Abstract
Key message
Our results showed that GbIRL1 belongs to the PCBER protein family. Besides, IRL1 gene was a novel gene regulating lignin change and also effecting the accumulation of flavonoids in Ginkgo.
Abstract
A cDNA encoding the IFR-like protein was isolated from the leaves of Ginkgo biloba L., designated as GbIRL1 (Accession no. KC244282). The cDNA of GbIRL1 was 1,203 bp containing a 921 bp open reading frame encoding a polypeptide of 306 amino acids. Comparative and bioinformatic analyses revealed that GbIRL1 showed extensive homology with IFLs from other gymnosperm species. Phylogenetic tree analysis revealed that GbIRL1 shared the same ancestor in evolution with other PCBERs protein and had a further relationship with other gymnosperm species. The recombinant protein was successfully expressed in E. coli strain with pET-28a vector. The vitro enzyme activity assay by HPLC indicated that recombinant GbIRL1 protein could catalyze the formation the TDDC, IDDDC from DDDC, DDC. Tissue expression pattern analysis showed that GbIRL1 was constitutively expressed in stem and roots, especially in the parts of the pest and fungal infection, with the lower expression being found in 1- or 2-year old stem. The increased expression of GbIRL1 was detected when the seedlings were treated with Ultraviole-B, ALA, wounding and ethephon, abscisic acid, salicylic acid. Correlation analysis between GbIRL1 activity and flavonoid accumulation during Ginkgo leaf growth indicated that GbIRL1 might be the rate-limiting enzyme in the biosynthesis pathway of flavonoids in Ginkgo leaves. Results of RT-PCR analysis showed that the transcription level of change in GbIRL1 power correlated with flavonoid contents, suggesting IRL1 gene as a novel gene regulating lignin change and also effecting the accumulation of flavonoids in Ginkgo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
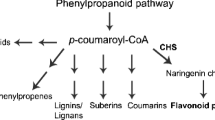
Plant produces a variety of secondary metabolites to counteract not only environmental stresses but also as part of normal growth and development processes (Gapper and Dolan 2006). As a major component of plant-specialized metabolism, phenylpropanoid biosynthetic pathways provide anthocyanins for pigmentation, flavonoids such as flavones for protection against UV photodamage, various flavonoids and isoflavonoid inducers of Rhizobium nodulation genes, polymeric lignin for structural support and assorted antimicrobial phytoalexins (Ferrer et al. 2008). Isoflavonoids, valuable secondary metabolites for important physiological processes, are produced primarily in leguminous plants and are rarely found in gymnosperms (Kim et al. 2010). Although isoflavonoid phytoalexins are confined primarily to legumes, IFR-like (abbreviated hereafter as IRL) proteins from non-leguminous plants are also part of the IFR family (Kim et al. 2010). At present, numerous studies dealing with the biosynthesis pathway to the lignans (Suzuki and Umezawa 2007) and isoflavonoids (Dixon 1999) have resulted in the isolation and characterization of various lignan reductases, namely pinoresinol lariciresinol reductases (PLRs) (Hano et al. 2006), phenylcoumaran benzylic ether reductases (PCBERs) (Turley 2008) and isoflavone reductases (IFRs) (Paiva et al. 1991). Several IRLs have been reported to be involved in responses to biotic or abiotic stress, such as Nicotiana tabacum NtIRL (Shoji et al. 2002), Zea mays ZmIRL (Petrucco et al. 1996), Oryza saliva OsIRL1 (Kim et al. 2003) and Fagopyrum cymosum FcIRL (Zhu et al. 2009). Phenylcoumaran benzylic ether reductase is a member of the IRL family (Karamloo et al. 2001), which catalyzes the non-enantio-specific NADPH-dependent reduction of the benzylic ether functionality in the 8-5′-linked lignans dehydrodiconiferyl alcohol and dihydro dehydrodiconiferyl alcohol (Gang et al. 1999). Because of the high sequence similarities, both PLR and PCBER are phylogenetically related to isoflavone reductase, a branch point enzyme in the biosynthesis of isoflavonoids (Paiva et al. 1991; Tiemann et al. 1991). Plant PCBER was reported to be up-regulated by abiotic stress (Nam et al. 2003), the circadian clock (Harmer et al. 2000), and reported to be strongly associated with phenylpropanoid biosynthesis in young xylem, phloem and differentiating xylem ray parenchyma cells (Vander Mijnsbrugge et al. 2000).
In the present time, Ginkgo biloba is one of the most popular functional plants, especially as medicinal plants. Extracts of G. biloba leaves contain active compounds such as flavonoids and terpene lactones (ginkgolides and bilobalide) and can, therefore, be used to increase peripheral and cerebral blood flow (Smith and Luo 2004; van Beek 2002). Flavonoids metabolism and lignans metabolic belong to phenylalanine metabolism path branch; lignans synthesis might affect the accumulation of G. biloba flavonoids. Therefore, to study the medicinal secondary metabolism, a cDNA library derived from high flavonoids producing leaves of G. biloba was constructed using SMART-RACE technique to isolate key genes related to biosynthesis of medical components. Here we report the cloning and characterization of a first GbIRL1 encoding a putative PCBER. We also discuss its potential function in secondary metabolism of G. biloba.
Materials and methods
Plant materials and treatments
Fourteen-year-old grafted G. biloba seedlings were grown in a greenhouse in Huanggang (E, 111°54′–112°19′, N, 30°6′–30°39′, Hubei province, central of China) were sampled as plant materials. For gene cloning and tissue expression, diverse tissues including young leaves, mature leaves, ovules, stamens, albumen, gynoecia, stems and roots were collected for DNA and RNA extraction as described by Xu et al. (2008b). Tissues were immediately frozen in liquid nitrogen and kept at −80 °C prior to total RNA extraction.
Total RNA was isolated from injured stem tissues of 14-year-old grafted G. biloba seedling after they were attacked by Rhizoctonia solani. For fungal infections blade, the total RNA was isolated from Ginkgo leaves cells punctured with an infected agent isolate of Phyllosticta mortoni Fairman. One-year-old cuttings from the same genotypic strain of G. biloba were subjected to treatments with UV-B, wounding (WOU), 5-aminolevulinic acid (ALA), abscisic acid (ABA), salicylic acid (SA) and ethephon (ETH). For UV-B treatment, seedlings were exposed to 1,500 μJ/m2 UV-B irradiation in a closed chamber, and the control cuttings were placed in a dark closed chamber. The edges of Ginkgo leaves were cut by about 0.6 cm with scissors for wounding treatment; the intact leaves of Ginkgo were as control. The ALA (100 μM), ABA (10 mM), SA (25 mM), and ETH (20 mM) were dissolved in 0.01 % Tween 20 and sprayed onto young leaves. The control leaves were sprayed with an equivalent volume of 0.01 % (v/v) Tween 20.
