Abstract
Key message
We cloned a novel salt stress-induced glycine-rich protein gene ( MsGRP ) from alfalfa. Its overexpression retards seed germination and seedling growth of transgenic Arabidopsis after salt and ABA treatments.
Abstract
Since soil salinity is one of the most significant abiotic stresses, salt tolerance is required to overcome salinity-induced reductions in crop productivity. Many glycine-rich proteins (GRPs) have been implicated in plant responses to environmental stresses, but the function and importance of some GRPs in stress responses remain largely unknown. Here, we report on a novel salt stress-induced GRP gene (MsGRP) that we isolated from alfalfa. Compared with some glycine-rich RNA-binding proteins, MsGRP contains no RNA recognition motifs and localizes in the cell membrane or cell wall according to the subcellular localization result. MsGRP mRNA is induced by salt, abscisic acid (ABA), and drought stresses in alfalfa seedlings, and its overexpression driven by a constitutive cauliflower mosaic virus-35S promoter in Arabidopsis plants confers salinity and ABA sensitivity compared with WT plants. MsGRP retards seed germination and seedling growth of transgenic Arabidopsis plants after salt and ABA treatments, which implies that MsGRP may affect germination and growth through an ABA-dependent regulation pathway. These results provide indirect evidence that MsGRP plays important roles in seed germination and seedling growth of alfalfa under some abiotic stress conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity stress is one of the primary causes of crop losses worldwide, but plants have evolved sophisticated mechanisms to adapt to salinity and other environmental stresses (Zhu 2002). Soil salinity is one of the most significant abiotic stresses for crop plants including legumes (Duzan et al. 2004; Golldack et al. 2011; Zahran 1999). Legumes are very important both ecologically and agriculturally since their roots are able to interact symbiotically with soil microorganisms to form nodules that fix atmospheric nitrogen. Several legumes, such as Medicago sativa (alfalfa) and the model Medicago truncatula, have a large diversity of cultivars that have adapted to soil salinity. Many M. sativa cultivars show high salinity tolerance (Munns and Tester 2008), especially the cultivar of Medicago sativa L. cv. Zhongmu-1 that we cultivated. Abiotic stresses, such as drought, salinity, and cold, regulate the expression of thousands of genes in plants at both the transcriptional and the posttranscriptional levels. The molecular mechanism of plant salt tolerance is very complex (Munns and Tester 2008; Zhu 2002). To date, little is known about the molecular links between morphological adaptations to salt stresses and endogenous signals. Suppressive subtractive hybridizations (SSH), array technologies, transcriptome high-throughput sequencing, and proteome difference analysis have become powerful and useful tools for identifying and analyzing the gene expression profiles of plants that are exposed to abiotic stresses such as drought, cold, and high salinity. We previously reported that an alfalfa cDNA library induced by salt stress was constructed using the SSH technique, which resulted in the identification of 82 expressed sequence tags (ESTs) induced by salt stress (Jin et al. 2010). In this study, we reported cloning and characterizing a novel glycine-rich protein (GRP) gene (MsGRP) using one of the EST sequences (GenBank accession No. FE896860) (Jin et al. 2010) that was previously identified from alfalfa.
