rubisco activase (spots 77 and 95) were significantly up-regulated (t test, p < 0.01) in Pitiúba leaves by salt stress (Figs. 5, 7; Online Resource 1) probably corresponding to different isoforms of this enzyme. Rubisco activase (EC 4. 1. 1. 39) removes inhibitory sugar phosphates such as ribulose-1,5-bisphosphate from the active sites of rubisco so that CO2 can activate the enzyme by carbamylation (Kang et al. 2012). Therefore, the increased abundance of this enzyme may be involved in the maintenance of CO2 assimilation under salt-stress conditions. Li et al. (2011) also reported a similar induction of rubisco activase in the halophyte Suaeda salsa, which was related to increased CO2 assimilation. Deeba et al. (2012), in contrast, reported down-regulation of this enzyme in cotton plants exposed to drought stress. We also found an isoform of rubisco activase (spot 93) that significantly decreased (t test, p < 0.05) in intensity under NaCl stress (Figs. 5, 7; Online Resource 1). Similarly, Kang et al. (2012) observed the up-regulation of two rubisco activase isoforms, while two others were down-regulated in wheat seedlings due to salt stress. According to Guo et al. (2012), the regulation of isoforms or isoproteins may represent a mechanism for plant adaptation to environmental changes.
We also found an enhanced abundance (t test, p < 0.001) of ribulose 5-phosphate kinase (Ru5PK), also known as phosphoribulokinase (EC 2. 7. 1. 19), which catalyzes the phosphorylation of ribulose-5-phosphate to ribulose-1,5-bisphosphate (RuBP), a key step in the Calvin cycle. The up-regulation of Ru5PK (spot 100) in Pitiúba indicated that the regeneration of RuBP could be maintained during salt stress. After the recovery period, the abundance of this protein returned to control levels (Figs. 5, 7; Online Resource 1). Consistent with our data, Deeba et al. (2012) reported up-regulation of Ru5PK in cotton leaves during progressive drought stress. In contradiction to our data, Xu and Huang (2010), investigating the proteomic responses to water stress induced by polyethylene glycol (PEG) in Agrostis stolonifera leaves, reported a down-regulation of this enzyme in both tolerant and sensitive cultivars. Another photosynthesis-related protein, oxygen-evolving enhancer protein 2 (OEE 2; spot 185), was found to be up-regulated (t test, p < 0.01) in response to salt stress (Figs. 5, 7; Online Resource 1). It is known that OEE 2 can be easily removed from the PSII complex in the presence of NaCl. The increased abundance of OEE 2 might be needed to repair protein damage caused by this dissociation and thus maintain oxygen evolution (Gazanchian et al. 2007). The up-regulation of OEE 2 in our report is in agreement with a similar study of the salt-stress response in seedling leaves of different salt-tolerant soybean genotypes, in which the OEE 2 was shown to increase under stress only with the salt-tolerant genotype (Ma et al. 2012).
Protein spot 184, identified as ATP synthase beta subunit, was significantly down-regulated (t test, p < 0.05) in salt-treated plants (Figs. 5, 7; Online Resource 1). ATP synthase (EC 3. 6. 3. 14) is the main enzyme for ATP biosynthesis in the presence of a proton gradient across the thylakoid membranes. The down-regulation of this protein might be specifically related to the inhibition of ATP synthesis by salt stress. In agreement with our results, the differential abundance of ATP synthase has also been reported in the proteomic studies of salt stress in other plant models as well (Caruso et al. 2008; Razavizadeh et al. 2009). However, the response to stress in these plants depended on the plant species and the type and intensity of stress. For instance, it was shown by Li et al. (2011) that ATP synthase was up-regulated in the halophyte S. salsa only by salt stress, but not by other stressors, such as heat shock or the combination of salt stress and heat shock. ATP synthase was down-regulated in salt-stressed soybean leaves (Sobhanian et al. 2010). On the other hand, this enzyme was not influenced by salt stress in watermelon seedlings (Yang et al. 2012).