Cloning of the full-length cDNA
Given species conservation of the functional domain of the IRLs protein and the Ginkgo EST database in GenBank, a pair of degenerate primers, IRF and IRR, was designed according to the conserved peptide sequences, LGGAGYR and PGSSILFT, to amplify a conservative fragment of the GbIRL1 gene. The purified PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian China), and the positive clone was confirmed by a BLASTN query of the NCBI Database to verify the homology of this cloned fragment with IRL sequences from other species.
Based on the sequence of cloned GbIRL1 fragments, the specific primer pairs (IR5R and IR3R) and the nested primer pairs (IR5 N and IR3 N) were designed to amplify the 5′ and 3′ end of GbIRL1 using the SMART™ RACE cDNA Amplification Kit (Clontech, USA). The PCR products were purified and cloned into the pMD18-T vector for sequencing. After comparing and aligning the sequence of 5′RACE, 3′RACE, and the internal fragment, the full-length cDNA sequence of GbIRL1 was obtained.
Amplification of the 5′-promoter region of GbIRL1
The promoter region was isolated by genomic walk strategy using a Universal Genomewalker™ kit (Clontech, USA) according to the manufacturer’s instructions. The first turn of PCR was carried out using AP1 and IR5R as primers, and constructed libraries as a template using the following protocol: 94 °C for 4 min; 30 cycles (98 °C for 10 s, 68 °C for 3 min) and 68 °C for 10 min. The products were 20-fold-diluted and used for nested PCR, which was performed using AP2 and IR5 N as primers under the following conditions: 94 °C for 4 min; 32 cycles (94 °C for 30 s, 56 °C for 50 s, 72 °C for 3 min); and 72 °C for 10 min. The PCR products were cloned into pMD18-T for sequencing (Sangon, China).
Relative quantification by QRT-PCR
The transcription levels of GbIRL1 were determined in different G. biloba tissues, as well as in young seedling leaf samples collected at different time points after stress and hormone treatments. QRT-PCR was carried out using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, American) with SYBR Green PCR Master Mix (Applied Biosystems, American), according to the manufacturers protocol. The G. biloba glyceraldehydes-3-phosphate dehydrogenase gene (GbGAPDH, L26924) (Jansson et al. 1994) was used as the reference gene as described by Xu et al. (2008b).
The gene-specific primers (IRT3, IRT4) and reference primers (GAPU, GAPD) for QRT-PCR were listed in Table 1. The QRT-PCR conditions were: 10 min at 95 °C, and 40 cycles (95 °C for 15 s, 60 °C for 1 min). Before performing QRT-PCR, primer efficiency was evaluated using both GbIRL1 and GbGAPDH at 100, 150, 200, 250 and 300 nM combinations. A 120 nM concentration was chosen as most suitable combination for both genes. For each plant sample, aliquots of 150 ng total RNA was analyzed for each gene and the two genes (GbIRL1 and GbGAPDH) were always analyzed, simultaneously. Each sample was amplified three times and all reactions were performed on ABI PRISM 7500 Sequence Detection System. With a housekeeping gene GbGAPDH, the relative amount of the GbIRL1 transcript is presented as 2(−ddCt) according to the C T method (Schmittgen and Livak 2008). When comparing the expression of GbIRL1 in different tissues or treatments, the relative expression of GbIRL1 was achieved by calibrating its transcription level to that of the reference gene, GbGAPDH.
Functional expression of the GbIRL1 in Escherichia coli
To clone GbIRL1 into an expression vector, a pair of primers, IRZF and IRZR, were designed and synthesized to amplify the coding region by RT-PCR with the incorporation of a restriction enzyme site and a protective base to simplify later vector construction. After confirmation by sequencing, the resulting recombinant plasmid was introduced into BL21 (DE3) by the heat shock method. A single colony of E. coli BL21 cells harboring the expression plasmid pET28a-GbIRL1 was inoculated at 37 °C in Luria–Bertani medium containing kanamycin (50 mg L−1) and were grown with shaking (150 rpm) at 37 °C until the optical density (OD600) reached about 0.6. For induction, IPTG was added at a final concentration of 1 mM and the cells were further cultured at 30 °C for 2 h. The cells were lysed by sonication for 10 s at 4 °C, and centrifuged at 7,000g for 15 min. Supernatants and pellets were analyzed by SDS-PAGE and Coomassie Brilliant Blue R250 staining. The recombinant GbIRL1 protein from induced cells was purified using Nickel-CL agarose affinity chromatography (Bangalore Genei) and used for in vitro enzyme assays.
Western blotting was carried out to verify expression of a GbIRL1 protein having a His-tag in the N-terminus. After electrophoresis, the proteins were electro-transferred onto a PVDF membrane and detected with antimouse RGS-His antibody (Santa Cruz, American), and a secondary antibody (goat anti-mouse IgG), conjugated to alkaline phosphatase (AP). Western Blue Stabilized Color Substrate for AP (Promega, USA) was used for the color reaction.
Assay of GbIRL1 activity
GbIRL1 activity was analyzed according to Gang et al. (1999) with a slight modification. The reaction mixture contained 10 mM potassium phosphate, pH 6.5, 2 mM DDC or DDDC, 4 mM NADPH, 5 mM dithiothreitol and the protein (8 μg for bacterial supernatants containing GbIRL1) in a total volume of 15 μl. The reaction was performed at 37 ◦C for 3 h, and stopped by boiling for 3 min. After centrifugation at 17,000×g for 3 min, an aliquot (12.5 μl) was mixed with 1.66 μl of acetonitrile and analyzed by reversed-phase HPLC under the same conditions as described in Gang et al. (1999), except that a C8-AP column (YMC, Kyoto) was used.
Purified GbIRL1 protein (275 ng) was incubated with 0.2 mM of either NADPH or NADH in 240 μl of 20 mM Tris–HCl pH 7.5 for 1 h at room temperature. The mixtures were concentrated to less than 50 μl on a Centricon-10 (Amicon, Beverly, USA) and then diluted with 350 μl of 20 mM Tris–HCl pH 7.5. After three concentration/dilution cycles, the protein-bound coenzyme was detected by measuring the absorption spectra of the protein solution.
