Abstract
Key message
The long-term proliferation of embryogenic cell suspensions of oil palm is associated with changes in both genomic methylation rates and embryogenic capacities.
Abstract
In the aim of exploring the relationship between epigenetic stability and the long-term in vitro proliferation of plant tissues, we have studied changes in genomic DNA methylation levels in embryogenic suspensions of oil palm (Elaeis guineensis Jacq.). Five embryogenic callus lines were obtained from selected hybrid seeds and then proliferated as suspension cultures. Each clonal line obtained from a single genotype was subdivided into three independent subclonal lines. Once established, cultures proliferated for 12 months and genomic DNA was sampled at 4 months intervals for the estimation of global DNA methylation rates through high performance liquid chromatography (HPLC) quantitation of deoxynucleosides. Our results show that in vitro proliferation induces DNA hypermethylation in a time-dependent fashion. Moreover, this trend is statistically significant in several clonal lines and shared between subclonal lines originating from the same genotype. Interestingly, the only clonal line undergoing loss of genomic methylation in the course of proliferation has been found unable to generate somatic embryos. We discuss the possible implications of genome-wide DNA methylation changes in proliferating cells with a view to the maintenance of genomic and epigenomic stability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The large-scale micropropagation of higher plants through embryogenic suspensions exploits their unrivaled capacity to generate large numbers of somatic embryos within a short period of time. This approach is thus of paramount interest for the large-scale propagation of elite plant genotypes. It has been successfully applied on many different species such as wheat (Vasil et al. 1990), soybean (Finer and Nagasawa 1988), cotton (Finer 1988), banana (Côte et al. 1996), coffee tree (Etienne and Bertrand 2001), date palm (Fki et al. 2003) and the oil palm (de Touchet et al. 1991; Kramut and Te-chato 2010). Embryogenic suspensions have also been considered as a choice material for the genetic engineering of higher plants either through biolistics (Schoëpke et al. 1996; Chen et al. 1998) or Agrobacterium-mediated technology (Andrade et al. 2009; Yang et al. 2011).
However, the proliferation of undifferentiated plant material over long periods of continuous subculturing has often been described as detrimental to both the genetic and epigenetic stability of the regenerant offspring (Etienne and Bertrand 2003; Smýkal et al. 2007; Smulders and de Klerk 2011; Miguel and Marum 2011; Neelakandan and Wang 2012). Indeed, the ability of such material to re-differentiate through somatic embryogenesis is either gradually lost or altered in the course of successive subcultures, and the mechanisms that underlie such instability are yet to be understood. Increasing the number of subcultures and their duration is also known to increase the emergence of somaclonal variations, especially in cell suspension and callus cultures (Reuveni and Israeli 1990; Rodriguez and Wetzstein 1998; Bairu et al. 2006; Bairu et al. 2010).
The stochastic nature of somaclonal variation poses a significant challenge as it hampers the commercial development of somatic embryogenesis-based plant propagation and hence the necessity to elucidate the mechanisms underlying this phenomenon (Jaligot et al. 2011).
Early on, it has been demonstrated that in most organisms DNA methylation rates and distribution patterns vary along with cell and tissue differentiation and with developmental changes (Suzuki and Bird 2008). Methylation changes have also been observed after the application of growth regulators or as a response to a variety of biotic and abiotic stresses, notably those associated with tissue culture processes (LoSchiavo et al. 1989; Chinnusamy and Zhu 2009; Verhoeven et al. 2010; Smulders and de Klerk 2011). For these reasons, it can be hypothesized that changes in genomic DNA methylation, as a component of the epigenetic mechanisms regulating gene expression, are involved in modulating the expression of the embryogenic capacity during tissue culture, as well as in the emergence of variants when the epigenetic marks are improperly erased or re-distributed (Wang and Wang 2012).
