Abstract
Key message
MdCRY2 was isolated from apple fruit skin, and its function was analyzed in MdCRY2 transgenic Arabidopsis. The interaction between MdCRY2 and AtCOP1 was found by yeast two-hybrid and BiFC assays.
Abstract
Cryptochromes are blue/ultraviolet-A (UV-A) light receptors involved in regulating various aspects of plant growth and development. Investigations of the structure and functions of cryptochromes in plants have largely focused on Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), pea (Pisum sativum), and rice (Oryza sativa). However, no data on the function of CRY2 are available in woody plants. In this study, we isolated a cryptochrome gene, MdCRY2, from apple (Malus domestica). The deduced amino acid sequences of MdCRY2 contain the conserved N-terminal photolyase-related domain and the flavin adenine dinucleotide (FAD) binding domain, as well as the C-terminal DQXVP-acidic-STAES (DAS) domain. Relationship analysis indicates that MdCRY2 shows the highest similarity to the strawberry FvCRY protein. The expression of MdCRY2 is induced by blue/UV-A light, which represents a 48-h circadian rhythm. To investigate the function of MdCRY2, we overexpressed the MdCRY2 gene in a cry2 mutant and wild type (WT) Arabidopsis, assessed the phenotypes of the resulting transgenic plants, and found that MdCRY2 functions to regulate hypocotyl elongation, root growth, flower initiation, and anthocyanin accumulation. Furthermore, we examined the interaction between MdCRY2 and AtCOP1 using a yeast two-hybrid assay and a bimolecular fluorescence complementation assay. These data provide functional evidence for a role of blue/UV-A light-induced MdCRY2 in controlling photomorphogenesis in apple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Light is one of the most important factors for plant growth and development. Light sensing occurs through multiple photoreceptors controlling complex cellular signaling pathways (Gyula et al. 2003). In general, there are three types of photoreceptors, including phytochromes, which are involved in red/far-red light signaling (Franklin et al. 2005); two classes of blue/UV-A light photoreceptors, phototropins and cryptochromes, which mediate various blue/UV-A light responses (Briggs and Christie 2002; Lin 2000); and the UV-B light receptor UVR8 (Rizzini et al. 2011; Christie et al. 2012; Wu et al. 2012). Cryptochromes are flavoprotein photoreceptors that evolved from photolyases, a class of blue light-activated microbial DNA-repair enzymes (Partch and Sancar 2005), which themselves do not possess photolyase activity (Lin and Shalitin 2003).
Plant cryptochromes have been characterized most thoroughly in the model species Arabidopsis, which possesses three cryptochromes (CRY1, CRY2, and CRY3) that function in blue/UV-A light perception (Lin and Shalitin 2003). The first cryptochrome gene was cloned through the molecular analysis of the T-DNA insertion mutant allele hy4 (Ahmad and Cashmore 1993), which encodes a protein of 681 amino acid residues, with a high degree of sequence homology to photolyase. Later, HY4 was designated as cryptochrome 1, CRY1 (Lin et al. 1995). The second member of the cryptochrome gene family, CRY2, was subsequently isolated by screening an Arabidopsis cDNA library using CRY1 as a probe (Lin et al. 1996). There is evidence for a third CRY (CRY3) in Arabidopsis, which belongs to the CRY-DASH clade of the photolyase/CRY superfamily and is known to act as a single-stranded DNA-repair enzyme (Kleine et al. 2003). It can also act as a dual-function photoreceptor in mitochondria and chloroplasts (Brudler et al. 2003). Since the discovery of the first cryptochrome in Arabidopsis, this type of photoreceptor has been widely found in organisms ranging from bacteria to human (Cashmore 2003; Partch and Sancar 2005).
Plant CRYs have conserved N-terminal photolyase-related domains as well as C-terminal DQXVP-acidic-STAES (DAS) domains, and they are distinguished mainly by their C-terminal extensions (Ahmad and Cashmore 1993; Lin et al. 1996). The N-terminal domain shares many features with microbial photolyases and includes regions that are proposed to bind the chromophores flavin adenine dinucleotide (FAD) and methenyltetrahydrofolate (MTHF). This region has been designated the photolyase-related or photolyase homologous region (PHR) domain (Lin and Shalitin 2003).
Cryptochromes regulate numerous aspects of growth and development in plants, animals, and prokaryotes. In plants, cryptochromes mediate the blue/UV-A light-dependent inhibition of hypocotyl elongation, de-etiolation responses, vegetative growth, the initiation of flowering, anthocyanin accumulation, the regulation of gene expression, and the maintenance of plant endogenous rhythms (Lin 2002). Studies of cry1 mutants have shown that CRY1 mediates de-etiolation responses under high-irradiance blue light (Ahmad and Cashmore 1993). CRY1 also appears to mediate hypocotyl inhibition in other plants. For example, overexpressing Arabidopsis CRY1 in tobacco resulted in exaggerated hypocotyl inhibition under blue light conditions (Lin et al. 1995), suggesting that the signal-transduction mechanism for CRY1 is conserved in different plants. CRY1 is also involved in regulating the circadian clock in Arabidopsis and anthocyanin accumulation in Brassica napus (Lin 2000; Chatterjee et al. 2006).