Lignin content analysis and histo-chemical lignin staining
Lignin content was quantitatively measured using the Klason method (Kirk and Obst 1988). Briefly, air-dried stem tissues were ground into powder and exhaustively extracted in a Soxhlet apparatus with toluene–ethanol (2:1, v/v), followed by 95 % ethanol and water. Samples were then vacuum-dried and 200 mg was hydrolysed in 3 ml of 72 % H2SO4 at 25 °C for 3 h with occasional stirring. The hydrolysate was diluted with the addition of 190 ml H2O and then autoclaved for 1 h. The sample was filtered through a fritted glass crucible, and then washed with hot water. The crucible was dried at 105 °C and weighed. The filtered solution was diluted to 500 ml and A 205 was determined spectrophotometrically using a 1 cm long cuvette. The Klason lignin was expressed as a percentage of the cell wall residue (CWR). The acid-soluble lignin was calculated by following formula: acid lignin (g l−1) = A 205/110.
For lignin staining observation, hand-cut sections of different sample were treated by 1 % phloroglucinol ethanol and 35 % HCl, and images were captured by Cannon camera (Lu et al. 2010).
Extraction and determination of flavonoid content
Flavonoids were extracted, identified and quantified as previously described by Xu et al. (2008b) with some modifications. Briefly, 100 mg of dried leaf powder was dissolved in 40 ml of acidified methanol. The samples solutions were then suitably diluted with methanol and filtered through 0.45 μM filter membranes (Millipore, Nylon) for high performance liquid chromatography analysis. Selected samples were analyzed by HPLC (model LC-10, Shimadzu, Kyoto, Japan) with a SPD-10AVP UV–Vis detector. Flavonoids were analyzed on a Shim-pack VP-ODS column (250 mm l × 4.6 mm), using a mobile phase of methanol: 0.4 % (v/v) phosphoric acid (60:40) for elution. The flow rate was 1.0 ml min−1 and injection volume was 20 μl. The UV detector was set at λ = 370 nm. Three extraction samples were prepared for HPLC analysis and each sample was injected three times. A standard mixture of quercetin, kaempferol and isorhamnetin was selected, because the many different flavonol glycosides that occur in ginkgo leaves are predominantly derivatives of these three flavonol aglycones. Based on the methods used to determine flavonoids concentrations described by van Beek (2002), flavonoid contents were calculated by multiplication of the total content of quercetin, kaempferol, and isorhamnetin by a factor 2.51 and were expressed as percentages.
Bioinformatics analysis and molecular evolution analyses
The obtained sequences were analyzed using bioinformatics tools at websites (http://www.ncbi.nlm.nih.gov and http://www.xpasy.org). Vector NTI Suite 10 was used for sequence alignment and analysis. The cis-acting element in the promoter region was predicted using PlantCARE (Lescot et al. 2002). Phylogenetic tree analysis of GbIRL1 and known IRLs from other plant species retrieved from GenBank were aligned with Mega 4.0 (Tamura et al. 2007). The phylogenetic tree was constructed by a neighbor-joining (NJ) method and measured by bootstrap analysis with 1,000 replicates. SPSS 17 was used for statistical analysis and graphing.
Results
Characterization of the GbIRL1 gene
A full-length cDNA of the IRL1 gene (Accession no. KC244282) was obtained from G. biloba leaf tissue by degenerate PCR and RACE methods. The cDNA sequence was 1,261 bp with a poly A tail, and suggested it contained a 921 bp ORF. A 115 bp 5′ untranslated region was upstream of the start codon, and the coding region was followed by a 225 bp 3′ untranslated region downstream of the stop codon. The 5′ untranslated region conformed to the predicted translation start site for eukaryotic genes (AXXATGG) (Kozak 1984; Lütcke et al. 1987), and two stop codons (TAA, positions 89–91 bp and TAG, 84–86 bp) found in the 3′ UTR proved that the cDNA contained the entire ORF. One potential polyadenylation signal AATAA was found at 43 bp position downstream from the stop codon. The G+C content of the coding region was 45.1 % similar to SDR family protein genes, such as VvIFL (46 %), PsIFR (40 %) and PoPCBER (46 %). The putative GbIRL1 was 306 aminoacid with a theoretical molecular weight of 33.28 kDa and a pI value of 6.18. Subcellular location prediction for GbIRL1 using SignalP 3.0 did not find any secretory pathway signal peptide (SP), and suggested that GbIRL1 was a cytoplasmic protein, as predicted for other members of the SDR family.
The 1,962 bp regulatory sequence upstream of the IRL gene was obtained by genome walking based on the obtained Ginkgo GbIRL1 sequences. The IRLP sequences were submitted to the PlantCARE server for analysis. The results indicated that the IRLP transcription site may be located at 249 bp upstream of the translation initiation site (CCGTCG, Fig. 2). Analysis of the upstream regulatory sequences showed that there was a TATA-box at 117 bp upstream of the transcription initiation site, and a CAAT-box cis element at 292 bp. It was speculated that this area may constitute the core sequences of the GbIRL1 promoter (Supplementary Figure 1S). The GbIRLP promoter also contained several environmental factors, the cell cycle inductive response elements and cis-acting elements associated with hormone induction.
Multiple sequence alignment with other plant PIPs and their homologs showed that GbIRL1 had higher similarities to PCBERs than to other PIPs (Supplementary Figure 2S). The amino acid sequence of the GbIRL1 shared 82.0 % identity with that of Tsuga heterophylla, 80.4 % with Pinus taeda, 80.1 % with Pinus pinaster, 74.2 % with Cryptomeria japonica, 70.9 % with Vitis vinifera, 69.5 % with Betula pendula, 68.6 % with Nicotiana tabacum, 68.3 % with Pyrus communis, 67.6 % with Glycine max, 66.7 % with Medicago truncatula, 65.1 % with Daucus carota, 65.0 % with Ricinus communis and 63.4 % with Lotus japonicus. In all of these protein sequences, the NADPH binding motif was conserved.
A phylogenetic tree generated by the neighbor-joining method based on the putative amino acid sequences of IRLs showed that all members of the IRL family could be sorted into five monophyletic groups. The results indicated that each kind of enzyme with distinct functions formed a separate clade, and the various IRLs fell into four different functional clades (PCBER, IFR, PLR and LAR) (Fig. 1). The PCBERs had a closer relationship with IFRs than with PLRs, and the LAR group was furthest from the PCBERs. The tree suggested that PCBER was among the earliest of these enzymes to arise, and that the IFR and PLR proteins evolved later, which was in agreement with the suggestions by Gang et al. (1999). GbIRL1 was grouped in the PCBER branch, indicating that GbIRL1 belonged to the class of PCBERs.