In the case of the clonally propagated oil palm, a significant genomic DNA hypomethylation has been consistently detected within both tissue cultures and adult organs of oil palms affected by the mantled somaclonal variation, irrespective of the genotype (Jaligot et al. 2000). By contrast, investigations of methylation changes at the level of individual sequences have shown either increased or decreased methylation in the abnormal material and a strong variability between clonal lines (Matthes et al. 2001; Jaligot et al. 2002; Kubis et al. 2003; Jaligot et al. 2004). Clearly, a large-scale perturbation of epigenetic mechanisms is at work in both the mantled variation-prone (tissue cultures) and the variation-expressing (adult tissues) material, although whether such methylation changes are the source or a by-product of somaclonal variation remains to be elucidated. In any case, the role played by micropropagation appears to be central since, depending on the protocol used, wide variations in the incidence of somaclonal variation can be obtained (Eeuwens et al. 2002; see Jaligot et al. 2011 for a review). In the aim of better understanding the impact of in vitro propagation on both genome stability and embryogenic properties, we monitored changes in genomic DNA methylation rates in five embryogenic cell suspension lines of oil palm cultivated over a 12-months period.
Materials and methods
Plant material
Mature zygotic embryos originating from elite hybrid seeds of oil palm (Elaeis guineensis Jacq) were used as primary explants. Seed material was produced by the INRAB-CRAPP Research Centre based in Pobè, Benin, West Africa. Seeds were obtained from 2nd cycle parents through a Reciprocal Recurrent Selection scheme (Cochard et al. 2005). The cross of origin (female parent × male parent) for all seeds was PO3174D × PO4747P, deriving from the [DA 115D self] × [LM5T self] crossing.
Experimental design
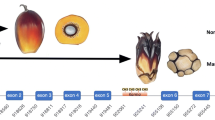
Dry seeds were rehydrated in sterile water supplemented with a fungicide solution (Dithane® M45 1 g.L−1) for 5 days at 27 °C in the dark under permanent stirring. The kernel endosperms were isolated by breaking the stone, then they were surface-sterilized using pure commercial bleaching solution (9.6 % active chlorine) for 30 min and rinsed 4 times with sterile water. Zygotic embryos (Fig. 1) were excised from the endosperm under aseptic conditions and cultivated in Pyrex tubes (25 mm diam. × 150 mm) for 4 months. Callogenesis was induced in the dark according to Pannetier et al. (1981) under the following culture conditions: temperature 24 ± 2 °C, relative humidity 45 ± 2 %. Suspension cultures were then generated from competent calli according to de Touchet et al. (1991). A fraction of these embryogenic calli was frozen in liquid nitrogen and stored at −80 °C until used for DNA extraction (see below). Five different clonal lines (each originating from a single zygotic embryo and therefore bearing a distinct genotype) were used for the present study, namely: 2071, 2074, 2154, 2334 and 2341. Each of these lines was subsequently divided into three subclonal lines (A, B and C) which were propagated independently (biological replicates). Embryogenic suspensions were isolated and proliferated under large-scale commercial conditions by SAS PalmElit in Colombia. Transfers (or subcultures) were performed on a monthly basis by re-suspending an aliquot of the suspension (250 ± 20 mg FW) into fresh liquid medium (20 mL in a 100 mL Erlenmeyer flask).
Experimental design for the estimation of DNA Global Methylation Rates in proliferating oil palm embryogenic suspensions. A, B and C are the subclonal lines, with A1, A2, A3 representing the three culture flasks sampled from the A subclone at a given time point. ze zygotic embryo, es endosperm, pk palm kernel, ec embryogenic callus, pem pro-embryogenic masses
Time zero (T0) of our experiment corresponds to the generation of a stabilized suspension, which is defined as a suspension with the ability to consistently generate a homogenous population of Pro-Embryogenic Masses (PEMs) upon every transfer. Once stabilization was achieved, a fraction of the cell culture was collected every 4 months (at T0, T4, T8, T12) by sieving through a 2 mm stainless steel-mesh filter. The harvested material was blotted dry on sterile paper, deep-frozen in liquid nitrogen and stored at −80 °C until further use. In parallel, plantlets were regenerated from each subclonal line at each time point according to the protocol described by de Touchet et al. (1991). Vitroplants were later field-planted for phenotypical evaluation.
Genomic DNA extraction
Genomic DNA was isolated from ca 2 g FW of frozen material (callus or cell suspension) using the DNeasy Plant Maxi Kit (Qiagen), according to the manufacturer’s instructions. DNA extractions were performed in the Agrobiotechnology Laboratory of the National University of Colombia in Bogotà, Colombia. The purified DNA was finally precipitated in 1 volume of absolute ethanol supplemented with 1/10th volume of 3 M sodium acetate, pH 5.2 before transportation to France, and stored at −20 °C upon arrival. For each of the five different genotypes monitored in the present study, DNA was extracted at each time point from the contents of three culture flasks for each of the three subclonal lines (Fig. 2). As a result, 45 DNA samples were obtained for each sampling date, which corresponds to a total of 180 DNA extracts over the whole experiment.