Arabidopsis CRY2 also contributes to blue/UV-A light-mediated de-etiolation; however, this contribution is relatively subtle compared with CRY1, as the CRY2 protein is subject to degradation under higher irradiances of blue light (Lin et al. 1998). CRY2 has a more prominent role in the photoperiodic control of flowering (Guo et al. 1998). In Arabidopsis, both CRY1 and CRY2 regulate primary root elongation in blue light; however, they act antagonistically in this response pathway (Canamero et al. 2006).
The CRY1 protein is localized in both the nucleus and cytosol. It has been shown that the nuclear CRY1 protein is responsible for the blue light inhibition of hypocotyl elongation, whereas the cytosolically localized CRY1 mediates the blue light stimulation of cotyledon expansion and root elongation (Wu and Spalding 2007). In contrast, CRY2 appears to complete its post-translational life cycle in the nucleus (Yu et al. 2007). The nuclear-localized CRY2 mediates floral initiation and hypocotyl inhibition. The blue light-dependent CRY2 phosphorylation and degradation also appear to occur only in the nucleus (Yu et al. 2007).
In Arabidopsis, both CRY1 and CRY2 interact with COP1 directly and inhibit its E3 ubiquitin ligase activity to prevent the degradation of downstream transcription factors (TFs), such as HFR1 and CO. Therefore, the CRYs stabilize the TFs to promote photomorphogenesis in Arabidopsis (Yang et al. 2005; Jang et al. 2008), symbolizing an important signal-transduction pathway in blue/UV-A light-mediated plant growth and development.
In plants, cryptochromes are mostly studied in lower and herbaceous crops. In this study, we isolated a blue light receptor gene, MdCRY2, from a woody plant, apple, analyzed the structure and relationship with CRYs from other species, and found it shows the highest similarity to the strawberry CRY protein. Functional complementation assays using an Arabidopsis cry2 mutant were performed to verify that MdCRY2 is a counterpart of AtCRY2. Furthermore, the analysis of MdCRY2 transgenic Arabidopsis has substantiated the role of MdCRY2 in regulating flowering time, plant height, root length, and anthocyanin accumulation. To deeply investigate the mechanism of how MdCRY2 possesses these functions, the interaction between MdCRY2 and AtCOP1 was tested by yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays, which may provide a new sight of MdCRY2-mediated plant growth and development in the blue/UV-A light signaling pathway in apple.
Materials and methods
Plant material and growth conditions
Columbia lines of Arabidopsis thaliana were used as the wild type (WT). The seeds were sown on MS medium, cold-treated for 3 days at 4 °C, and then transferred to controlled environment cabinets under either long days (LDs, 16 h light/8 h dark) or short days (SDs, 8 h light/16 h dark) at 22 °C. Plants were all grown in LDs or SDs of white light unless stated otherwise. Experiments involving dark or different light treatments were performed in a controlled environment chamber using the blue, red or white tubes at 22 °C.
Apple callus ‘Orin’ were grown on MS medium with 0.5 mM 2,4-D and 1.5 mM 6-BA, and grown in controlled environment cabinets in the dark at 22 °C unless stated otherwise.
Bagged ‘Red Delicious’ (Malus domestica Borkh.) apples for experiments were harvested from an orchard in late September. The bags were then removed and the apples treated with appointed light for the expression analysis.
Isolation of full-length cDNA of MdCRY2 by rapid amplification of cDNA ends (RACE)
According to an MdCRY2 EST sequence approximately 1,000 bp in length obtained from (National Center for Biotechnology Information, NCBI), the gene-specific primers CRY1F (5′-GACCGCATCAATAATCCAC-3′) and CRY1R (5′-GGGTTGTTCTTGGCAGGT-3′) were designed. Then, the EST cDNA of MdCRY2 was isolated though reverse transcription-PCR (RT-PCR) with the RNA extracted from apple skin. To obtain a full-length gene, 5′- and 3′-RACE methods were used. The 5′-RACE primers 5Race-CRY2R1 (5′-CGGGAAGTTAAAACATAGATACCG-3′) and 5Race-CRY2R2 (5′-GCCCACAGTATCTGCTTCATTCG-3′) and the 3′-RACE primers 3Race-CRY2F1 (5′-CGGCACAGATGAAGTTGTTGTTG-3′), and 3Race-CRY2F2 (5′-CCTCTCCTTGCCCTACATACTCC-3′) were designed on the basis of the MdCRY2 EST sequence. The PCR products of expected sizes were purified, cloned into the pMD18-T vector (Invitrogen, San Diego, CA, USA), and sequenced. Then, the putative 3′- and 5′-RACE cDNAs and the EST sequence were over-lapped with DNAMAN to form a cDNA contig, which was used to determine the putative initiating translation codon (ATG) and open reading frame (ORF). To obtain a full-length cDNA of MdCRY2, a pair of full-length primers MdCRY2F (5′-GTGAAGTTTGATACTGAATTTTG-3′) and MdCRY2R (5′-GGTGTTCATCTTCCACGGCTCC-3′) were designed according to the contig. The full length of MdCRY2 was then obtained by RT-PCR using the full-length primers.