Plant GRPs are a group of proteins characterized by their high content and repetitive sequences of glycine residues. A number of GRP-coding genes have been isolated from various plant species and can be divided into three main classes according to their amino acid sequences. The first GRP class consists of cell wall component proteins that contain signal peptide sequences that indicate protein export into the apoplastic space (Ringli et al. 2001). The second GRP class contains RNA-binding proteins that contain RNA recognition motifs (RRMs) at the N-terminal and a glycine-rich region at the C-terminal (Burd and Dreyfuss 1994). The third GRP class contains dehydrins, which are homologous to stress and/or developmentally regulated proteins and are thought to protect plasma membranes during stresses that involve increased osmotic pressure. Some structural cell wall GRPs have glycine contents that comprise up to 60 or 70 % of all amino acid residues (Ringli et al. 2001). Their role has yet to be elucidated. Bean GRP1.8 is a structural cell wall GRP evidenced to locate in the cell wall (Keller et al. 1988). Glycine-rich RNA-binding proteins have been described in plants including maize (Gomez et al. 1988), Arabidopsis thaliana (Carpenter et al. 1994; van Nocker and Vierstra 1993), barley (Dunn et al. 1996; Molina et al. 1997), and alfalfa (Ferullo et al. 1997). Some of these proteins are suggested to play a role in stress responses, as their mRNA levels increased following exposure to a variety of environmental stresses including cold, injury, water stress, and heavy metals (Sachetto-Martins et al. 2000). At least one GRP has been found in each higher plant species that has been examined. This finding suggests that these proteins are involved in essential plant functions, but not study to date has provided clear evidence of their functional roles. This leads us to hypothesize the possible involvement of these proteins in plant responses to changing environmental conditions.
The functions and importance of GRP in stress responses are speculative since the in vivo functional data collected so far remains limited. MsGRP is predicted to be the ortholog of one GRP gene (AT4G21620) in Arabidopsis thaliana due to their high amino acid sequence similarity. The function of AT4G21620 is currently unknown. In this paper, we demonstrate the subcellular location of MsGRP in the onion epidermis as well as its expression patterns under salt, ABA, and drought stresses. We also demonstrate its in vivo function in salt and ABA sensitivity by analyzing MsGRP-overexpressing transgenic Arabidopsis plants (35S::MsGRP). Our results suggest that MsGRP can negatively affect seed germination and seedling growth of transgenic Arabidopsis plants after salt and ABA treatments.
Materials and methods
Plant material and treatments
Medicago sativa L. cultivated variety Zhongmu-1 was gown in hydroponic culture of Hoagland’s solution in a growth chamber at 25 °C under a 16 h light/8 h dark photoperiod and 75 % relative humidity. The solution was changed every 5 days. To investigate the expression pattern of MsGRP under different stresses, 30-day-old Zhongmu-1 seedlings were transferred into Hoagland’s solution of 300 mM NaCl, 0.1 mM ABA, and 20 % PEG-6000. Shoot and root of the seedlings were harvested separately at different time points (0, 2, 4, 8, 12, and 24 h) and frozen in liquid nitrogen for further analysis. To investigate the expression pattern of MsGRP in different tissues, the leaf, stem, and root of 30 days old Zhongmu-1 seedlings grown in normal Hoagland’s solution were harvested separately for further analysis.
Arabidopsis thaliana ecotype Col-0 was used in transgene experiment. Arabidopsis thaliana plants were grown at 22 °C under long-day conditions (16 h light/8 h dark). Seeds harvested from individual plants grown under identical environmental conditions were used for germination and seedling growth assays. Germination assays were carried out on three replicates of 80–100 seeds. Seeds were sown on MS medium supplemented with 3 % sucrose, and the plates were placed at 4 °C for 2 days in the dark prior to germination. To determine the effect of salt stress on germination, the medium was supplemented with 100, 200 mM NaCl. The effect of ABA on germination was measured on medium supplemented with 5 and 10 μM ABA. To determine the effect of salt stress on seedling growth, 10-day-old seedlings germinated in normal medium were transferred to medium supplemented with 200 mM NaCl, and the chlorophyll a and b content of the seedlings was measured after treating for 10 days. To test the effect of ABA stress on seedling growth, the seeds were allowed to germinate under normal growth conditions and then transferred to medium supplemented with 10 μM ABA. And the length of the roots was measured at the indicated times after transfer to medium with ABA for 10 days.
RNA isolation and reverse transcription
Total RNA was extracted using Trizol (Invitrogen) and then dissolved in ddH2O that was treated with diethylpyrocarbonate. Prior to the reverse transcription process, the total RNA was treated with RNase-free DNase I (MBI Fermentas) for 30 min at 37 °C. The cDNA was synthesized using PrimeScript reverse transcriptase (Takara). The harvested complementary cDNA was stored at −20 °C until use.