Aphylogenetic tree based on deduced amino acid sequences of various IRLs. Amino acid sequences were analyzed using the CLUSTALW program. The numbers at each node represented the bootstrap value, with 1,000 replicates. The bars represent evolutionary distance. The polymerization results are divided into five categories, IRL isoflavone reductase-like protein; IFR isoflavone reductase; LAR leucoanthocyanidin reductase; PCBER phenylcoumaran benzylic ether reductase; PLR pinoresinol lariciresinol reductase. The star indicates the gene studied in this paper. The sequence (accession numbers) are as followed: AAL85023IFR Arabidopsis thaliana, BAF34843PLR Lotus japonicus, AAC24001IRL Pyrus communis, AEY79728IRL Daucus carota, CAI56335IRL Vitis vinifera, BAG84267PCBER Nicotiana tabacum, AAC05116IFR Betula pendula, CAA06708PCBER Populus trichocarpa, ACA60729 PCBER Linum corymbulosum, XP 003537595IRL Glycine max, NP 001236037IFR Glycine max, BAF34847IFR Lotus japonicus, CAA41106IFR Medicago sativa, AAB31368IFR Pisum sativum, AAK27264IRL Cryptomeria japonica, AF242497PCBER Tsuga heterophylla, ABF39004PCBER Pinus strobus, CCC55425PCBER Pinus pinaster, AAC32591PCBER Pinus taeda, CCC55424PLR Pinus pinaster, AF242506PLR Thuja plicata, ABM68630PLR Linum perenne, BAI66418PLR Linum album, CAH60857PLR Linum album, CAI56321LAR Pinus taeda, AAU45392LAR Lotus uliginosus, EEF06163LAR Populus trichocarpa, ADD51358LAR Theobroma cacao
The activity of GbIRL1 in vitro
To express GbIRL1 in E. coli, we cloned the coding sequence of GbIRL1 into pET-28a, an expression vector with the T7 promoter and a His-tag, yielding pET28a-GbIRL1. The expression construct was checked for in-frame fusion by DNA sequencing (Sangon, Shanghai, China). Upon induction by IPTG, GbIRL1 was expressed as a major soluble protein product (Fig. 2). Figure 2, Lane 6 shows the insoluble proteins after IPTG induction for 2.0 h. The molecular weight of the expressed recombinant protein was estimated as a 34 kDa band with a His-tag: this size was in agreement with the value predicted by bioinformatics methods. The recombinant GbIRL1 protein was purified using Nickel-NTA agarose affinity chromatography and the purified protein showed the expected size (Fig. 2, lane 7). Western blotting of the purified recombinant GbIRL1 protein confirmed its specific immune reactivity to anti-His antibodies (Fig. 2, lane Western blot).
SDS-PAGE gel and Western blot analysis of GbIFR1 expressed in E. coli BL21 (DE3). After IPTG induction, E. coli BL21 cells containing pET28a-IFR were grown at 30 °C for 2 h. lane 1 protein of total cells without IPTG induction, lane 2 protein of total cells with IPTG induction for 30 min, 3 protein of total cells with IPTG induction for 60 min, 4 induction for 1.5 h, 5 soluble proteins with IPTG induction for 2 h, 6 purified recombinant GbIFR1 protein with Nickel-CL agarose affinity chromatography and used for enzyme activity assay; M molecular marker; Lane Western blot western blotting of the purified recombinant GbIFR1 protein with an anti-His-tag primary antibody probe
The GbIRL1 protein shared considerable amino acid sequence homology to PCBERs from Pinus taeda L. and poplar. To test whether GbIRL1 has PCBER activity, recombinant GbIRL1 was expressed in E. coli and assayed for PCBER activity. GbIRL1 was further purified to homogeneity through chromatographic steps (Fig. 3a). Two different reductase reactions in the presence of NADPH were measured by reversed-phase HPLC: the formation of tetrahydrodehydrodiconiferyl alcohol (TDDC) from dihydrodehydrodiconiferyl alcohol (DDDC), and isodihydrodehydrodiconiferyl alcohol (IDDDC) from dehydrodiconiferyl alcohol (DDC). The protein extract from E. coli expressing the fusion protein showed clear PCBER activity, with either DDC or DDDC as the substrate. GbIRL1 reduced DDDC to TDDC, and DDC to IDDDC, this agreed well with results reported previously (Gang et al. 1999). Negative controls in which empty vectors were expressed, or in which boiled enzyme preparations were used, produced no reaction products (Fig. 3b). These results indicated that GbIRL1 is a Ginkgo PCBER family protein involved in lignan biosynthesis.
a GbIRL activities of BL21 pET28a recombinant proteins. b HPLC profiles of the products of the control reaction. a(I) Reactions of IRL in lignan biosynthesis. DDC and DDDC were enzymatically reduced to IDDDC and TDDC by GbIRL, respectively. Both reactions require NADPH as the hydride donor. a(II) reaction products (4. IDDDC) were monitored by absorbance at 280 nm. a(III) another products (1. TDDC) were monitored by absorbance at 280 nm. Chromatographic peaks were identified by comparing their retention times with those of authentic standards. b(I) Standard chromatogram (280 nm). b(II) The extract of bacteria harboring the empty vector was use for the protein, the reaction with DDC as the substrate. b(III) The reaction with DDDC
The purified GbIRL1 protein was also incubated with NADPH or NADH and excess unbound cofactors were removed by filtration through a size-exclusion membrane. Absorption spectra of the protein fractions showed that GbIRL1 selectively bound NADPH but not NADH (Fig. 4). These results suggested that GbIRL1 has a typical PCBER activity, and it catalyzes a distinct NADPH-dependent oxidoreductase reaction.
Expression profile of the GbIRL1 gene in different tissues
To determine whether the GbIRL1 gene was related to lignin or flavonoid synthesis in Ginkgo, its expression patterns were analyzed by qRT-PCR. We found that the transcription level of GbIRL1 varied according to tissues, growth conditions and tissue developmental stage. The results of qRT-PCR analysis are shown in Fig. 5. GbIRL1 was expressed most abundantly in fungal-infected blades, followed by damaged or fungus-infected stem segments. The expression levels also varied significantly in different development stages in the stem. The highest level of transcription was found in the third-year stems. The next highest levels of GbIRL1 were observed in fourth-, and second-year stems, while the lowest level of expression was found in first-year stems.
The expression of GbIRL1 and lignin contents in different organs. In the top panel, hand-cut sections of different sample were stained with phloroglucinol-HCl method. The lignin content of the tissue was determined by modified Kalson method in the middle panel. Bottom panel, mRNA levels was measured by QRT-PCR, Expression pattern of GbIRL1 gene in different tissues with GAPDH gene as control, Data are mean values of triplicate tests ± SD a annual stem, b biennial stem; c triennial stem, d Four years stem, e petiole, f root, g leaf veins of the longitudinal cutting, h endosperm. i sclerotesta. j injured stems; k diseases stems, l fungal infections leaves. The arrow indicates histo-chemical lignin staining
The data obtained were analyzed by linear regression. The results showed that lignin content (Y) had a linear correlation with the transcription level of GbIRL1 (X). The correlation coefficient was as follows R 2 = 0.517 (Supplementary Table 1S). Thus, changes in IRL1 expression could affect the lignin content in different tissues of Ginkgo.