Analysis of MdCRY2 sequence
The full-length amino acid sequence of MdCRY2 was used to search homologous sequences via the BLASTP method in NCBI. Multiple alignments of amino acid sequences were performed between apple and other plants using MEGA4.1 and ClustalW. A phylogenetic relationship tree was then constructed using the neighbor-joining (NJ) method.
RNA extraction, semiquantitative, and real-time quantitative RT-PCR analysis
Apple fruit skin or callus were harvested, frozen in liquid nitrogen, and then ground under RNase-free conditions. The RNA was extracted using the TRizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions and then treated with DNase I at 37 °C for 30 min.
The RNA was then reverse transcribed using the PrimeScript™ First Strand cDNA Synthesis Kit (Takara, Otsu, Japan), following the manufacturer’s instructions. Ten microliters of cDNA was diluted to a final volume of 100 μl with water.
Semiquantitative RT-PCR was carried out in 25-μl reactions with 5 ng diluted cDNA template. The PCR profile was 94 °C for 3 min, 32 cycles of 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 30 s, with a 5 min extension at 72 °C. The primer sequences used were 5′-GAAGACACGAGCTCTACTGC-3′ and 5′-GAGCTAGTCATACCAAAAGGAC-3′. Md18S cDNA amplification was used as the external control. PCR products were electrophoresed on a 1.5 % agarose gel and viewed under UV light after standard staining with ethidium bromide.
For real-time quantitative RT-PCR analysis, the specific primer sequences used were 5′-AAGATGTGGGAAATGGAAGCA-3′ and 5′-TAGAGGAGTATGTAGGGCAAGGAGA-3′ used for PCR analysis. Md18S gene was used as loading controls. Fluorescence-quantitative PCR reactions were repeated three times.
Construction of transformation vectors and production of transgenic plants
To generate Arabidopsis MdCRY2 overexpressing lines, the full length of MdCRY2 cDNA was amplified by PCR using the primers MdCRY2F and MdCRY2R. The amplified cDNA was cloned into the expression vector pBI121 under the control of the cauliflower mosaic virus (CaMV)-35S promoter. Then, the plasmids were transformed into WT and cry2 mutant Arabidopsis separately, mediated by Agrobacterium GV3101 using the floral dip method (Clough and Bent 1998).
Hypocotyl and root measurements
For hypocotyl and root growth inhibition experiments, hypocotyl and root lengths of at least 30 Arabidopsis seedlings grown in appointed conditions were measured. Experiments were performed in at least three independent biological replicates.
Measurement of the total anthocyanin concentration
Total anthocyanin was extracted according to methanol–HCl method, in which the samples were extracted independently overnight in 5 ml methanol and 0.1 % HCl with extraction at room temperature. The absorbance of each extract was measured at 530, 620 and 650 nm with a spectrophotometer (UV-1600, Shimadzu, Kyoto, Japan).
Bimolecular fluorescence complementation (BiFC)
The full-length MdCRY2 and AtCOP1 cDNAs were cloned into pYFP-N (1–155) and pYFP-C (156–239) vectors and sequenced. Onion epidermal cells were transiently transformed using the Agrobacterium infection method with different combinations of these constructs. YFP-dependent fluorescence was detected 24 h after transfection using a confocal laser scanning microscope (Carl Zeiss; LSM 510 Meta).
Yeast two-hybrid assays
Yeast two-hybrid assays were performed using the Matchmaker GAL4-based two-hybrid system as recommended (Clontech, Palo Alto, CA, USA). Full-length MdCRY2 and AtCOP1 cDNA fragments were cloned into pGAD424 and pGBT9 (Clontech, Palo Alto, CA, USA). All constructs were transformed into the yeast strain AH109 by the lithium acetate method and yeast cells were grown on minimal medium/−Leu/−Trp according to the manufacturer’s instructions (Clontech, Palo Alto, CA, USA). Transformed colonies were plated onto minimal medium/−Leu/−Trp/−His/−Ade and dyed with X-gal to test for possible interactions.
Results
Full-length cDNA cloning of MdCRY2
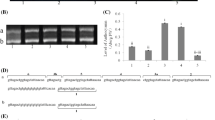
To isolate the full-length cDNA sequence of the apple cryptochrome gene MdCRY2, an apple expressed sequence tag (EST) clone on NCBI with similarity to Arabidopsis cryptochrome 2 was identified, isolated from apple fruit skin and was approximately 1,000 bp in length. The fragment was used as a probe to carry out 3′ and 5′ RACE-PCR to obtain the missing sequence of MdCRY2. We obtained a 700-bp fragment upstream of the EST sequence and an 800-bp fragment downstream. The EST sequence and the two fragments were then combined using DNAMAN software by matching to obtain the full-length sequence of MdCRY2. Special primers based on the sequence we obtained were designed to isolate the full-length MdCRY2. Finally, we obtained the full-length cDNA sequence of MdCRY2, which is 2,137 bp in length and contains a 1,944 bp ORF, which encodes a predicted protein of 648 amino acids with a calculated mass of 73.5 kDa. The deduced protein is basic with an isoelectric point of 5.88. In addition, we obtained the genomic DNA (MDP0000141094) encompassing the coding sequences of MdCRY2 from the apple genomic database FEM-IASMA Computational Biology Web Resources by BLAST using the cDNA sequence we obtained.