Rapid amplification of cDNA ends (RACE) of the MsGRP 3′ and 5′ ends
A SSH cDNA library of Zhongmu-1 alfalfa induced by salt was previously constructed and screened. An EST (GenBank accession No. FE896860) of a novel gene was isolated. RACE was performed to amplify its unknown 3′ and 5′ ends. Total RNA was used to synthesize 5′-RACE-Ready-cDNA and 3′-RACE-Ready-cDNA according to the manufacturer’s recommendation of BD SMARTTM RACE cDNA amplification kit (Clontech). Based on the EST sequence already obtained, we designed gene-specific primers P1 (5′-CGGAACGGCTGAATGAGGAGAAACACT-3′) to amplify the 5′-cDNA end, and P2 (5′-CAAATGGAGGACACTCTAAAAATGGCA-3′) to amplify the 3′-cDNA end. The polymerase chain reaction (PCR) conditions were as follows: pre-denaturalization at 94 °C for 5 min, 30 cycles of thermocycling at 94 °C for 30 s, 68 °C for 1 min, and an additional polymerization step at 72 °C for 10 min. The PCR products were detected using electrophoresis on a 1 % agarose gel stained with ethidium bromide and then purified using the DNA gel extraction kit (Takara). The products were cloned into the pMD-19T vector (Takara) and then transformed into E. coli DH5α. And recombinant plasmids were sequenced by Beijing Genome Institute.
We obtained the full-length cDNA sequence of MsGRP by comparing and aligning the sequences of the known EST, 5′RACE, and 3′RACE sequences. MsGRP and its putative encoding protein was subsequently analyzed for molecular characterization purposes such as the conserved motifs, sequence homology, secondary structure, hydropathicity analysis, phylogenetic analysis, subcellular localization, and expression patterns under different environmental stresses.
Bioinformatics analysis
The secondary structure of the putative protein was analyzed using the HNN SECONDARY STRUCTURE PREDICTION METHOD (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_hnn.html). The specifically functional sites of the putative protein were analyzed using the SMART (http://smart.embl-heidelberg.de/). Sequences homology analysis was performed against the nucleotide and protein database of GenBank using the BLAST (NCBI) program. The phylogenetic relationship between the amino acid sequences was analyzed using the ClustalX 2.0 and MEGA 5.0 programs using the neighbor joining method. A potential signal peptide cleavage site was identified using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/). The pI/MW of the putative protein was predicted using the ExPaSy website (http://us.expasy.org/tools/pi-tool.htm).
Subcellular localization of MsGRP
To observe the subcellular localization of MsGRP, an MsGRP–GFP fusion was constructed. Primers P3 (5′-ctcgagATGAAAACTATGACCACCTTTCT-3′) and P4 (5′-gtcgacACAATAGGCACTGCACTTCTT-3′) were designed to amplify the MsGRP-coding sequence. The PCR product was cloned into a pMD19T vector. The recombinant vector pMD19T-MsGRP and transient expression vector pA7-GFP were digested by XhoI and SalI restriction endonucleases. The digested pA7-GFP fragment and the MsGRP gene fragment were linked with T4-ligase to obtain the fusion construct pA7-MsGRP-GFP. MsGRP-GFP was driven by a 35S promoter. The pA7-MsGRP-GFP vector was transformed into onion inner epidermis using a particle gun (Bio-Rad) and visualized under laser scanning confocal microscopy (Nikon).
Real-time fluorescent quantitative PCR
The 30-day-old alfalfa seedlings were transferred into Hoagland’s solution with salt (300 mM), ABA (0.1 mM), and PEG-6000 (20 %) for 0, 2, 4, 10, and 24 h. Seedlings grown on normal Hoagland’s solution were used as controls. The roots and shoots of the seedlings were harvested. Total RNA of each was reverse transcribed, and the synthesized cDNA was used as a template in real-time PCR. The real-time fluorescent quantitative PCR was analyzed by ABI-7500 (ABI). Primers P5 (5′-CAAATGGAGGACACTCTAAAAATGGC-3′) and P6 (5′-GGAACGGCTGAATGAGGAGAAACA-3′) were used for amplification of MsGRP. β-actin gene was used as housekeeping gene to normalize the target gene quantities. The amplified fragment of MsGRP was 122 bp. P7 (5′-TGGGCTGCCACAGAACATTTGA-3′) and P8 (5′-GCTGTGGTTGCTTTTTTGGTGTCTC-3′) were used to amplify the actin gene. The amplified fragment of actin gene was 117 bp. The PCR annealing temperature of all primers was 60 °C.