UV-B, wounding and hormones regulate the transcription of GbIRL1
UV-B and plant hormones can induce flavonoid content changes in Ginkgo leaves. To study the expression pattern of the GbIRL1 gene under UV-B, ultraviolet light of 260 nm, 1,500 Lx light intensity was used to process Ginkgo and sample it in different lighting time for RT-PCR analysis. In the low-dose UV-B treatment, the trend of the transcription level of GbIRL1 fell after rising and it reached its maximum expression quantity at about 48 h (Fig. 6a). Damage treatment significantly induced GbIRL1 expression. The transcription level rose slowly in the first 8 h after cut damage treatment, and then increased rapidly and reached its maximum level at 12 h (Fig. 6b).
Relative quantities of GbIRL1 mRNA at various time points post-treatment with UV-B (a), wounding (b), SA (c), ETH (d), ALA (e) and ABA (f). Each sample was individually assayed in triplicate. Values shown represent the mean reading from three treated plants and the error bars indicated the standard errors of the mean
In the anabolism of Ginkgo leaf flavonoids, endogenous hormones are correlated with flavonoid content changes. The hormones ABA, ALA, SA and ETH were selected for spraying treatments, after which RT-PCR was used to test the transcription of GbIRL1.
Salicylic acid is a phenolic hormone that can regulate plant growth and development. It is also involved in plant defense against pathogen as endogenous signal and belongs to the phenylalanine metabolic pathways like flavone and flavodoxin (Ferrer et al. 2008; Hayat et al. 2007). Previous researches had indicated that salicylic acid treatment can significantly increase the expression level of Ginkgo flavonoid-related genes (Xu et al. 2008a, b). Spraying Ginkgo leaf with low concentrations of SA induced the GbIRL1 gene rapidly, and the transcription level reached 3.8 times that of the control at 4 h after treatment. It reached its maximum level at the 8 h of sampling point and then declined slightly, but was still maintained at about five times the level of the control (Fig. 6c).
Changes in flavonoid content, GbIRL1 expression level during growth of ginkgo leaves. In the annual change of flavonoid in Ginkgo leaf, the flavonoid content and GbIRL1 expression level showed power-related. Data are mean values of triplicate tests and the error bars indicated the standard errors of the mean
Previous research has indicated that ethephon treatment can effectively increase the accumulation of Ginkgo leaf flavonoids, the basis of which is the increased expression of key flavonoid metabolic genes (Cheng et al. 2004, 2005, 2011; Wang and Cheng 2002; Xu et al. 2008a, b). We found in Ginkgo leaf, which was sprayed with the same concentration of ethephon, that the GbIRL1 transcription level also increased significantly, but it responded faster than to the other hormones and reached its maximum at 4 h, before declining rapidly to the control level at about 48 h (Fig. 6d).
ALA is not a plant hormone, but it has been reported that spraying ALA appropriately can deepen the color of apples (Wang et al. 2004b). Previous research has indicated that 100 mg/L ALA can significantly improve the enzymatic activity of Ginkgo PAL, CHS and CHI. Ginkgo leaf was treated with the same concentration of ALA (100 mg/L) in this study. The GbIRL1 transcription level increased slowly up to 24 h and reached a maximum at 48 h of 2.3 times the control level, then started to decline slowly (Fig. 6e).
ABA is formed by carotenoid degradation and can prevent the promotive growth effects of gibberellic acid and cytokinin. It is also correlated with leaf aging and fruit abscission. Generally, stresses such as drought, cold, hyperthermia, salt and flooding can all make the ABA levels in plants increase rapidly, which enhances plant resistance (Shen et al. 2006). In the GbIRL1 metabolic pathway, ABA could induce GbIRL1 transcription, but it responded slower than the other ABA-responsive processes. After 4 h of treatment, the GbIRL1 transcription level had increased significantly compared with the control. It continued to rise and reached a maximum at 48 h of about 12 times the level of the control. It then started to decrease, but was still maintained at eight times the control level at 96 h (Fig. 6f).
The relationship between GbIRL1 expression and flavonoid accumulation during Ginkgo leaf growth
PCBERs belong to the phenylalanine metabolic pathway branch and have a parallel position to flavonoid metabolism. Thus, changes in the IRL expression level may cause changes of flavonoid content in Ginkgo leaves. To study the relationship between flavonoid content in Ginkgo leaf and the GbIRL1 gene expression level, the annual changes in GbIRL1 gene expression level and flavonoid content were measured during the growth of Ginkgo leaves (Fig. 7). Grafted Ginkgo leaves were collected at 15 different sampling points (May 3, 18; June 13; July 4, 18, 27; August 1, 13, 30; September 11, 29; October 17; November 1, 14 and 29) for RT-PCR and the flavonoid content measurement.
It can be seen in (Fig. 7) that the annual changes in flavonoid content showed several peak values. Some obvious peak values were found on July 4, September 29, and November 1, and the corresponding contents were 0.8039, 0.9383 and 1.0994, respectively. The highest level was at the sampling point of November 1. The main trend was an increase in flavonoid content. The GbIRL1 gene expression levels showed two main peak values during the growth of the Ginkgo leaves. The first appeared on July 18, which was close to the time of the first peak in flavonoid content. It descended to a nadir on August 13, and then increased continuously to the highest peak of the year on October 17, when the relative transcription level was 37.77. Thereafter, the expression level decreased to about 16.33.
SPSS17 was used to analyze the relationship between the GbIRL1 transcription level and seasonal variations in flavonoid content (Supplementary Table 2S). Curve fit was conducted according to the test results, among which the goodness of fit of the S-shaped curve, power curve and square curve was high. All three regression models were statistically significant. Taking the goodness of fit and the model’s degree of conciseness together, the power curve was most appropriate. According to the results, in the annual changes of flavonoids in Ginkgo leaf, the flavonoid content (Y) and the GbIRL1 expression level (X) were power-related, the F statistic = 208.188, concomitant probability value p = 0.000 < 0.001, correlation coefficient R 2 = 0.610. Although the GbIRL1 expression level and the accumulation of flavonoids were not completely synchronous, the results of curve fitting showed that they were closely related to each other.
Discussion
The GbIRL1 (isoflavone reductase-like protein, IRL) gene from Ginkgo was cloned through degenerate PCR and RACE techniques. IRL genes have been identified in several flowering plants and their functions have been analyzed. However, to date, especially for gymnosperms, there are few reports on IRL genes in medicinal plants that have high flavonoid contents.