Through sequence comparison between the cDNA sequence and the genomic DNA sequence, we found that the coding region of MdCRY2 has three introns and four exons, similar to AtCRY2 (Fig. 1). However, AtCRY2 has a characteristic intron in the 5′ UTR, like the gene PsCRY2b (Xu et al. 2009), which does not exist in the MdCRY2 sequence (Fig. 1). We also found that the last two exons of MdCRY2 were longer than the front two exons, similar to AtCRY2 and PsCRY2b. However, the first intron of MdCRY2 is much longer than the last two, which differs from AtCRY2 and PsCRY2 (Fig. 1). It has been reported that the cryptochrome genes are well conserved in different plants species (Zhang et al. 2008) and perform different functions mainly through their divergent C-terminal extension (Lin and Shalitin 2003). In this study, there is a massive change in the first intron, suggesting that the MdCRY2 we cloned from apple may have some special functions compared with the cryptochrome genes from Arabidopsis and other plants.
Relationships with cryptochromes from other species and sequence analysis of MdCRY2
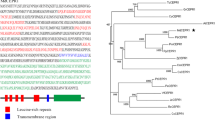
To analyze the phylogenetic relationship between MdCRY2 and CRYs from other plants, we compared the MdCRY2 sequence with 18 other representative plant cryptochromes (Fig. 2a). The phylogenetic analysis indicates that MdCRY2 is more closely related to CRY proteins from pea CRY2a (AAS79665.1) and CRY2b (AAO23972.1), clover blossom CRY2 (XP_003590036.1), and soybean CRY2 (BAI47558.1), as they clustered into the same clade. In contrast, MdCRY2 is most distantly related to the CRY2 of the monocotyledon plants rice (CAC82538.1) and wheat (ABX58030.1). Figure 2a also shows the dicotyledon plants CRY2 and monocotyledon plants CRY2 formed into one clade separately, while plant CRY1 formed into another group, indicating the regularity of plant evolution.
Phylogenetic relationship of the MdCRY2 protein. a The tree was constructed using MEGA4.1 and ClustalW. The scale bar represents 0.05 substitutions per site. b Comparison of the predicted MdCRY2 protein sequence with other CRY2 proteins. The DQXVP and STAES domains are highlighted in solid lines. The abbreviations used are as follows: Ps Pisum sativum, Mt Medicago truncatula, Gm Glycine max, Md Malus domestica, Sl Solanum lycopersicum, At Arabidopsis thaliana, Bj Brassica juncea, Bn Brassica napus, Os Oryza sativa, Ta Triticum aestivum, Sb Sorghum bicolor, Zm Zea mays
Sequence analysis by BLASTP in the NCBI database revealed that the amino acid sequence of MdCRY2 shared high identity similarities with known CRYs from other plant species, such as Fragaria vesca (ACP27856.1, 83 %), Glycine soja (BAK61591.1, CRY2a, 73 %; BAK61598.1, CRY2b, 67 %; BAK61601.1, CRY2c, 80 %), Glycine max (BAI47558.1, 73 %), Pisum sativum (AAS79665.1, CRY2a, 72 %; AAO23972.1, CRY2b, 70 %), Nicotiana sylvestris (ABB36797.1, 70 %), Solanum lycopersicum (NP_001234245.1, 69 %), and Arabidopsis thaliana (AAT80618.1, 68 %). The alignment of the putative amino acid sequences from these sequences showed that MdCRY2 contains the well-conserved N-terminal photolyase-related domain and the FAD-binding domain, as well as the C-terminal DAS domain (Fig. 2b), and it is also distinguished mainly by its C-terminal extensions from other plants like Arabidopsis.
Expression analysis of MdCRY2
To check the tissue-specific expression pattern of MdCRY2, we performed semiquantitative and real-time quantitative RT-PCR analysis using total RNAs obtained from different apple tissues. As shown in Fig. 3a, e, the gene expression of MdCRY2 was detected in all of the tissues we used, and varied expression levels among the different tissues were observed. MdCRY2 was expressed at the highest level in the root and leaf, followed by the flower, whereas weaker signals could be detected in the stem and fruit.
Expression analysis of MdCRY2 in different tissues and different blue-light irradiation times. a, e MdCRY2 transcript level in apple roots, stems, leaves, flowers and fruits using semiquantitative and real-time quantitative RT-PCR analysis; b MdCRY2 transcript level in dark, blue (20 μmol s−1 m−2), and red (20 μmol s−1 m−2) light conditions; c, f MdCRY2 transcript level of bagged uncolored fruits treated with different blue-light irradiation times using semiquantitative and real-time quantitative RT-PCR analysis; d MdCRY2 transcript level of apple callus treated with different blue-light irradiation times
To claim the expression of MdCRY2 is induced by light, bagged apple fruits were treated by dark, blue, and red light conditions for 120 h. MdCRY2 expression level was analyzed by semiquantitative RT-PCR. As shown in Fig. 3b, the expression of MdCRY2 was obviously induced by light, especially by blue light.