Overexpression of MsGRP in Arabidopsis
The full-length coding region of MsGRP was cloned into the XbaI and BamHI site of the pBI121 vector to create pBI121-MsGRP that over-expresses MsGRP and GUS (uidA) reporter gene under control of the CaMV 35S promoter (35S::MsGRP). Primers P9 (5′-tctagaATGAAAACTATGACCACCTTTCT-3′) and P10 (5′-ggatccACAATAGGCACTGCACTTCTT-3′) were designed to amplify the coding sequence of MsGRP. The construction process of the overexpression vector pBI121-MsGRP was performed using the construction method of the subcellular localization vector above. The pBI121-MsGRP vector was transformed into A. tumefaciens strain GV3101. Transformation of Arabidopsis was performed by the vacuum infiltration method (Bechtold and Pelletier 1998) using A. tumefaciens GV3101. The seeds were harvested and plated on kanamycin (50 μg/mL) selection medium for identification of the transgenic plants. After further selection of the transgenic lines using a 3:1 segregation ratio, T3 homozygous lines were used for the phenotypic investigation. MsGRP overexpression in the transgenic plants was verified using RT-PCR analysis. GUS activity was visualized in the leaf tissue as described previously (Jefferson et al. 1987). Chlorophyll a and b were measured after extraction in 80 % acetone (Lichtenthaler 1987).
Results
Cloning of the full-length MsGRP cDNA
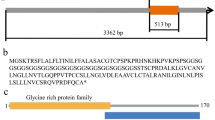
In the 5′ RACE PCR reaction, a 486-bp fragment was obtained using primers P1 and UPM (contained in the BD SMARTTM RACE cDNA amplification kit). A 344-bp fragment was isolated using the 3′ RACE primers P2 and UPM. A full-length 631-bp sequence was obtained by assembling the known EST sequence with the 3′ and 5′ end sequences. The full length contained a 492-bp open reading frame (ORF) that encoded a protein containing 163 amino acids (26 % glycine, 10 % serine) (Fig. 1). The molecular weight of the putative protein was approximately 16.02 kDa, while the pI was 9.67. It was tentatively named MsGRP for its high glycine content. (Its nucleotide accession number is JQ340083 and the coded protein accession number is AFA45120.)
Analysis result of the putative MsGRP protein
The secondary structure of the MsGRP protein was predicted to contain 11.6 % alpha helix, 6.13 % extended strand, and 82.21 % random coil using the hierarchical neural network secondary structure prediction method. According to the prediction results obtained by SMART, there was a Structural Classification of Proteins (SCOP) domain of d1a0tp from Lys40 to Ser103, which acts as a transmembrane domain in the membrane and in cell surface proteins and peptides. There was a glycine-rich region from Gly46 to Gly150 (Fig. 1). A signal peptide was predicted to be present in MsGRP by SignalP 4.0. Based on the amino acid sequence, a phylogenetic tree was constructed by using the neighbor joining method. The results showed that MsGRP was predicted to be ortholog of some GRPs in some other plants, such as NP_001190788 (AT4G21620) of A. thaliana (Fig. 2).