Another interesting protein in Pitiúba is vicilin (spot 140), which was found to be down-regulated (t test, p < 0.01) by salt stress (Figs. 5, 7; Online Resource 1). The vicilins (7S globulins) are the major cowpea storage proteins and belong to a highly heterogeneous multigene family. In addition, previous findings have shown that vicilins bind to chitin-containing structures. This association could potentially explain the detrimental effects of variant vicilins, found in some cowpea varieties, on the development and survival of the storage pest insect Callosobruchus maculatus, as well as on spore/conidia germination and on the development of some phytopathogenic fungi (Gomes et al. 1997; Sales et al. 2001).
We also observed that peptidyl-prolyl cis–trans isomerase (PPIase; spot 212) was significantly up-regulated (t test, p < 0.05) under salinity conditions and then returned to control abundance levels after recovery from stress (Figs. 2, 7; Online Resource 1). PPIase (EC 5. 2. 1. 8), a widely distributed enzyme, catalyzes the cis–trans isomerization of proline imidic peptide bonds in oligopeptides (Huang et al. 2006). PPIase may function as a chaperone to prevent and reverse incorrect interactions between proteins and to facilitate correct protein folding under stress conditions (Debez et al. 2012; Deeba et al. 2012). In addition, PPIase may also be involved in signal transduction and thus plays a crucial role in stress responsiveness (Kottapalli et al. 2009). Microarray analysis revealed that the transcript level of the gene encoding PPIase was up-regulated in tomato plants subjected to salt stress (Zhou et al. 2007).
We observed that glycine decarboxylase (spot 230) was significantly up-regulated (t test, p < 0.01) in Pitiúba leaves by salt stress (Figs. 5, 7; Online Resource 1). This H-protein is part of the glycine decarboxylase (GDC; EC 2.1.2.10) multienzyme complex, and catalyzes the interconversion of glycine to serine, an integral part of the photorespiratory pathway (Bauwe and Kolukisaoglu 2003). Up-regulation of this protein in Pitiúba leaves suggests the possible function of photorespiration as an electron sink to minimize the production of reactive oxygen induced by salt-stress conditions. Caruso et al. (2008) also reported a similar induction of GDC in wheat leaves in response to salt stress.
One protein (spot 149) was found to be related to the plant defense system and identified as lectin precursor. The quantitative increase in lectin levels is one of the universal nonspecific mechanisms of plant protection against the influence of unfavorable environmental conditions, although its function(s) in salt stress is not known. Differential regulation of specific lectins has previously been reported in response to both abiotic and biotic stresses (Zhang et al. 2000; Babosha 2008; Ma et al. 2012). In our study, however, this protein was significantly down-regulated (t test, p < 0.05) in salt-treated plants (Figs. 2, 7; Online Resource 1). Aghaei et al. (2009) reported a similar down-regulation of lectins in response to salt stress in soybean hypocotyls and roots.
Stress-responsive proteins in TVu
Two spots representing proteins involved in photosynthesis, ribulose 5-phosphate kinase (Ru5PK; spot 60) and ATP synthase beta subunit (spot 172), experienced salt-induced changes (t test, p < 0.05) in TVu leaves. As previously reported in NaCl-treated Pitiúba leaves, an increased abundance of these enzymes was also present in TVu plants exposed to salt stress (Figs. 6, 7; Online Resource 1). In our study, the marked accumulation of Ru5PK and ATP synthase during salt stress and their mobilization during subsequent recovery from salt stress clearly supported such a role for these proteins in the stress acclimation of cowpea.