In addition, the expression levels of MdCRY2 treated with continuous blue-light for different times using uncolored bagged fruits were analyzed by semiquantitative and real-time quantitative RT-PCR. As shown in Fig. 3c, f, the expression of MdCRY2 increased with different blue light irradiation times; interestingly, during the first 48 h, the expression pattern of MdCRY2 represents a circadian rhythm, just as the observation in previous studies that the transcript levels of CRY genes show a nearly 24 h oscillation period (Toth et al. 2001; Platten et al. 2005).
Because the apples we used in the expression analysis above were grown in general field conditions, in order to remove the environmental influences on the expression levels of MdCRY2, we also analyzed the expression pattern of MdCRY2 using apple callus cultured indoors and treated with the same light conditions (Fig. 3d) to confirm the result of light-regulated MdCRY2 expression in apple fruits. We also found the rhythm of the MdCRY2 expression pattern; however, the rhythm cycle was within 12 h.
Functional complementation assay of MdCRY2 in Arabidopsis cry2 mutant
To investigate the function of MdCRY2 in plants, MdCRY2 was connected to the pbI121 expression vector driven by the CaMV-35S promoter and transferred to the cry2 mutant and WT Arabidopsis. The results demonstrated that we successfully obtained the transgenic Arabidopsis lines (Fig. 4a). The two transgenic lines, Re-1 and Line-2, with high levels of MdCRY2 expression were chosen to detect the leaf number of blotting, root length, hypocotyl elongation, and anthocyanin accumulation, to compare the phenotype of WT, cry2 mutant, MdCRY2 transgenic in cry2 mutant, and MdCRY2 overexpressing lines.
Expression levels of MdCRY2 and comparison of flowering time and anthocyanin content of WT, cry2 mutant, MdCRY2 transgenic in cry2 mutant background, and MdCRY2 transgenic in WT background Arabidopsis. a Expression levels of MdCRY2 in WT, cry2 mutant, three MdCRY2 transgenic lines in cry2 mutant background (Re-1, Re-2, and Re-3), and three MdCRY2 transgenic lines in WT background (Line-1, Line-2, and Line-3). b Comparison of leaf numbers and days to flower of the four types of Arabidopsis under long day (LD) and short day (SD) conditions. c Comparison of anthocyanin content of the four types Arabidopsis. Values shown are means of 30 Arabidopsis seedlings of each line. Error bars represent the standard deviation of the data, three replicates were performed
Previous studies demonstrated that AtCRY2 has prominent roles in the promotion of floral initiation (Guo et al. 1998). Therefore, this function was first examined in the four types Arabidopsis. As shown in Fig. 4b, the cry2 mutant plants exhibited a late-flowering phenotype on LDs compared with the WT, while the MdCRY2 transgenic in cry2 mutant exhibit a reduced late-flowering phenotype, which flowered earlier than the cry2 mutant seedlings according to both the number of leaves at bolting and days to flowering (Fig. 4b). This result indicated that MdCRY2 also has the ability to promote floral initiation under LD conditions. On SDs, the MdCRY2 transgenic plants in cry2 mutant exhibited no significant differences from the WT; however, the MdCRY2 overexpression lines showed an early-flowering phenotype. The results for both LDs and SDs suggest that MdCRY2 has the same function of promoting floral initiation as CRY2 in Arabidopsis.
To ensure the ability of MdCRY2 to inhibit hypocotyl elongation, the hypocotyl length was measured in the four types Arabidopsis. The results showed that the phenotype of reduced inhibition of hypocotyl elongation in the cry2 mutant was recovered by the transgenic MdCRY2 under blue light. Also, the MdCRY2 overexpressing lines showed the shortest hypocotyl length compared with the other three lines (Fig. 5a, c), although the inhibition ability was not as strong as AtCRY1 (Lin and Shalitin 2003).
Comparison of primary root and hypocotyl length of WT, cry2 mutant, MdCRY2 transgenic in cry2 mutant background, and MdCRY2 transgenic in WT background Arabidopsis. a Comparison of hypocotyl length of the four types Arabidopsis. b Comparison of primary root of the four types Arabidopsis. Values shown are means of 30 Arabidopsis seedlings of each line. Error bars represent the standard deviation of the data, three replicates were performed. The scale bar represents 5 mm
In addition to promoting floral initiation, it has been shown that AtCRY2 functions to inhibit root elongation under blue light conditions, while cry2 mutant seedlings show increased root elongation (Canamero et al. 2006). Therefore, the role of root elongation inhibition of MdCRY2 was investigated using the four types of Arabidopsis lines we used above. The seedlings were treated with different qualities of light for 8 days when cultured on MS medium, after which the root length was measured. As shown in Fig. 5b, c, the cry2 mutant seedlings exhibited a lengthened root phenotype under blue light conditions compared with WT and this phenotype disappeared in MdCRY2 transgenic in cry2 mutant lines. This result suggests that the MdCRY2 also plays roles in the inhibition of root elongation similar to AtCRY2. Additionally, in accordance with our hypothesis, the MdCRY2 overexpressing lines exhibited a more reduced root elongation phenotype under both blue and white light conditions.