Phylogenetic tree analysis of MsGRP and other proteins. The phylogenetic tree was constructed using the neighbor-joining method and employed amino acid sequences. The amino acid sequences were obtained from NCBI with the following accession numbers: AFA45120 from Medicago sativa (MsGRP), XP_003593472 from Medicago truncatula, NP_001167690 from Zea mays, NP_001190788 from Arabidopsis thaliana, NP_001050889 from Oryza sativa, XP_002317234 from Populus trichocarpa, NP_001240061 from Glycine max, XP_002521418 from Ricinus communis, XP_002442605 from Sorghum bicolor, and XP_003561364 from Brachypodium distachyon. Tree reliability was verified by bootstrap analysis with 1,000 replicates
Subcellular localization of MsGRP
The online ProtComp version 9.0 program (http://www.softberry.com) was used to predict the subcellular localization. The result predicted by the integral prediction method showed that MsGRP is most likely localized in the extracellular space. (The prediction score was approximately 8.68.) To observe the subcellular localization of MsGRP, the MsGRP-GFP fusion was transformed into onion inner epidermis using a particle gun. In cells where the pA7-GFP vector was introduced alone (Fig. 3), fluorescence was visualized throughout those cells with no clear preference for localization. However, in cells expressing the MsGRP-GFP fusion, fluorescence was found to be clearly localized near the plasmalemma or cell wall (Fig. 3).
Subcellular localization of MsGRP in the onion inner epidermis. The pA7-MsGRP-GFP and pA7-GFP vectors were transformed into onion epidermal cells using a particle gun. The fluorescence signals were examined using confocal laser scanning microscopy. The GFP fluorescence from cells expressing MsGRP-GFP fusion protein was localized near the plasmalemma (G1–G3). The GFP fluorescence was distributed throughout the cells expressing pA7-GFP vector (C1–C3). The photographs were taken in superposition of bright and dark vision (G1 and C1), dark field vision (G2 and C2), and bright light vision (G3 and C3)
Expression of MsGRP in different environmental stresses
A real-time PCR analysis was carried out to investigate MsGRP gene expression in alfalfa under different environmental stresses. The alfalfa seedlings were treated with 300 mM NaCl, 0.1 mM ABA, and 20 % PEG-6000 for 2, 4, 10, and 24 h. Seedlings grown on Hoagland’s solution without treatment were used as controls. The relative expression of MsGRP was calculated as follows: ratio = \( 2^{{ - \Delta \Delta C_{{\text{t}}} }} = 2^{{ - [C_{{{\text{t,t}}}} - C_{{{\text{t}},{\text{r}}}} ]}} \) (C t cycle threshold, C t,t C t of the target gene, C t,r C t of the control gene β-actin). The expression levels of MsGRP in different alfalfa tissues were also detected using real-time PCR.
The expression level of MsGRP in different time periods after 300 mM NaCl treatment is showed in Fig. 4c. Compared with control (0 h), the transcripts of MsGRP in the roots and shoots increased significantly after 4-h treatment, while the MsGRP expression level in the roots increased more significantly than that in the shoots (Fig. 4c). After treatment with 0.1 mM ABA, the relative expression level of MsGRP in the roots began to increase significantly after 4-h treatment, while the MsGRP mRNA level in the shoots increased obviously after 2-h treatment (Fig. 4b). After 2-h treatment with 20 % PEG, the relative expression level of MsGRP mRNA increased significantly. At 10 h, the expression level of MsGRP in the roots reached its maximum, while the expression level of MsGRP in the shoots reached its maximum at 4 h (Fig. 4a). Of the three detected tissues (root, stem, and leaf) without any stress, the transcripts of MsGRP in the leaf were the highest (Fig. 4d).
Relative expression levels of MsGRP in Zhongmu-1 alfalfa roots and shoots under different stresses. a–c The relative expression levels of MsGRP in Zhongmu-1 alfalfa treated with 20 % PEG-6000, 0.1 mM abscisic acid, or 300 mM NaCl. The y-axis indicates the relative fold increased expression level compared to 0 h. d The expression level of MsGRP in Zhongmu-1 alfalfa roots, leaves, and stems under normal conditions. The y-axis indicates the relative fold increased expression level in the root
Analysis of the transgenic Arabidopsis plants
To investigate the functional role of the MsGRP gene, a complete ORF (Fig. 1) was overexpressed in Arabidopsis plants using Agrobacterium-mediated transformation. Preliminary screening of kanamycin-resistant putative Arabidopsis transformants was performed using a GUS assay (Fig. 5). The presence of MsGRP was confirmed in the GUS-positive plants by PCR using primers P9 and P10 (data not shown). Sixteen independently transformed T 0 transgenic plants were selected and grown to maturity to obtain homozygous lines. Two T 3 homozygous lines, G1 and G5, were randomly selected and used throughout the study.