The oxygen-evolving enhancer proteins (OEEs) consist of three subunits: OEE 1, OEE 2, and OEE 3. These are nuclear-encoded chloroplast proteins and are peripherally bound to photosystem II (PSII) on the lumenal side of the thylakoid membrane (Sugihara et al. 2000). Our results showed that OEE 1 (spot 139) was down-regulated (t test, p < 0.01) in the TVu cultivar under the condition of salt stress (Figs. 6, 7; Online Resource 1), while an OEE 2 (spot 185) was induced (t test, p < 0.01) by salt stress in the Pitiúba cultivar (Figs. 5, 7; Online Resource 1). These findings suggest that the stress-sensitive cultivar (TVu) might have experienced greater dissociation of this complex from the PSII compared to Pitiúba. Interestingly, Pang et al. (2010) observed that OEE 1 was significantly down-regulated in Thellungiella halophila, a halophyte, under salt stress. For these authors, the physiological significance of these protein changes was intriguing. In our research, PSII stability/assembly factor HCF136 (spot 80), a structural element of PSII, and two proteins involved in chloroplast electron transport, FAD/NAD(P)-binding oxidoreductase (spot 143) and plastocyanin (spot 263), were found to be up-regulated (t test, p < 0.01) under salt stress (Figs. 6, 7; Online Resource 1). The same was not true for manganese-stabilizing protein-II (spot 199), whose abundance was reduced (t test, p < 0.01) by salt stress. Kang et al. (2012) reported an up-regulation of the PSII stability/assembly factor HCF136 in response to salt stress in wheat seedlings. In addition, manganese-stabilizing protein was required for the stability of PSII in higher plant chloroplasts and participated in the splitting of water to liberate O2 (Yi et al. 2005). Previous experiments showed that a decrease in the photosynthetic rate in both salt-tolerant and salt-sensitive cowpea plants appeared to affect both stomatal and non-stomatal behavior after treatment with NaCl (Silva et al. 2003; Praxedes et al. 2010). Therefore, an increase or a decrease caused by salt stress in abundance of the proteins associated with photosynthetic electron transport may cause the inhibition of photosynthesis and growth.
Carbonic anhydrase (EC 4. 2. 1. 1) is a zinc-containing metalloenzyme that catalyzes the reversible interconversion of CO2 and HCO3 −, and it has been shown to function in a variety of biological processes, including pH regulation, CO2 transfer, ion exchange, respiration, biosynthesis, and photosynthetic CO2 fixation (Yu et al. 2007). According to Caruso et al. (2008), an increased abundance of carbonic anhydrase may influence the rate of the reversible reaction HCO3 − + H+ ↔ CO2 + H2O, facilitate CO2 diffusion and thus may affect the buffering capacity of plant cells with high concentrations of HCO3 − and \({\text{CO}}_{ 3}^{2 - }\). Therefore, under CO2-limiting conditions, such as those imposed by various types of environmental stresses, carbonic anhydrase can prevent photorespiration in plants. In the present study, we found a significant down-regulation (t test, p < 0.05) of this enzyme in TVu plants under salt stress (spot 163; Figs. 6, 7; Online Resource 1). In contrast, Aghaei et al. (2008), studying the relative salt tolerance of two potato cultivars, the salt-tolerant Kennebec and salt-sensitive Concord, reported that salt stress led to a decrease in the carbonic anhydrase abundance in both cultivars.
The Rubisco large subunit (spot 195) was also observed to be down-regulated (t test, p < 0.001) by salt stress in TVu leaves (Figs. 6, 7; Online Resource 1). As the key enzyme involved in the Calvin photosynthesis cycle in C3 plants, rubisco (EC 4. 1. 1. 39) catalyzes the fixation of CO2 to ribulose-1,5-bisphosphate (RuBP). It is widely accepted to be the ultimate rate-limiting step in photosynthetic carbon fixation and, therefore, plays a key role in the production of agriculture crops (Yang et al. 2012). In addition, in our study, the abundance of rubisco after recovery from stress remained lower than in control plants (Figs. 6, 7; Online Resource 1). These results suggest that this reduced level of rubisco might be insufficient to maintain CO2 assimilation in this cowpea cultivar. In cotton leaves, the down-regulation and/or inhibition of photosynthesis under water stress was related to a reduction in Rubisco subunits levels (Deeba et al. 2012). However, an increased abundance of Rubisco under salt stress has also been reported in proteomic studies of other plant species (Zörb et al. 2004; Razavizadeh et al. 2009).
Another enzyme involved in the Calvin cycle, sedoheptulose-1,7-bisphosphatase (SBP; spot 79), was found to be up-regulated (t test, p < 0.05) in response to salt stress (Figs. 6, 7; Online Resource 1). Our results are in disagreement with Deeba et al. (2012), who found a significant reduction in SBP abundance in drought-stressed cotton leaves. The authors attributed the down-regulation of this enzyme to diminished photosynthetic activity under drought stress. Our data also showed that the abundance of SBP returned to the control level after recovery treatment, suggesting that this protein may play an important role in maintaining CO2 assimilation. In S. salsa plants, high CO2 assimilation under stress conditions was explained by an increase in the Rubisco content, although SBP was down-regulated by salt stress (Li et al. 2011).