Evolution over a 12-months culture time of DNA Global Methylation Rates in embryogenic cell cultures of oil palm. Each genotypically distinct clonal line (rows) is represented by three independently propagated subclonal lines (columns). GMRs are measured every 4 months for each subclonal line (see Materials and methods for details). P values followed by one, two or three asterisks are statistically significant at the 0.05, 0.01 or 0.001 level, respectively
Enzymatic hydrolysis of DNA
The purified DNA pellets were re-suspended in 50–100 μL of sterile water, and the concentration and the purity were estimated using a NanoDrop ND-100 microvolume spectrophotometer (Thermo Scientific). The enzymatic hydrolysis of genomic DNA was performed according to a modified version of our previously published protocol (Jaligot et al. 2000). Twenty micrograms of DNA were hydrolysed into nucleosides during 3 h at 37 °C by the addition of 5 μl of a 0.5 U/μl solution of nuclease P1 (EC number 3.1.30.1, Sigma N8630) and 17.5 μl of a 0.017 U/μl solution of alkaline phosphatase (EC number 3.1.3.1, Sigma P4252) in a reaction volume adjusted to 100 μl with the digestion buffer (30 mM NaCH3, 0.1 mM ZnCl2, pH 5.3). Digestions were performed in duplicate (technical replicates) for each DNA extract, and in triplicate whenever permitted by the amount of DNA available. The reaction was stopped by the addition of 245 μl of absolute ethanol, and the digestion products were centrifuged at 11,000×g for 15 min. The supernatant was transferred to a new tube, vacuum-dried and nucleosides were re-suspended with 500 μl of sterile water. The extracts were finally filtered (0.2 μm syringe filters, Nalgene) prior to HPLC analysis.
HPLC estimation of global methylation rates
An isocratic elution method was followed according to Monteuuis et al. (2008), using a modified version of the buffer described by Gehrke et al. (1984): 50 mM KH2PO4, 8 % (V/V) methanol, pH 3.5 (instead of 4.4), through a Supelcosil LC-18S reverse-phase column (SUPELCO Inc.; column length 25 cm; diameter 4.6 mm; particle diameter: 5 μm), with a flow rate of 0.8 ml/min and a run time of 30 min. These conditions allowed a clear separation of the peaks corresponding to dC and Uracil (as identified by their respective retention time and UV absorption spectrum). The effluent was monitored at the wavelength of 285 nm with a photodiode array detector (Beckman Coulter). Global Methylation Rates (GMRs) were calculated as percentages using the following formula: GMR = 100 × (5mdC)/[(dC) + (5mdC)], where (5mdC) and (dC) are the respective concentrations of the methylated and unmethylated forms of deoxycytidine. Concentrations were calculated through peak integration from the calibration curves obtained with external standards of known concentrations, which were monitored simultaneously with samples: 2′desoxycytidine (Sigma D3897), 5-methyl-2′desoxycytidine (MP Chemicals 198883), 2′desoxyguanosine (Sigma D7145), thymidine (Fluka 89270) and 2′desoxyadenosine (Sigma D7400). In order to identify any residual RNA contaminant, the following nucleosides were also used as standards: cytidine (Fluka 30270), uridine (Sigma U3750), guanosine (Sigma G6752) and adenosine (Sigma A9251).
Statistical analyses
GMR data were analyzed for each subclonal line using linear regression, the duration of proliferation being considered as a quantitative variable. Statistical analyses were undertaken using the Statistica computing programme (Statsoft) for linear regression and ANOVA, followed by the Newman and Keuls test. A probability level of P < 0.05 was considered as significant.
Results and discussion
Changes in DNA GMRs were monitored in five different clonal lines subdivided into three independent subclonal lines each, which were propagated during 12 months.