The wild-type and 35S::MsGRP plants were compared for their germination and seedling growth on medium with NaCl and ABA concentrations. The germination rates of the transgenic plant and wild-type seeds were not significantly different in normal MS medium (Fig. 6). And all the germination retarded by a few days in NaCl or ABA MS medium compared with that in normal MS medium. In NaCl or ABA medium, the germination rate of the transgenic plant seeds was lower than that of the wild-type (Figs. 7a, b, 8a, b). The inhibition of transgenic plant germination was more significant as the concentration of NaCl or ABA increased. In the MS medium with ABA, >90 % of the wild-type seeds germinated by day 10. The germination rates of G1 and G5 were approximately 70 % in 5 µM ABA and 50 % in 10 μM ABA (Fig. 6a). In the MS medium with 100 mM NaCl, the germination rates of the transgenic plants and wild-type seeds were >90 % at day 10, while the germination rate of the wild-type was a little higher than those in lines G1 and G5 (Fig. 6b). The germination rate of the wild-type was significantly higher than that of the transgenic plants in 200 mM NaCl stress from day 5 (Fig. 6b). These results clearly indicate that MsGRP retarded seed germination under salt and ABA conditions.
The growth influences of salt and ABA conditions were also tested for the wild-type plant and 35S::MsGRP plants. Seeds of wild-type and 35S::MsGRP plants were allowed to fully germinate on normal MS medium, and 10-day-old seedlings were transferred to MS medium containing 200 mM NaCl or 10 μM ABA. The two 35S::MsGRP lines were observed to be more sensitive to NaCl and ABA than the wild-type plants (Figs. 7c, d, 8c, d). The seedlings grown in 10 μM ABA or 200 mM NaCl were monitored by measuring the content of chlorophyll (Fig. 9). The chlorophyll contents of the transgenic lines were similar with the content of the wild-type plant under normal conditions, but the chlorophyll contents of the transgenic lines were significantly lower than those of the wild-type plants under 10 μM ABA or 200 mM NaCl treatment. These results indicate that MsGRP overexpression retarded seed germination of Arabidopsis plants and retarded the seedling growth under salt or ABA treatments.
Chlorophyll content of the leaves of wild-type, G1, and G5 plants. Several 10-day-old transgenic G1, G5, and wild-type plants were transferred to MS medium with 10 μM abscisic acid (ABA) or 200 mM NaCl, and the chlorophyll contents of these plants were measured after 10-day treatment. The different letters in each column indicate significant differences (p < 0.05) (Student’s t test)
Discussion
We are interested in studying the molecular mechanism of salt stress on plant gene expression. To begin this work, we isolated a cDNA that is inducible in alfalfa (M. sativa L. cv. Zhongmu-1) tissues by salt stress. This cDNA was predicted to encode a glycine-rich protein (MsGRP). The subcellular localization analysis experiment showed that the MsGRP protein is localized near the membrane, and we had predicted it to be a porin on the membrane or an extracellular structural component of the cell wall. The GRP group members have been found in the cell walls of many higher plants and form a third group of structural protein components of the wall in addition to extensins and proline-rich proteins. Each cell wall GRP is localized mainly in the vascular tissue of the plant. For instance, the major cell wall GRP of the French bean is localized at the ultrastructural level in the modified primary cell wall of protoxylem. Immunological studies have shown that these proteins form a major part of these highly extensible and specialized cell walls (Ringli et al. 2001). Each GRP is characterized by its high glycine residue content. However, a GRP is not necessarily a structural protein, as RNA-binding proteins also have glycine-rich domains (Burd and Dreyfuss 1994; Kim et al. 2008; Sachetto-Martins et al. 2000). Protein structure and domain analysis results suggest that MsGRP contains a glycine-rich domain and a SCOP domain (SCOP is a basic domain of some membrane and cell surface porins) (Fig. 1) but does not contain any RNA-binding motifs. MsGRP also contains a (Gly-X) n repetitive motif of some extracellular GRP proteins in the cell wall (Fig. 1).