Calreticulin (spot 262) was also identified as a protein up-regulated (t test, p < 0.05) by salt stress in TVu leaves (Figs. 3, 7; Online Resource 1). Calreticulin is an important calcium-binding protein with chaperone functions and plays a pivotal role in regulating calcium homeostasis and protein folding in the endoplasmic reticulum of plants (Wang et al. 2004). Differential abundance of calreticulin has previously been reported in response to salt and osmotic stresses (Aghaei et al. 2008; Zang and Komatsu 2007; Nam et al. 2012). Although its function(s) in salt stress remains unclear, calreticulin expression appears to be involved in Ca2+ storage in response to environmental-stress signal transduction (Li et al. 2008; Sobhanian et al. 2010). Shaterian et al. (2005) reported that calreticulin expression appeared to be involved in ABA-induced salt tolerance in potato clones and that both salt tolerance and the trigger for calreticulin expression are regulated by the roots. Such results suggest the participation of calreticulin in stress and hormone signaling.
The present study demonstrated that the salt stress imposed was enough to affect protein abundance levels. We also observed, for the first time, that homeostatic mechanisms responded to new conditions by returning the levels of some proteins to control levels during recovery from salt stress, suggesting a role for these proteins in mediating the response of the cowpea to salt stress.
Our current proteome data indicate that the majority of the identified proteins in cowpea leaf were implicated in photosynthesis and energy metabolism and were most likely a part of a general stress response to help plants survive in saline conditions. We could speculate, in view of the data presented here, that tolerance may be related to the maintenance of an appropriate enzymatic-protein level required for an active energy metabolism, such as photosynthesis in salinity and/or recovery. Therefore, increases of abundance in the Rubisco activase isoforms, Ru5PK, glycine decarboxylase and OEE protein 2 in Pitiúba might be involved in the relative salt tolerance of this cowpea cultivar. However, a down-regulation of OEE protein 1, Mn-stabilizing protein-II, carbonic anhydrase and rubisco in TVu could result from metabolic inhibition in the plant and a consequent reduction in plant growth.
Abbreviations
- ACN:
-
Acetonitrile
- cTP:
-
Chloroplast transit peptides
- DTT:
-
Dithiothreitol
- EDTA:
-
Ethylenediamine tetraacetic acid
- EST:
-
Expressed sequence tag
- GDC:
-
Glycine decarboxylase
- IEF:
-
Isoelectric focusing
- MW:
-
Molecular weight
- MS:
-
Mass spectrometry
- mTP:
-
Mitochondrial targeting peptide
- OEE:
-
Oxygen-evolving enhancer
- PEG:
-
Polyethylene glycol
- pI:
-
Isoelectric point
- PVPP:
-
Polyvinylpolypyrrolidone
- PMSF:
-
Phenylmethylsulfonyl fluoride
- PPIase:
-
Peptidyl-prolyl cis–trans isomerase
- PSII:
-
Photosystem II
- RuBP:
-
Ribulose-1,5-bisphosphate
- Ru5PK:
-
Ribulose-5-phosphate kinase
- SBP:
-
Sedoheptulose-1,7-bisphosphatase
- SP:
-
Signal peptide
- TCA:
-
Trichloroacetic acid
- 2-DE:
-
Two-dimensional electrophoresis
References
Aghaei K, Ehsanpour AA, Komatsu S (2008) Proteome analysis of potato under salt stress. J Proteome Res 7:4858–4868. doi:10.1021/pr800460y
Aghaei K, Ehsanpour AA, Shah AH, Komatsu S (2009) Proteome analysis of soybean hypocotyl and root under salt stress. Amino Acids 36:91–98. doi:10.1007/s00726-008-0036-7
Babosha AV (2008) Inducible lectins and plant resistance to pathogens and abiotic stress. Biochemistry 73:812–825. doi:10.1134/S0006297908070109