When considering each of the culture flasks sampled in the course of this study, individual GMR measurements performed in proliferating oil palm embryogenic suspensions ranged from 14.85 to 25.23 % (overall average 20.49), depending on the clonal and subclonal origin of the analyzed material and on the duration of proliferation. These percentages fall within the same range as those of the embryogenic calli from which these suspensions were generated (individual GMR values comprised between 18.75 and 23.53 %; overall average value 21.07 %). Such values are in accordance with those previously obtained by Jaligot et al. (2000) in oil palm leaves (average GMR 22 % for somatic embryo-derived palms) and in embryogenic calli (23 %).
The size of the diploid oil palm genome has previously been determined to be 3.4 Gb by flow cytometry (Rival et al. 1997). The HPLC measurements performed in the present study allowed us to estimate the average total deoxycytidine base (dC + 5mdC) content of the oil palm genome: 19.52 %. As a result, it can be inferred that the oil palm genomic DNA includes a total of approximately 663,000 dCs and 5mdCs. Therefore, at the start of this experiment (T0) the number of 5mdCs in our embryogenic suspensions ranges from a clone-based average of 127,600 (19.24 % GMR out of 663,000 deoxycytidine bases) to 133,600 (20.15 % GMR) (Table 1).
In four out of five clonal lines studied, GMRs in cell suspensions showed a time-dependent increase and this trend towards hypermethylation was statistically significant in three of them, namely 2071, 2154 and 2334 (Fig. 2). Interestingly, within each of these clones, all three subclonal lines displayed a comparable trend in terms of DNA methylation changes. At the end of this 12-months proliferation period, the average GMR values of the three “hypermethylating” lines have increased by approximately 10 % (respectively, 9.72, 13.31 and 11.66 %; Table 1), with per-subclone extreme variations comprised between +5 % (clone 2334, line C) and +16 % (clone 2154, line A).
A slight DNA hypermethylation has also been observed for clone 2341 (average GMR increased by 4.75 %, Table 1), although this variation has not been found statistically significant (Fig. 2).
In our experimental set-up, the behavior of clone 2074 stands out as a singularity: in contrast with the four other clonal lines under study, GMR values obtained for all three subclonal lines from this genotype decreased throughout the 12 months proliferation period with an average variation rate of −4.89 % (Table 1). This variation is, however, significant only in subclonal line A (Fig. 2). It is worth noticing that, among the five cell suspensions used in the present work, clone 2074 is the only one from which we were unable to re-differentiate somatic embryos after the termination of the proliferation period (de Touchet et al. 1991). On the opposite, the four other lines retained their full embryogenic capacity, and vitroplants have been successfully regenerated. Indeed, the productivity of suspensions reached 438, 882, 145 and 280 vitroplants month−1 L−1 for clonal lines 2074, 2154, 2334 and 2441, respectively.
Among the five oil palm cell lines under study, we can estimate that a change in methylation status (from 5mdC to dC or the other way round) has occurred at a number of dCs ranging from 6,232 (clones 2074 and 2341) to 17,238 (clone 2154) throughout the genome during the time span of this experiment (Table 1). According to our calculations, this should correspond to a methylation change affecting, respectively, 0.94 to 2.60 % of the 663,000 dCs in the oil palm genome. However, it must be kept in mind that HPLC analyses only allow the quantitation of DNA methylation from hydrolysed samples. As a consequence, it is not possible to determine from our results which proportion the observed dC methylation shift affects gene-containing regions of the oil palm genome. Even though it has been announced that the sequencing of the Elaeis guineensis genome has been completed (Rival and Jaligot 2011), the data are yet to be released to the public. Thus, basic information such as the relative proportion between gene and non-gene space in the oil palm genome is still missing to the investigators.
The consequences of long-term in vitro propagation on both the genetic and epigenetic stability of cultured plant cells have been the subject of several investigations. Studying de novo shoot regeneration in Arabidopsis thaliana, Li et al. (2011) recently reported that DNA methylation and histone modifications are able to regulate regeneration through modulating WUSCHEL transcription factor expression and auxin signaling.
Smýkal et al. (2007) assessed the genetic and epigenetic stability of a multiple shoot culture of pea (Pisum sativum), which has been propagated in vitro for 24 years. Their HPCE measurements showed no significant differences in GMR between in vitro-derived (either from newly established or long-term cultures) and seed-derived plants. These authors also detected low level of polymorphisms through the AFLP technique and its methylation-sensitive counterpart MSAP (11 and 18 %, respectively) among samples. Not surprisingly in this highly stable background, no evidence of TE reactivation could be demonstrated.