Enzyme and recombinant protein analysis
PCBER catalyzes the corresponding reduction reaction in lignans with an 8-5′ connection (Min et al. 2003). The analysis of enzyme activity in vitro shows that PCBER depends on NADPH and can catalyze several 7-O-4′ reduction reactions of benzyl coumarin lignan related to defense and produce relevant dihydric phenols, such as catalyzing DDC and DDDC to produce corresponding IDDDC and TDDC (Ferrer et al. 2008; Gang et al. 1999). Although the catalysis of the reaction is less efficient, the two isomer forms of the substrate can be recognized by the enzyme and involved in the reaction. PCBER does not catalyze spatial specific reaction of the substrate, but the catalytic substrates of PLRs and IFRs do have spatial specificity. From this it can be predicted that the PCBER protein is evolutionarily older than PLRs and IFRs (Gang et al. 1999). Because lignin-like material is mostly found in ancient plants such as the fern, Ceratophyllum demersum L, gymnosperms and early angiosperms, isoflavones exist only in leguminous plants and few are seen in non-leguminous plants. Therefore, it appears that the metabolic pathway of lignans is older than that of isoflavones (Dinkova-Kostova et al. 1996). Although PLR and IFR catalyze similar reactions, IFR is an evolution of PLR according to evolutionary analysis and substrate specificity inference (Ferrer et al. 2008). The Ginkgo IRLs evolutionary tree shows that GbIRL1 and ancient gymnosperm enzymes are of the same species, and the IRL1 sequence alignment shows that GbIRL1 closely matches the conserved sites of PCBER and lacks the insertion sequences. It was confirmed by analyzing the enzyme activity that its recombinant protein possesses the characteristics of PCBER enzymes, which depend on NADPH, and its catalytic function is close to PCBER, Thus, it can be deduced that GbIRL1 belongs to the PCBER protein family.
The expression patterns of the GbIRL1 gene and accumulation of flavonoids in different tissues and leaf developmental stages
Vander Mijnsbrugge et al. (2000) found that the poplar shoot and root secondary xylem synthesized large amounts of PCBER, suggesting that it was related to protective lignan formation in the xylem, and lignans may be involved in the relevant timber growth metabolism during plant development. The PCBER transcription level in cotton testa is high, and its expression pattern is very important in cotton seed development as it can not only protect the seed structure, but also functions in defending against external infection. Deletion mutations can easily affects the synthesis of xylary fiber lines (Turley 2008). We found from expression analysis of different tissues in Ginkgo that GbIRL1 is highly transcribed in leaves infected by fungi, as well as in injured and insect-damaged shoot segments, suggesting that GbIRL1 may be involved in lignification in Ginkgo during its defense against pest and disease damage and to protect non-infected tissues. However, whether it forms a lignin protective barrier or pest-defensive lignans requires further investigation. Its expression in the stem showed that besides involvement in adapting to biotic stress, GbIRL1 is also related to wooden formation in different tissues as part of its regular expression. Thus, GbIRL1 is a multi-function gene during Ginkgo growth and development.
Studies have indicated that the reason ultraviolet light can induce flavonoid synthetic gene expression is that flavonoids can reduce UV damage to plants; this response is mediated by ultraviolet responsive elements in the upstream promoters of genes related to flavone metabolism (Bashandy et al. 2009; Cheng et al. 2011). UV-B was shown to induce high level expression of an IRL gene in grape. This grape IRL was simultaneously induced by injury and pest and disease damage (Lers et al. 1998). A Pomelo IRL gene has also been found to be involved in the response to ultraviolet radiation (Min et al. 2003). So, these results suggested that the high level of GbIRL1 expression after UV-B induction may be related to the accumulation of flavonoids (Xu et al. 2008a).
After Pyricularia grisea inoculation of rice, the OsIRL gene was induced with an expression pattern similar to MeJA induction (Kim et al. 2003). Studies of the Coffea arabica CaIRL gene promoter found a damage response element in 0.8 kb upstream of the coding region. However, the damage induction regulation function ceased after deleting the sequence about 0.4 kb upstream of the promoter (in which there was a W-box and an ACCTACC-motif element) (Brandalise et al. 2009). There are several cis-acting elements, including a W-box and a WRKY element, in the upstream regulatory sequence of GbIRL1 (Supplementary 1S). The W-box is associated with damage induction and ERF responses caused by damage (Nishiuchi et al. 2004). WRKY elements are correlated with pest defense, disease damage induction and abscisic acid signal regulation (Eulgem et al. 1999; Xie et al. 2005). These results offer possible evidence for injury-induced expression of the GbIRL1 gene in G. biloba.
When plants encounter osmotic stress or any other adversity, ABA can regulate relevant genes to adapt to the environmental change (Skriver and Mundy 1990). ABA treatment will improve the transcription level of the Ginkgo leaf flavonoid synthesis genes FLS (Xu et al. 2012), ANS (Xu et al. 2008b), CHI (Cheng et al. 2011) in the flavonoid metabolic pathway. GbIRL1 is involved in lignan metabolism, which is a branch of phenylalanine metabolism, and has a parallel position to flavonoid metabolism. Thus, the significantly increased transcriptional level induced by ABA could be seen as raising the downstream metabolic level by promoting the phenylalanine metabolic pathway.
ETH can promote fruit maturation and coloring, regulate the relevant gene expression of flavonoids metabolism, and is associated with the accumulation of flavone in Ginkgo (Cheng et al. 2011; Xu et al. 2008a, b, 2012). Ethephon can induce CHS gene expression in Brazilian rubber (Kush et al. 1990) and exogenous ETH can induce both CHS and F3H gene expression in avocado (Ardi et al. 1998). The GbIRL1 expression level was not obviously increased in Ginkgo sprouts treated with ETH; expression increased slightly in the beginning but then declined rapidly to the control level. To date, there have been few reports about IRL genes that are induced by ETH, but we found ETH induction is not obvious compared with other flavonoids metabolic genes.
5-aminolevulinic acid (ALA) is a key precursor of all porphyrin compound biosynthesis in vivo. The production increase can reach 11–15 % if ALA is used in the wheat growth period or rice panicle and flowering periods (Hotta et al. 1997). A lot of research has proven that a low concentration of exogenous ALA can promote photosynthesis in plants, and photosynthesis can in turn promote lignin synthesis (Nishihara et al. 2001, 2003; Wang et al. 2004a). Moreover, treatment of 100 mg/L ALA can increase the activity of Ginkgo PAL, CHS and CHI (Xu 2008). Therefore, the increased transcription level of Ginkgo IRL in response to ALA we observed may be related to the increased level of substrate in this pathway.