Bauwe H, Kolukisaoglu Ü (2003) Genetic manipulation of glycine decarboxylation. J Exp Bot 54:1523–1535. doi:10.1093/jxb/erg171
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:246–254. doi:10.1016/0003-2697(76)90527-3
Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P, Laganà A (2008) Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal Bioanal Chem 391:381–390. doi:10.1007/s00216-008-2008-x
Cavalcanti FR, Lima JPMS, Ferreira-Silva SL, Viégas RA, Silveira JAG (2007) Roots and leaves display contrasting oxidative response during salt stress and recovery in cowpea. J Plant Physiol 164:591–600. doi:10.1016/j.jplph.2006.03.004
Chourey K, Ramani S, Apte SK (2003) Accumulation of LEA proteins in salt (NaCl) stressed young seedlings of rice (Oryza sativa L.) cultivar Bura Rata and their degradation during recovery from salinity stress. J Plant Physiol 160:1165–1174. doi:10.1078/0176-1617-00909
Costa PHA, Silva JV, Bezerra MA, Enéas-Filho J, Prisco JT, Gomes-Filho E (2003) Crescimento e níveis de solutos orgânicos e inorgânicos em cultivares de Vigna unguiculata submetidos à salinidade. Rev Bras Bot 26:289–297. doi:10.1590/S0100-84042003000300002
Debez A, Braun H-P, Pich A, Taamalli W, Koyro H-W, Abdelly C, Huchzermeyer B (2012) Proteomic and physiological responses of the halophyte Cakile maritima to moderate salinity at the germinative and vegetative stages. J Proteomics 75:5667–5694. doi:10.1016/j.jprot.2012.08.012
Deeba F, Pandey AK, Ranjan S, Mishra A, Singh R, Sharma YK, Shirke PA, Pandey V (2012) Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol Biochem 53:6–18. doi:10.1016/j.plaphy.2012.01.002
Du C-X, Fan H-F, Guo S-R, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71:1450–1459. doi:10.1016/j.phytochem.2010.05.020
Echevarría-Zomeño S, Ariza D, Jorge I, Lenz C, del Campo A, Jorrín JV, Navarro RM (2009) Changes in the protein profile of Quercus ilex leaves in response to drought stress and recovery. J Plant Physiol 166:233–245. doi:10.1016/j.jplph.2008.05.008
Ehlers JD, Hall AE (1997) Cowpea (Vigna unguiculata L. Walp). Field Crop Res 53:187–204. doi:10.1016/S0378-4290(97)00031-2
Gazanchian A, Hajheidari M, Sima NK, Salekdeh GH (2007) Proteome response of Elymus elongatum to severe water stress and recovery. J Exp Bot 58:291–300. doi:10.1093/jxb/erl226
Gomes VM, Mosqueda MI, Blanco-Labra A, Sales MP, Fernandes KVS, Cordeiro RA, Xavier-Filho J (1997) Vicilin storage proteins from Vigna unguiculata (Legume) seeds inhibit fungal growth. J Agric Food Chem 45:4110–4115. doi:10.1021/jf960942g
Guo G, Ge P, Ma C, Li X, Lv D, Wang S, Ma W, Yan Y (2012) Comparative proteomic analysis of salt response proteins in seedling roots of two wheat varieties. J Proteomics 75:1867–1885. doi:10.1016/j.jprot.2011.12.032
Huang F, Fulda S, Hagemann M, Norling B (2006) Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6:910–920. doi:10.1002/pmic.200500114
Ingle RA, Schmidt UG, Farrant JM, Thomson JA, Mundree SG (2007) Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Cell Environ 30:435–446. doi:10.1111/j.1365-3040.2006.01631.x
Kang G, Li G, Zheng B, Han Q, Wang C, Zhu Y, Guo T (2012) Proteomic analysis on salicylic acid-induced salt tolerance in common wheat seedlings (Triticum aestivum L.). Biochim Biophys Acta 1824:1324–1333. doi:10.1016/j.bbapap.2012.07.012
Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. J Proteomics 74:1301–1322. doi:10.1016/j.jprot.2011.02.006
Kottapalli KR, Rakwal R, Shibato J, Burow G, Tissue D, Burke J, Puppala N, Burow M, Payton P (2009) Physiology and proteomics of the water-deficit stress response in three contrasting peanut genotypes. Plant Cell Environ 32:380–407. doi:10.1111/j.1365-3040.2009.01933.x