In micropropagated hop (Humulus lupulus), Peredo et al. (2009) found that the majority of the epigenetic variation was attributable to in vitro culture conditions per se. These authors also demonstrated that additional variation accumulates along the 12 successive subculturing cycles performed throughout the 2 years of the study. In Taxus media cell cultures propagated over 5 years, HPLC quantitation showed a time-dependent increase of GMRs (from 17 to 25.7 % on average), whereas MSAP analyses revealed a number of locus-specific methylation gains and losses throughout the period (Fu et al. 2012). Another woody perennial, Malus xiaojinensis, underwent 20 cycles (spanning a 27-months period) of either nodal stem segments or adventitious buds culture, but it was only after the 13th subculturing event that the propagated material displayed a significantly decreased GMR (from 28.4 and 31.1 % at the start of the experiment to 22.4 and 20.5 %, respectively) (Huang et al. 2012). For both Taxus and M. xiaojinensis culture systems, the alteration of genomic methylation over time was paralleled by the emergence of somaclonal variations.
The stability of the cell suspension epigenome has been studied by Tanurdzic et al. in Arabidopsis (2008). When compared to seedlings, the cell suspension genome displayed a global reshuffling of the epigenetic marks: dC methylation was decreased in the heterochromatic regions (triggering the reactivation of different TE families) and increased in euchromatic, gene-rich regions, with the longer in vitro propagation time yielding the more drastic changes.
Overall, this overview demonstrates that genome-wide epigenetic changes are common in long-term in vitro cultures and that changes accumulate in a time-dependent fashion. Nevertheless, the various plant species investigated differ greatly in terms of the direction of the DNA methylation alterations (i.e. increase or decrease) or in terms of outcome of these modifications (reactivation of TEs, onset of phenotypic variation).
Since the pioneering work of Peschke et al. (1987), increased genetic and epigenetic variability arising from the in vitro culture-induced reactivation of Transposable Elements (TEs) has been well-documented. The first demonstration of the activation of a plant retrotransposon by tissue culture was made by Hirochika (1993) on the Tto family in tobacco (Nicotiana tabacum), where the copy number of Tto1 was found to increase 10-fold in cultivated cell lines. Ozeki et al. (1997) later showed that the insertion of TEs is one of the mechanisms that can cause variation of plant cell cultures during repeated subculture.
In parallel, the DNA methylation within both the repetitive portion of plant genomes (Kaeppler et al. 2000) and individual TE sequences (Peschke et al. 1987) has been found to change as a consequence of in vitro culture. It is, therefore, thought that, similarly to other sources of stress, the primary effect of tissue culture on genome stability and expression is through the release of TEs from the DNA methylation-dependent repression of both their transcription and their mobility (Grandbastien 1998; Lippman et al. 2004; Ngezahayo et al. 2009; Rigal and Mathieu 2011).
The present work complements previous papers published by our group (Jaligot et al. 2000; Jaligot et al. 2002; Jaligot et al. 2004) and others (Matthes et al. 2001; Kubis et al. 2003). Taken together, these different studies have contributed to demonstrate through a variety of methods that genomic DNA methylation could be related with the occurrence of the mantled somaclonal variation in the oil palm. Our work adds fuel to the assumption that GMR could be modulated by the environmental conditions imposed on in vitro cultivated cells, a long series of subcultures inducing a sizeable increase in GMRs in most lines.