SA is a signaling molecule for plants to react to external environmental stresses and damage from infection by pests and disease (Gaffney et al. 1993). When a plant is damaged by external pests or disease infection, the expression level of flavonoid metabolism genes is improved significantly (Fofana et al. 2002; Meer et al. 1993). Previous studies have indicated that SA treatment can significantly increase the expression level of the Ginkgo PAL gene (Xu et al. 2008a). In this study, SA could significantly induce the transcription of the GbIRL1 gene. Moreover, the increased expression level of GbIRL1 when leaves were damaged by the outside wounding showed that GbIRL1 is involved in the SA signaling response pathway induced by pest and disease damage.
Flavonoids such as flavone can protect plants from ultraviolet light and various flavonoids and isoflavone can induce the expression of genes related to root nodules and offer protection for nitrogen-fixing of the plant. The formation of lignin provides structural support for higher plant evolution, and lignans provide protection for the normal growth of plants as antibacterial agents and antitoxins (Ferrer et al. 2008). Ginkgo has strong resistance to adverse environments and very high esthetic value, and the flavones in Ginkgo leaf have high medicinal value. All of these are closely related to the vigorous phenylalanine metabolic pathway. After the production of coumaroyl-CoA by 4CL catalysis, the flavone synthetic pathway and the lignin and lignan synthetic pathway start to differentiate. In the flavone anabolic pathway, the gene expression levels directly related to Ginkgo flavone metabolism have a significant positive correlation with the correlating enzyme activity and flavone accumulation (Cheng et al. 2011). We found that changes in Ginkgo GbIRL1 expression have some power correlation with the flavone content in the leaf. This result suggests that the carbon flow regulation by the lignan metabolic pathway with GbIRL1′s involvement influences the flavone metabolic pathway. Thus, flavone accumulation is not only regulated by key genes in the flavone metabolic pathway itself, but is also related to parallel pathway metabolisms such as the anabolism of lignins and lignans.
Abbreviations
- bp:
-
Base pair
- HPLC:
-
High performance liquid chromatography
- IPTG:
-
Isopropyl b-d-thiogalactoside, C9H18O5S
- IFR:
-
Isoflavone reductase
- IRL:
-
Isoflavone reductase-like protein
- IRLP:
-
IRLgene promoter
- ORF:
-
Open reading frame
- PCBER:
-
Phenylcoumaran benzylic ether reductases
- PCR:
-
Polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- QRT-PCR:
-
Real-time quantitative PCR
- SD:
-
Standard deviation
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- UV:
-
Ultraviolet
References
Ardi R, Kobiler I, Jacoby B, Keen N, Prusky D (1998) Involvement of epicatechin biosynthesis in the activation of the mechanism of resistance of avocado fruits to Colletotrichum gloeosporioides. Physiol Mol Plant P 53(5–6):269–285
Bashandy T, Taconnat L, Renou JP, Meyer Y, Reichheld JP (2009) Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol Plant 2(2):249–258
Brandalise M, Severino FE, Maluf MP, Maia IG (2009) The promoter of a gene encoding an isoflavone reductase-like protein in coffee (Coffea arabica) drives a stress-responsive expression in leaves. Plant Cell Rep 28(11):1699–1708
Cheng S, Wang Y, Fei Y, Zhu G (2004) Studies on the effects of different treatments on flavonoids contents in Ginkgo biloba leaves and their regulating mechanism. J Fruit Sci 21(2):116–119 (in Chinese)
Cheng S, Wang Y, Liu W, Chen K (2005) Effects of plant growth regulators on phenylalanine ammonia-lyase activities in leaves of Ginkgo biloba in vitro. J Plant Resour Environ 14(1):20–22 (in Chinese)
Cheng H, Li L, Cheng S, Cao F, Wang Y, Yuan H (2011) Molecular cloning and function assay of a chalcone isomerase gene (GbCHI) from Ginkgo biloba. Plant Cell Rep 30(1):49–62
Dinkova-Kostova AT, Gang DR, Davin LB, Bedgar DL, Chu A, Lewis NG (1996) (+)-pinoresinol/(+)-lariciresinol reductase from Forsythia intermedia. J Biol Chem 271(46):29473–29482
Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends Plant Sci 4(10):394–400
Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J 18(17):4689–4699
Ferrer JL, Austin M, Stewart C Jr, Noel J (2008) Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Bioch 46(3):356–370
Fofana B, McNally DJ, Labbé C, Boulanger R, Benhamou N, Séguin A, Bélanger RR (2002) Milsana-induced resistance in powdery mildew-infected cucumber plants correlates with the induction of chalcone synthase and chalcone isomerase. Physiol Mol Plant P 61(2):121–132
Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261(5122):754–756
Gang DR, Kasahara H, Xia ZQ, Vander Mijnsbrugge K, Bauw G, Boerjan W, Van Montagu M, Davin LB, Lewis NG (1999) Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinol lariciresinol and isoflavone reductases. J Biol Chem 274(11):7516–7527
Gapper C, Dolan L (2006) Control of plant development by reactive oxygen species. Plant Physiol 141(2):341–345
Hano C, Martin I, Fliniaux O, Legrand B, Gutierrez L, Arroo RRJ, Mesnard F, Lamblin F, Lainé E (2006) Pinoresinol–lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 224(6):1291–1301
Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290(5499):2110–2113
Hayat S, Ali B, Ahmad A (2007) Salicylic acid: biosynthesis, metabolism and physiological role in plants, Springer publication, Netherlands, pp 1–14
Hotta Y, Tanaka T, Takaoka H, Takeuchi Y, Konnai M (1997) Promotive effects of 5-aminolevulinic acid on the yield of several crops. Plant Growth Regul 22(2):109–114
Jansson S, Meyer-Gauen G, Cerff R, Martin W (1994) Nucleotide distribution in gymnosperm nuclear sequences suggests a model for GC-content change in land-plant nuclear genomes. J Mol Evol 39(1):34–46
Karamloo F, Wangorsch A, Kasahara H, Davin LB, Haustein D, Lewis NG, Vieths S (2001) Phenylcoumaran benzylic ether and isoflavonoid reductases are a new class of cross-reactive allergens in birch pollen, fruits and vegetables. Eur J Biochem 268(20):5310–5320
Kim ST, Cho KS, Kim SG, Kang SY, Kang KY (2003) A rice isoflavone reductase-like gene, OsIRL, is induced by rice blast fungal elicitor. Mol Cells 16(2):224–231