Li Z, Cao Y, Zhang J, Chen S (2008) Characterization of Arabidopsis calreticulin mutants in response to calcium and salinity stresses. Prog Nat Sci 18:1219–1224. doi:10.1016/j.pnsc.2008.02.013
Li W, Zhang C, Lu Q, Wen X, Lu C (2011) The combined effect of salt stress and heat shock on proteome profiling in Suaeda salsa. J Plant Physiol 168:1743–1752. doi:10.1016/j.jplph.2011.03.018
Liu Y, Du H, He X, Huang B, Wang Z (2012) Identification of differentially expressed salt-responsive proteins in roots of two perennial grass species contrasting in salinity tolerance. J Plant Physiol 169:117–126. doi:10.1016/j.jplph.2011.08.019
Ma H, Song L, Shu Y, Wang S, Niu J, Wang Z, Yu T, Gu W, Ma H (2012) Comparative proteomic analysis of seedling leaves of different salt tolerant soybean genotypes. J Proteomics 75:1529–1546. doi:10.1016/j.jprot.2011.11.026
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–467. doi:10.1111/j.1365-3040.2009.02041.x
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663. doi:10.1111/j.1469-8137.2005.01487.x
Murillo-Amador B, Troyo-Dieguez E, García-Hernández JL, López-Aguilar R, Ávila-Serrano NY, Zamora-Salgado S, Rueda-Puente EO, Kaya C (2006) Effect of NaCl salinity in the genotypic variation of cowpea (Vigna unguiculata) during early vegetative growth. Sci Hortic 108:423–431. doi:10.1016/j.scienta.2006.02.010
Nam MH, Huh SM, Kim KM, Park WJ, Seo JB, Cho K, Kim DY, Kim BG, Yoon IS (2012) Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci 10:25. doi:10.1186/1477-5956-10-25
Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262. doi:10.1002/elps.1150090603
Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophile. J Proteome Res 9:2584–2599. doi:10.1021/pr100034f
Praxedes SC, Lacerda CF, DaMatta FM, Prisco JT, Gomes-Filho E (2010) Salt tolerance is associated with differences in ion accumulation, biomass allocation and photosynthesis in cowpea cultivars. J Agron Crop Sci 196:193–204. doi:10.1111/j.1439-037X.2009.00412.x
Razavizadeh R, Ehsanpour AA, Ahsan N, Komatsu S (2009) Proteome analysis of tobacco leaves under salt stress. Peptides 30:1651–1659. doi:10.1016/j.peptides.2009.06.023
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2:1131–1145. doi:10.1002/1615-9861(200209)2:9<1131:AID-PROT1131>3.0.CO;2-1
Sales MP, Pimenta PP, Paes NS, Grossi-de-Sá MF, Xavier-Filho J (2001) Vicilins (7S storage globulins) of cowpea (Vigna unguiculata) seeds bind to chitinous structures of the midgut of Callosobruchus maculatus (Coleoptera: Bruchidae) larvae. Braz J Med Biol Res 34:27–34. doi:10.1590/S0100-879X200100010000
Shaterian J, Georges F, Hussain A, Waterer D, De Jong H, Tanino KK (2005) Root to shoot communication and abscisic acid in calreticulin (CR) gene expression and salt-stress tolerance in grafted diploid potato clones. Environ Exp Bot 53:323–332. doi:10.1016/j.envexpbot.2004.04.008
Shen S, Matsubae M, Takao T, Tanaka N, Komatsu S (2002) A proteomic analysis of leaf sheaths from rice. J Biochem 132:613–620
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi:10.1038/nprot.2006.468
Silva FAS, Azevedo CAV (2002) Versão do programa computacional assistat para o sistema operacional windows. Rev Bras Prod Agroind 4:71–78
Silva JV, Lacerda CF, Costa PHA, Enéas Filho J, Gomes-Filho E, Prisco JT (2003) Physiological responses of NaCl stressed cowpea plants grown in nutrient solution supplemented with CaCl2. Braz J Plant Physiol 15:99–105. doi:10.1590/S1677-04202003000200005