At each transfer, a small aliquot of cultured plant cells is sampled and used to generate a new suspension culture. This fraction of the original cell population, therefore, undergoes a brutal change in culture conditions which could be assimilated to a stress event (Grafi et al. 2011; Wang and Wang 2012). Through several successive transfers, this phenomenon is likely to result in the progressive selection of the best adapted sub-population of plant cells, i.e. those combining continuous division with the ability to overcome the environmental constraints associated with in vitro culture. We propose that both these criteria for a successful adaptation rely at least partially on the preservation of a “reservoir” of genomic plasticity in the cultured cells, and that this protective function is dependent on their capacity to dynamically redistribute their DNA methylation. Indeed, the extreme genomic plasticity of plant cells allows them, under appropriate conditions, to recover their totipotency, which is the aptitude to generate a complete individual. This de-differentiation process is thought to be achieved through the erasing of some, if not all, of the pre-existing epigenetic marks (and among them, DNA methylation) across the genome (Bourc’his and Voinnet 2010; Feng et al. 2010; Jacob and Martienssen 2011). As a matter of fact, the ability of plant cells to modulate genome plasticity through epigenetic mechanisms has been pointed as a possible origin for generic stress tolerance and adaptation by an increasing number of papers (Bruce et al. 2007; Chinnusamy and Zhu 2009; Boyko and Kovalchuk 2011; Mirouze and Paszkowski 2011; Richards 2011; Rival and Jaligot 2011; Yaish et al. 2011). It has also been postulated that the better adaptability of a certain cell populations to in vitro culture through the acquisition of the appropriate epigenetic modifications, combined with the pre-existing natural epigenetic variation between different explants or different cells within a callus, could contribute to the emergence of somaclonal variant phenotypes associated with epigenetic alterations (Wang and Wang 2012).
In the present study, we observed that most of our clonal lines retained the ability for re-differentiating somatic embryos after 12 months in propagation. In this view, it is plausible that the substantial increase in GMRs that we have shown in these cell lines could reflect the relocation of dC methylation between euchromatic, gene-rich and heterochromatic, TE-rich regions of the genome (Tanurdzic et al. 2008), and that this remodeling conditions the expression of the embryogenic potential. In support to this idea of embryogenicity as an epigenetically-based capacity, it has been demonstrated in Arabidopsis that the transition of embryonic genes from an heterochromatic to a euchromatic state prompted the formation of somatic embryos (Tang et al. 2012), and that the generation of embryogenic calli from leaf explants involved the genome-wide redistribution of a repressive chromatin mark with an increased accumulation in TE sequences (He et al. 2012). Moreover, the ability to efficiently silence TE sequences has been shown to play a beneficial role in the adaptation of the host genome to various biotic and abiotic stresses, and this phenomenon is dependent on the small RNA-based connection between the distinct DNA methylation and/or chromatin remodeling pathways targeting, respectively, repetitive or gene sequence. This cross-talk has been illustrated lately through independent works conducted by Ito et al. (2011), McCue et al. (2012), Dowen et al. (2012) and Yao et al. (2012).
As for the divergent behavior observed in one of our clonal lines, combining a decreasing GMR and the loss of the embryogenic capacity might reflect in this particular genotype the insufficiency or inadequacy of the DNA methylation remodeling mechanisms involved in genome plasticity. A recent study by Fraga et al. (2012) showed that the use of the hypomethylating drug 5-Azacytidine at moderate doses stimulated somatic embryo formation in Acca sellowiana, but yielded a decreased embryo-to-plantlet conversion. The authors attribute this result to the genome-wide dysregulatory effects of the drug treatment on the DNA methylation patterns changes associated with the re-differentiation process. Similarly, the occurrence of a deficient DNA methylation machinery in certain cell lines, as a consequence of either natural diversity in the initial explant cells or deleterious accumulation of in vitro-induced variation, would be likely to impair the cells’ morphogenetic capacity (Wang and Wang 2012).
The present work addressed the impact of long-term proliferation on GMR in plant cells. Results showed that DNA hypermethylation parallels the long-term in vitro proliferation of oil palm embryogenic suspensions.
Different parameters such as the cell type, the developmental age of the tissues and the physical age of the plant are known to affect the efficiency of the reprogramming process (Grafi 2004). Also, the synthetic auxin analogues that are necessary for the production of dedifferentiated calli or cell suspensions have been frequently suspected to compromise genomic stability through the promotion of aberrant DNA methylation coupled with modifications in gene expression (LoSchiavo et al. 1989; Bairu et al. 2006; Zeng et al. 2007; Alexandre et al. 2009; Krogan and Long 2009; Roowi et al. 2010). Such perturbations might interfere with the large-scale redistribution of epigenetic marks occurring as a part of the in vitro multiplication process, leading to the impairment of the embryogenic potential in the cultured cells and ultimately to the generation of somaclonal variations in the regenerant offspring. Regarding the oil palm, it has been previously shown that the expression of several auxin-responsive genes is altered in the variant-producing in vitro material (Morcillo et al. 2006), and in a more recent study, the expression of one such gene has been positively correlated with the embryogenic capacity of the cultured tissues (Ooi et al. 2012). Interestingly, the recent report by Zhang et al. (2012) on the characterization of the rice (Oryza sativa) mutant rpl1 provides evidence for a functional interplay between pathways, respectively, in charge for environmentally-induced epigenetic modifications, response to growth regulators and phenotypic plasticity. The functional characterization of a pineapple (Ananas comosus) receptor kinase gene by Ma et al. (2012) led the authors to the similar conclusion that the signaling cascades driven by abiotic constraints or exogenous hormones were interconnected and participated to the acquisition of the embryogenic competence. Thus, it would be most interesting to get information on the floral phenotype of the regenerant oil palms which will be obtained from the different embryogenic cell lines used in this work. Somatic embryos were regenerated from these lines in the aim of establishing large-scale field trials, involving over 100 palms per clonal line, in Latin America. Considering the long life cycle of the oil palm, two more years are necessary before we are able to monitor the floral phenotype of these individuals, and thereby evaluate the impact of the proliferation time on the mantled somaclonal variation.