Kim SG, Kim ST, Wang Y, Kim SK, Lee CH, Kim KK, Kim JK, Lee SY, Kang KY (2010) Overexpression of rice isoflavone reductase-like gene (OsIRL) confers tolerance to reactive oxygen species. Physiol Plantarum 138(1):1–9
Kirk TK, Obst JR (1988) Lignin determination. Method Enzymol 161(5):87–101
Kozak M (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 12(2):857–872
Kush A, Goyvaerts E, Chye ML, Chua NH (1990) Laticifer-specific gene expression in Hevea brasiliensis (rubber tree). Proc Natl Acad Sci USA 87(5):1787
Lers A, Burd S, Lomaniec E, Droby S, Chalutz E (1998) The expression of a grapefruit gene encoding an isoflavone reductase-like protein is induced in response to UV irradiation. Plant Mol Biol 36(6):847–856
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Lu X, Liu Y, An J, Hu H, Peng S (2010) Isolation of a cinnamoyl CoA reductase gene involved in formation of stone cells in pear (Pyrus pyrifolia). Acta Physiol Plant 10(7):1–7
Lütcke H, Chow K, Mickel F, Moss K, Kern H, Scheele G (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6(1):43–49
Meer I, Stuitje A, Mol J, Verma D (1993) Control of plant gene expression: regulation of general phenylpropanoid and flavonoid gene expression, CAB Direct, United Kingdom, pp 125–155
Min T, Kasahara H, Bedgar DL, Youn B, Lawrence PK, Gang DR, Halls SC, Park HJ, Hilsenbeck JL, Davin LB (2003) Crystal structures of pinoresinol–lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases. J Biol Chem 278(50):50714–50723
Nam MH, Heo EJ, Kim JY, Kim SI, Kwon KH, Seo JB, Kwon O, Yoo JS, Park YM (2003) Proteome analysis of the responses of Panax ginseng CA Meyer leaves to high light: use of electrospray ionization quadrupole-time of flight mass spectrometry and expressed sequence tag data. Proteomics 3(12):2351–2367
Nishihara E, Takahashi K, Nakata N, Tanaka K, Watanabe K (2001) Effect of 5-aminolevulinic acid (ALA) on photosynthetic rate, hydrogen peroxide content, antioxidant level and active oxygen-scavenging enzymes in spinach (Spinacia oleracea L.). J Jpn Soc Hortic Sci 70(3):346–352
Nishihara E, Kondo K, Parvez MM, Takahashi K, Watanabe K, Tanaka K (2003) Role of 5-aminolevulinic acid (ALA) on active oxygen-scavenging system in NaCl-treated spinach (Spinacia oleracea). J Plant Physiol 160(9):1085–1091
Nishiuchi T, Shinshi H, Suzuki K (2004) Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves. J Biol Chem 279(53):55355–55361
Paiva NL, Edwards R, Sun Y, Hrazdina G, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L.). Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol Biol 17(4):653–667
Petrucco S, Bolchi A, Foroni C, Percudani R, Rossi GL, Ottonello S (1996) A maize gene encoding an NADPH binding enzyme highly homologous to isoflavone reductases is activated in response to sulfur starvation. Plant Cell 8(1):69–80
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3(6):1101–1108
Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443(7113):823–826
Shoji T, Winz R, Iwase T, Nakajima K, Yamada Y, Hashimoto T (2002) Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol Biol 50(3):427–440
Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2(6):503
Smith J, Luo Y (2004) Studies on molecular mechanisms of Ginkgo biloba extract. Appl Microbiol Biot 64(4):465–472
Suzuki S, Umezawa T (2007) Biosynthesis of lignans and norlignans. J Wood Sci 53(4):273–284
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Tiemann K, Inzé D, Van Montagu M, Barz W (1991) Pterocarpan phytoalexin biosynthesis in elicitor-challenged chickpea (Cicer arietinum L.) cell cultures. Purification, characterization and cDNA cloning of NADPH: isoflavone oxidoreductase. Eur J Biochem 200(3):751–758
Turley R (2008) Expression of a phenylcoumaran benzylic ether reductase-like protein in the ovules of Gossypium hirsutum. Biol Plantarum 52(4):759–762
van Beek TA (2002) Chemical analysis of Ginkgo biloba leaves and extracts. J Chromatogr A 967(1):21–55
Vander Mijnsbrugge K, Beeckman H, De Rycke R, Van Montagu M, Engler G, Boerjan W (2000) Phenylcoumaran benzylic ether reductase, a prominent poplar xylem protein, is strongly associated with phenylpropanoid biosynthesis in lignifying cells. Planta 211(4):502–509
Wang Y, Cheng S (2002) Studies on the effects of regulating measures on the flavonoids contents in Ginkgo biloba leaves. Hubei Agric Sci 25(5):103–105 (in Chinese)
Wang LJ, Jiang WB, Huang BJ (2004a) Promotion of 5-aminolevulinic acid on photosynthesis of melon (Cucumis melo) seedlings under low light and chilling stress conditions. Physiol Plantarum 121(2):258–264
Wang LJ, Wang ZH, Li ZQ, Liu H, Liu WQ, Chen ZY, Yan P, Sun DQ (2004b) Effect of 5-aminolevulinic acid on enhancing apple fruit coloration. J Fruit Sci 21(6):512–515 (in Chinese)
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137(1):176–189
Xu F (2008) Cloning and expression of GbPAL and GbANS genes and effect of ALA on the content of flavonoids in Ginkgo biloba. Doctor. Shandong Agricultural University, Shandong
Xu F, Cai R, Cheng S, Du H, Wang Y (2008a) Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr J Biotechnol 7(6):721–729
Xu F, Cheng H, Cai R, Li LL, Chang J, Zhu J, Zhang FX, Chen LJ, Wang Y, Cheng SH (2008b) Molecular cloning and function analysis of an anthocyanidin synthase gene from Ginkgo biloba, and its expression in abiotic stress responses. Mol Cells 26(6):536–547
Xu F, Li L, Zhang W, Cheng H, Sun N, Cheng S, Wang Y (2012) Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol Biol Rep: 1–12
Zhu Q, Guo T, Sui S, Liu G, Lei X, Luo L, Li M (2009) Molecular cloning and characterization of a novel isoflavone reductase-like gene (FcIRL) from high flavonoids-producing callus of Fagopyrum cymosum. Acta Pharmacol Sin 44(7):809–819
Acknowledgments
This work was supported by the Natural Science Foundation of China (30971974 and 31270717), Economic Forest Germplasm Improvement and Comprehensive Utilization of Resources of Hubei Key Laboratories (20011BLKF238 and 2011BH0030), and University-industry Cooperation Fund of Hubei Educational Office (CXY2009B009).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by H. Judelson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hua, C., Linling, L., Feng, X. et al. Expression patterns of an isoflavone reductase-like gene and its possible roles in secondary metabolism in Ginkgo biloba . Plant Cell Rep 32, 637–650 (2013). https://doi.org/10.1007/s00299-013-1397-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1397-2