Because MdCRY2 shows similar functions in both promoting floral initiation and inhibiting hypocotyl elongation and root elongation in Arabidopsis seedlings, it is reasonable to believe that it also has some other functions that are similar to those of AtCRY2, such as promoting anthocyanin accumulation (Lin 2002). To test this possibility, the anthocyanin content of the four types of Arabidopsis seedlings treated with continuous light for 8 days was measured. As shown in Fig. 4c, the overexpression of MdCRY2 in WT increased anthocyanin accumulation significantly. In addition, the overexpression of MdCRY2 in cry2 mutant seedlings also recovered the phenotype of reduced anthocyanin accumulation. These results suggest that MdCRY2 functions to promote anthocyanin accumulation in plants under blue light conditions.
MdCRY2 interacts with AtCOP1
Previous studies on the blue light receptors AtCRY1 and AtCRY2 regarding the light signal transduction molecular mechanisms have suggested that, in Arabidopsis both CRY1 and CRY2 perform their functions largely through the direct interaction with COP1 (Yang et al. 2001; Wang et al. 2001). Because MdCRY2 has such high sequence homology with AtCRY2 and similar functions as AtCRY2 in the regulation of plants growth, the question is presented as to whether the MdCRY2 protein also interacts with the COP1 protein to transfer the light signals it accepts. To answer this question preliminarily, we examined the interaction between MdCRY2 and AtCOP1 based on the extremely high similarly of AtCRY2 and MdCRY2 sequences. Yeast two-hybrid and BiFC assays were used to investigate the interaction. As shown in Fig. 6, in both yeast and onion epithelial cells, MdCRY2 and AtCOP1 interact with each other, which may provide a molecular mechanism involving MdCRY2 in the signal-transduction pathway and provide further evidence suggesting that MdCRY2 is a counterpart of AtCRY2.
Discussion
Studies conducted in the past 20 years indicate that cryptochromes are most likely the most widely spread photoreceptors in nature and that they play various important biological functions across all three major evolutionary lineages, from bacteria, to plants, to animals (Yu et al. 2010). In this study, we cloned the apple cryptochrome gene MdCRY2 and preliminary proved its function as a blue light receptor similar to AtCRY2.
Gene structure and sequence analysis of MdCRY2
The CRY apoprotein contains two domains: the N-terminal PHR domain and the C-terminal cryptochrome C-terminal extension (CCE) domain (Liu et al. 2011). All cryptochromes share sequence similarity at their N-terminal PHR domains with DNA photolyases (Yu et al. 2010). In this study, we found that the MdCRY2 also has conserved domains, such as the N-terminal photolyase-related domain and the FAD-binding domain as well as the C-terminal DAS domain. Comparing the genomic and cDNA sequences of MdCRY2 with AtCRY2, we found that they resemble each other, including in the number of introns and exons (Xu et al. 2009). All of these analyses suggest that the MdCRY2 we cloned is an AtCRY2 homolog that may have similar functions regarding the regulation of plant growth and development.
Comparing the gene structure of the two genes, we found that AtCRY2 has a characteristic intron in the 5′ UTR which does not exist in the MdCRY2 sequence. We also found that the first intron of MdCRY2 was longer than that of AtCRY2 (Fig. 1). These differences indicate that MdCRY2 is likely to have some distinct expression patterns and functions compared with Arabidopsis or other plant cryptochrome genes because the UTR regions and introns of a gene mostly function in regulating gene expression (Wray et al. 2003). Considering that the apple is a perennial woody plant and that its blossoming and fruit bearing should pass the young period and break dormancy, MdCRY2 may have some function in this special process; therefore, more regulators are required to regulate its expression and the expression pattern of genes in the apple must be distinct from that of the model plant Arabidopsis.
Expression analysis of MdCRY2
Previous studies showed that the expression of cryptochrome genes in Arabidopsis was induced by light, as expected, and exhibited a 24-h photoperiod (Toth et al. 2001; Platten et al. 2005). In our light-treated experiments using apple fruits and calluses, we found the expression pattern of MdCRY2 to be similar to that of AtCRY2.
In addition, we investigated the expression pattern of MdCRY2 in different tissues of apple and found it was constitutively expressed, similar to AtCRY2 in Arabidopsis (Ahmad and Cashmore 1993; Lin et al. 1998; Toth et al. 2001); this is in accordance with the various functions of cryptochrome genes in different tissues (Yu et al. 2010). These results of the MdCRY2 expression analysis implicate the function of MdCRY2 in the photo-response and preliminarily prove that MdCRY2 may be a blue/UV-A light receptor in the apple.