GRP expression has been shown to be developmentally regulated in some cases. For example, the grp1.8 and grp1.8-like gene(s) are mainly expressed in young tissue, whereas the expression in older stages of development is strongly reduced (Condit 1993; Ye et al. 1991). In addition to developmental regulation, grp gene expression has been shown to be altered by external influences such as injury, hormone treatment, low temperatures, and water or ozone stress (Bergeron et al. 1994; Condit 1993; de Oliveira et al. 1990; Keller et al. 1988; Laberge et al. 1993; Lei and Wu 1991; No et al. 1997; Showalter et al. 1992). In fact, some grp genes were cloned in differential hybridization experiments performed with the goal of isolating genes induced by drought, water, or low-temperature stress (Chang et al. 1996; Laberge et al. 1993; No et al. 1997). Thus, the GRP proteins are not only required in certain stages of development but are often also stress-induced. Transcriptional expression experiments using real-time PCR showed that MsGRP gene expression is not tissue-specific as shown by the accumulation of its transcript in roots, leaves, and stems. It also showed that MsGRP mRNA was induced by NaCl and ABA as well as significantly by PEG 6000 (Fig. 4). These results suggest that MsGRP gene expression is dependent upon the water status of plants and that ABA may act as a signal in this response.
We have shown that MsGRP affects seed germination and seedling growth of transgenic Arabidopsis plants under salt stress and ABA. The germination rate of 35S::MsGRP Arabidopsis seeds was restrained more obviously in salt and ABA stress conditions. The growth of 35S::MsGRP Arabidopsis seedlings was more sensitive to NaCl and ABA stresses. Although the molecular mechanisms underlying the roles of MsGRP under stress conditions are not clearly understood, the induction by salt and ABA as well as the overexpressing transgenic Arabidopsis showing more sensitivity to ABA and salt suggest that MsGRP may play a critical role in salt stress regulation and may cause an anaphylactic response in plants after salt and ABA treatments. It is interesting that, despite being induced by salt, ABA and PEG, MsGRP plays a negative role in salt and ABA stresses since our results showed that the 35S::MsGRP Arabidopsis became more sensitive than the wild-type under those stresses.
The function of MsGRP is similar to that of Arabidopsis GRP7 reported previously (Kim et al. 2008). However, these two genes have many differences. Arabidopsis GRP7 is an RNA-binding protein in the nucleus, but MsGRP does not contain an RRM domain and probably localizes in the plasmalemma or cell wall. Based on the results of subcellular localization and bioinformatics analysis (Fig. 3), we predict that MsGRP probably plays a role in the porins on the membrane and participates in iron or water transport. Further studies are needed to investigate the molecular mechanism of MsGRP in the plant salt stress response. The knockout of MsGRP in alfalfa will be another way to investigate its function in future studies.
In conclusion, the present work provides novel information that increases our current knowledge of the functional roles of GRP in response to environmental stresses. The phenotypic analysis of MsGRP-overexpressing transgenic plants implies that MsGRP plays an important role on germination and seedling growth under salt and ABA conditions. The proposition that MsGRP negatively affects plant tolerance under salt and ABA stresses and has a negative impact on seed germination and seedling growth under salt and ABA stresses encourages further investigations into the cellular functions of MsGRP in stress responses. Since our knowledge regarding the cellular functions of the GRP family in stress responses remains far from sufficient, further studies might help us discover more of their functional roles in stress conditions.