Sobhanian H, Razavizadeh R, Nanjo Y, Ehsanpour AA, Jazil FR, Motamed N, Komatsu S (2010) Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci 8:20. doi:10.1186/1477-5956-8-19
Sobhanian H, Aghaei K, Komatsu S (2011) Changes in the plant proteome resulting from salt stress: toward the creation of salt-tolerant crops? J Proteomics 74:1323–1337. doi:10.1016/j.jprot.2011.03.018
Sugihara K, Hanagata N, Dubinsky Z, Baba S, Karube I (2000) Molecular characterization of cDNA encoding oxygen evolving enhancer protein 1 increased by salt treatment in the mangrove Bruguiera gymnorrhiza. Plant Cell Physiol 41:1279–1285. doi:10.1093/pcp/pcd061
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527. doi:10.1093/aob/mcg058
Türkan I, Demiral T (2009) Recent developments in understanding salinity tolerance. Environ Exp Bot 67:2–9. doi:10.1016/j.envexpbot.2009.05.008
Veeranagamallaiah G, Jyothsnakumari G, Thippeswamy M, Chandra Obul Reddy P, Surabhi G-K, Sriranganayakulu G, Mahesh Y, Rajasekhar B, Madhurarekha Ch, Sudhakar C (2008) Proteomic analysis of salt stress responses in foxtail millet (Setaria italica L. cv. Prasad) seedlings. Plant Sci 175:631–641. doi:10.1016/j.plantsci.2008.06.017
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132. doi:10.1016/j.copbio.2005.02.001
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252. doi:10.1016/j.tplants.2004.03.006
Xu C, Huang B (2010) Differential proteomic responses to water stress induced by PEG in two creeping bentgrass cultivars differing in stress tolerance. J Plant Physiol 167:1477–1485. doi:10.1016/j.jplph.2010.05.006
Yang Y, Wang L, Tian J, Li J, Sun J, He L, Guo S, Tezuka T (2012) Proteomic study participating the enhancement of growth and salt tolerance of bottle gourd rootstock-grafted watermelon seedlings. Plant Physiol Biochem 58:54–65. doi:10.1016/j.plaphy.2012.05.026
Yi X, McChargue M, Laborde S, Frankel LK, Bricker TM (2005) The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J Biol Chem 280:16170–16174. doi:10.1074/jbc.M501550200
Yu S, Zhang X, Guan Q, Takano T, Liu S (2007) Expression of a carbonic anhydrase gene is induced by environmental stresses in Rice (Oryza sativa L.). Biotechnol Lett 29:89–94. doi:10.1007/s10529-006-9199-z
Zang X, Komatsu S (2007) A proteomics approach for identifying osmotic-stress-related proteins in rice. Phytochemistry 68:426–437. doi:10.1016/j.phytochem.2006.11.005
Zhang W, Peumans WJ, Barre A, Astoul CH, Rovira P, Rougé P, Proost P, Truffa-Bachi P, Jalali AA, van Damme EJ (2000) Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210:970–978. doi:10.1007/s004250050705
Zhou S, Wei S, Boone B, Levy S (2007) Microarray analysis of genes affected by salt stress in tomato. Afr J Environ Sci Technol 1:14–26
Zörb C, Schmitt S, Neeb A, Karl S, Linder M, Schubert S (2004) The biochemical reaction of maize (Zea mays L.) to salt stress is characterized by a mitigation of symptoms and not by a specific adaptation. Plant Sci 167:91–100. doi:10.1016/j.plantsci.2004.03.004
Acknowledgments
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and the Instituto Nacional de Ciência e Tecnologia em Salinidade (INCTSal/CNPq).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Menossi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Abreu, C.E.B., Araújo, G. ., Monteiro-Moreira, A.C.O. et al. Proteomic analysis of salt stress and recovery in leaves of Vigna unguiculata cultivars differing in salt tolerance. Plant Cell Rep 33, 1289–1306 (2014). https://doi.org/10.1007/s00299-014-1616-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1616-5