As a photoreceptor that senses light signals directly, MdCRY2 may be expressed highly and have more functions in organs aboveground, which perceive light in normal conditions. It is, therefore, reasonable that there is a high level of MdCRY2 expression in leaves (Fig. 3a, e). MdCRY2 may receive more light signals in the large leaf area and then transfer these signals to other tissues and organs; therefore, it regulates plant development using a dynamic approach. However, surprisingly, MdCRY2 is also expressed in roots at high levels (Fig. 3a, e) and inhibited the primer root elongation (Fig. 5b) from the tissue-specific expression analysis and functional complementation assay. The root of the apple is underground, which cannot perceive light in normal conditions; therefore, how can MdCRY2 expression be induced in the root? Previous studies in Arabidopsis have demonstrated that all three types of photoreceptors, including red/far-red light receptors PHYA and PHYB, blue/UV-A light receptors phototropins and cryptochromes, are expressed in Arabidopsis root tissues (Sakamoto and Briggs 2002; Toth et al. 2001). The first consideration was that the relatively dim light gradients that may penetrate soil, as well as light guiding, could be sufficient to activate the three major plant photosensory receptors (Mandoli et al. 1990). However, later research in Arabidopsis found that the functions of PHYA and PHYB in root tissues may involve a signal transmission from the shoot related with some hormones (Correll and Kiss 2005). Moreover, the functions of AtCRY1 and AtCRY2 in regulating the primary root elongation were also related to a hormone transport (auxin) from the aboveground part to root tissues (Canamero et al. 2006).
Following that idea, we propose a hypothesis that both the high levels of expression and the function in inhibiting the primary root elongation of MdCRY2 may be induced by some environmental signals perceived aboveground and may involve hormone signaling. In other words, the expression of MdCRY2 in the root might be induced by hormones in the apple. For example, we found that the expression of MdCRY2 is induced by ABA treatment (data not shown). However, more evidence is required to confirm this hypothesis. In addition, it is reasonable to deduce that MdCRY2 has more important functions in root development, such as regulating the primary and lateral root elongation and the portion of primary and lateral roots quantities, which play quite important roles in apple tree growth and development.
Functional complementation assay on Arabidopsis cry2 mutant
The function analysis of MdCRY2 on the promotion of floral initiation, inhibition of root and hypocotyl elongation, and anthocyanin accumulation indicated that MdCRY2 could recover the phenotype of the Arabidopsis cry2 mutant (Figs. 3, 4). All these results suggest that MdCRY2 is a counterpart of AtCRY2 and are in accordance with the expression analysis. Surprisingly, MdCRY2 has the function of promoting anthocyanin accumulation, even though its expression level in apple fruits is lower than the other tissues we used (Fig. 3a, e).
Anthocyanin concentration is an important determinant for the coloration of organs, such as flowers, leaves, and fruits (Quattrocchio et al. 1999; Allan et al. 2008). In this study, MdCRY2 has an effect on the promotion of anthocyanin accumulation; therefore, why is its expression in apple fruits so low? To answer this question, we formed two hypothesizes. First, we consider that the light signal can be transmitted between tissues, such as leaves to apple fruits, via hormone signaling, just like we mentioned in the roots (Correll and Kiss 2005; Canamero et al. 2006). Second, MdCRY2 may simply act as an initial promoter for anthocyanin accumulation and function at the translational or post-translational level in the regulation of anthocyanin content. It may directly or indirectly induce the expression of some downstream genes, including TFs, which regulate anthocyanin accumulation (Hong et al. 2008; Li et al. 2012). We regard the second hypothesis as more plausible because the uncolored bagged apple fruits, which have parted from apple trees, can obtain good coloration after treating with light conditions (Li et al. 2012).
Interaction between MdCRY2 and AtCOP1
Previous studies in Arabidopsis indicated that both AtCRY1 and AtCRY2 regulate plant development by the direct interaction with AtCOP1 (Yang et al. 2001; Wang et al. 2001). Both of them are activated by blue light, after which their protein construction is altered (Yu et al. 2009; Liu et al. 2011). The activated CRYs interact with COP1 to prevent its E3 ligase activity from the ubiquitination and degradation of TFs, such as HFR1, LAF1, CO, HY5, and BIT1, to regulate the expression of downstream photomorphogenic genes (Yang et al. 2005; Seo et al. 2003; Liu et al. 2008; Osterlund et al. 2000; Hong et al. 2008). In this study, we examined the transcription level of the downstream TFs and the anthocyanin-associated structural genes in both WT and MdCRY2 overexpressing Arabidopsis. We found overexpressing of MdCRY2 had little influence on the expression of the TFs, such as AtPAP1, AtBIT1, and AtCO, while expression of AtHY5 was higher in transgenic lines than in WT seedlings. Expression of the structural genes, such as AtCHS and AtDFR, increased obviously in MdCRY2 overexpressing lines compared to the WT (Fig. S1). Therefore, we propose, in most case, CRY2 regulates TFs stability in translational or post-translational level, and regulates downstream genes expression in transcript level.
Previous study showed MdCOP1s negatively regulate anthocyanin accumulation and fruit coloration in apple and Arabidopsis (Li et al. 2012). To investigate the molecular mechanism of MdCRY2 in promoting anthocyanin accumulation, we examined the interaction between MdCRY2 and AtCOP1. The positive result supports our hypothesis that MdCRY2 is simply an initial promoter of the regulation of anthocyanin accumulation and demonstrated that there should be one or more proteins, similar to AtCOP1, which interact with MdCRY2 directly in apple and transfer the light signal MdCRY2 receives to the genes downstream to regulate plant growth and development.