References
Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82:259–266
Bergeron D, Boivin R, Baszczynski CL, Bellemare G (1994) Root-specific expression of a glycine-rich protein gene in Brassica napus. Plant Sci 96:87–98
Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615–621
Carpenter CD, Kreps JA, Simon AE (1994) Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol 104:1015–1025
Chang SJ, Puryear JD, Dias MADL, Funkhouser EA, Newton RJ, Cairney J (1996) Gene expression under water deficit in loblolly pine (Pinus taeda): isolation and characterization of cDNA clones. Physiol Plant 97:139–148
Condit CM (1993) Developmental expression and localization of petunia glycine-rich protein 1. Plant Cell 5:277–288
de Oliveira DE, Seurinck J, Inzé D, Van Montagu M, Botterman J (1990) Differential expression of five Arabidopsis genes encoding glycine-rich proteins. Plant Cell 2:427–436
Dunn MA, Brown K, Lightowlers R, Hughes MA (1996) A low-temperature-responsive gene from barley encodes a protein with single-stranded nucleic acid-binding activity which is phosphorylated in vitro. Plant Mol Biol 30:947–959
Duzan HM, Zhou X, Souleimanov A, Smith DL (2004) Perception of Bradyrhizobium japonicum Nod factor by soybean [Glycine max (L.) Merr.] root hairs under abiotic stress conditions. J Exp Bot 55:2641–2646
Ferullo JM, Vezina LP, Rail J, Laberge S, Nadeau P, Castonguay Y (1997) Differential accumulation of two glycine-rich proteins during cold-acclimation alfalfa. Plant Mol Biol 33:625–633
Golldack D, Luking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30:1383–1391
Gomez J, Sanchez-Martinez D, Stiefel V, Rigau J, Puigdomenech P, Pages M (1988) A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature 334:262–264
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jin H, Sun Y, Yang Q, Chao Y, Kang J, Li Y, Margaret G (2010) Screening of genes induced by salt stress from Alfalfa. Mol Biol Rep 37:745–753
Keller B, Sauer N, Lamb C (1988) Glycine-rich cell wall proteins in bean: gene structure and association of the protein with the vascular system. EMBO J 7:3625
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh CH, Woo YM, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466
Laberge S, Castonguay Y, Vézina LP (1993) New cold- and drought-regulated gene from Medicago sativa. Plant Physiol 101:1411
Lei M, Wu R (1991) A novel glycine-rich cell wall protein gene in rice. Plant Mol Biol 16:187–198
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Molina A, Mena M, Carbonero P, Garcia-Olmedo F (1997) Differential expression of pathogen-responsive genes encoding two types of glycine-rich proteins in barley. Plant Mol Biol 33:803–810
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
No EG, Flagler RB, Swize MA, Cairney J, Newton RJ (1997) cDNAs induced by ozone from Atriplex canescens (saltbush) and their response to sulfur dioxide and water-deficit. Physiol Plant 100:137–146
Ringli C, Keller B, Ryser U (2001) Glycine-rich proteins as structural components of plant cell walls. Cell Mol Life Sci 58:1430–1441
Sachetto-Martins G, Franco LO, de Oliveira DE (2000) Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta 1492:1–14
Showalter AM, Butt AD, Kim S (1992) Molecular details of tomato extensin and glycine-rich protein gene expression. Plant Mol Biol 19:205–215
van Nocker S, Vierstra RD (1993) Two cDNAs from Arabidopsis thaliana encode putative RNA binding proteins containing glycine-rich domains. Plant Mol Biol 21:695–699
Ye ZH, Song YR, Marcus A, Varner JE (1991) Comparative localization of three classes of cell wall proteins. Plant J 1:175–183
Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989, table of contents
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 30970409).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Benvenuto.
Rights and permissions
About this article
Cite this article
Long, R., Yang, Q., Kang, J. et al. Overexpression of a novel salt stress-induced glycine-rich protein gene from alfalfa causes salt and ABA sensitivity in Arabidopsis. Plant Cell Rep 32, 1289–1298 (2013). https://doi.org/10.1007/s00299-013-1443-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1443-0