In addition, previous studies on apple have shown that the TF MdMYB1 plays a crucial role in apple fruit coloration; it binds to the promoters of the structural genes MdUFGT and MdDFR to regulate anthocyanin accumulation (Espley et al. 2007). We demonstrated that MdCOP1 regulates the stability of the MdMYB1 protein by ubiquitination and degradation (Li et al. 2012). Here, we tested the interaction of MdCRY2 and MdCOP1 by yeast two-hybrid assay using both full length of MdCRY2 and MdCOP1; unfortunately, we did not find the interaction in yeast cells. In Arabidopsis, there is augmentation of the interaction between AtCRY2 and AtCOP1 in yeast cells (Yang et al. 2001; Wang et al. 2001). However, by coimmunoprecipitation AtCRY2 and AtCOP1 were shown to interact in Arabidopsis extracts (Wang et al. 2001). In this study, transgenic of MdCRY2 recovered the phenotype of Arabidopsis cry2 mutant, and the expression of COP1-mediated photomophorgenetic genes in light signaling pathway, such as AtHY5, AtCHS, and AtDFR, increased in MdCRY2 overexpressing lines compared to WT (Fig. S1). Therefore, it is reasonable to deduce that MdCRY2 should interact with MdCOP1 directly or indirectly to mediate the stability of the MdMYB1 protein in a similar way as described in previous studies (Hong et al. 2008); this interaction also explains the function of MdCRY2 in promotion anthocyanin accumulation.
Combining the studies on the Arabidopsis light signal transduction mechanism and our results in apple about the functions of MdCRY2, we try to propose a light signal transduction mechanism in apple. First, on integral level of the apple, MdCRY2 induces the generation of some hormones after it receives a light signal. The hormones can be transported to other organs of the apple tree, such as the fruit and root, to induce the expression of some relative genes. MdCRY2 regulates apple growth and development on an overall level in this way. In addition, after MdCRY2 is activated by light, its protein conformation will be changed and it can interact with some special proteins to regulate the expression of downstream genes or the stability of some TFs. This may also be the manner in which MdCRY2 induces the generation of hormones.
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7(5):204–210
Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED (2003) Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11:59–67
Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y, Ahmad M (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224:995–1003
Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114:537–543
Chatterjee M, Sharma P, Khurana JP (2006) Cryptochrome1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol 141:61–74
Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, Jenkins KI, Getzoff ED (2012) Plant UVR8 photo-receptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335:1492–1496
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Correll MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46(2):317–323
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Franklin KA, Larner VS, Whitelam GC (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49:653–664
Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363
Gyula P, Schafer E, Nagy F (2003) Light perception and signaling in higher plants. Curr Opin Plant Biol 6:446–452
Hong SH, Kim HJ, Ryu JS, Choi H, Jeong S, Shin J, Choi G, Nam HG (2008) CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J 55:361–371
Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288
Kleine T, Lockhart P, Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35(1):93–103
Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5(8):337–342
Lin C (2002) Blue light receptors and signal transduction. Plant Cell 14:207–225
Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54:469–496
Lin C, Ahmad M, Gordon D, Cashmore A (1995) Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc Natl Acad Sci USA 92:8423–8427
Lin C, AhmadM Chan J, Cashmore AR (1996) CRY2, a second member of the Arabidopsis cryptochrome gene family. Plant Physiol 110:1047
Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690
Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306
Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16(12):684–691
Mandoli DF, Ford GA, Waldron LJ, Nemson JA, Briggs WR (1990) Some spectral properties of several soil types: implications for photomorphogenesis. Plant Cell Environ 13:287–294
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Partch CL, Sancar A (2005) Cryptochromes and circadian photoreception in animals. Methods Enzymol 393:726–745
Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL (2005) Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol 139:1472–1482
Quattrocchio F, Wing J, Woude K, Souer E, Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11(8):1433–1444
Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, Ulm R (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106
Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin1. Plant Cell 14(8):1723–1735
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999
Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616
Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158
Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20(9):1377–1419
Wu G, Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA 104:18813–18818
Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, Deng XW, Shi Y (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484:214–219
Xu P, Xiang Y, Zhu H, Xu H, Zhang Z, Zhang C, Zhang L, Ma Z (2009) Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol 149:760–774
Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–2587
Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821
Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104:7289–7294
Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21:118–130
Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue-light receptors. The Arabidopsis book 8:e0135. doi:10.1199/tab.0135
Zhang Q, Li H, Li R, Hu R, Fan C, Chen F, Wang Z, Liu X, Fu Y, Lin C (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc Natl Acad Sci USA 105:21028–21033
Acknowledgments
We thank Prof. Hongquan Yang of Shanghai Jiao Tong University, China, for providing Arabidopsis cry2 mutant. This work was supported by National Basic Research Program of China (2011CB100600), National Natural Science Foundation of China (31272142), and Program for Changjiang Scholars and Innovative Research Team in University (IRT1155).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2013_1387_MOESM1_ESM.tif
Figure S1. Expression of AtPAP1, AtBIT1, AtCO, AtHY5, AtCHS, and AtDFR in WT and MdCRY2 overexpressing Arabidopsis. AtACTIN was used as the external control (TIFF 112 kb)
Rights and permissions
About this article
Cite this article
Li, YY., Mao, K., Zhao, C. et al. Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep 32, 555–566 (2013). https://doi.org/10.1007/s00299-013-1387-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1387-4