Meanwhile, experiments are under way in our lab to monitor the TE activity in relationship with changes in DNA methylation, during the long-term proliferation of oil palm embryogenic suspensions.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- 5mdC:
-
5-Methyldeoxycytidine
- dC:
-
Deoxycytidine
- FW:
-
Fresh Weight
- Gb:
-
Gigabases
- GMR:
-
Global Methylation Rate
- HPLC:
-
High Performance Liquid Chromatography
- PEM:
-
Pro-Embryogenic Mass
- PGR:
-
Plant Growth Regulator
- TE:
-
Transposable Element
References
Alexandre C, Moller-Steinbach Y, Schonrock N et al (2009) Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant 2:675–687
Andrade GM, Nairn CJ, Le HT, Merkle SA (2009) Sexually mature transgenic American chestnut trees via embryogenic suspension-based transformation. Plant Cell Rep 28:1385–1397
Bairu MW, Fennell CW, van Staden J (2006) The effect of plant growth regulators on somaclonal variation in Cavendish banana (Musa AAA cv. “Zelig”). Sci Hortic 108:347–351
Bairu MW, Aremu AO, Van Staden J (2010) Somaclonal variation in plants: causes and detection methods. Plant Growth Regul 63:147–173
Bourc’his D, Voinnet O (2010) A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330:617–622
Boyko A, Kovalchuk I (2011) Genome instability and epigenetic modification—heritable responses to environmental stress? Curr Opin Plant Biol 14:260–266
Bruce TJA, Matthes MC, Napier JA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173:603–608
Chen L, Zhang S, Beachy RN, Fauquet CM (1998) A protocol for consistent, large-scale production of fertile transgenic rice plants. Plant Cell Rep 18:25–31
Chinnusamy V, Zhu J-K (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12:133–139
Cochard B, Amblard P, Durand-Gasselin T (2005) Oil palm genetic improvement and sustainable development. Oléagineux Corps gras Lipides 12:141–147
Côte FX, Domergue R, Monmarson S et al (1996) Embryogenic cell suspensions from the male flower of Musa AAA cv Grand nain. Physiol Plant 97:285–290
de Touchet B, Duval Y, Pannetier C (1991) Plant regeneration from embryogenic suspension cultures of oil palm (Elaeis guineensis Jacq.). Plant Cell Rep 10:529–532
Dowen RH, Pelizzola M, Schmitz RJ et al (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109:E2183–E2191
Eeuwens CJ, Lord S, Donough CR et al (2002) Effects of tissue culture conditions during embryoid multiplication on the incidence of “mantled” flowering in clonally propagated oil palm. Plant Cell Tiss Organ Cult 70:311–323
Etienne H, Bertrand B (2001) Trueness-to-type and agronomic characteristics of Coffea arabica trees micropropagated by the embryogenic cell suspension technique. Tree Physiol 21:1031–1038
Etienne H, Bertrand B (2003) Somaclonal variation in Coffea arabica: effects of genotype and embryogenic cell suspension age on frequency and phenotype of variants. Tree Physiol 23:419–426
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330:622–627
Finer JJ (1988) Plant regeneration from somatic embryogenic suspension cultures of cotton (Gossypium hirsutum L.). Plant Cell Rep 7:399–402
Finer JJ, Nagasawa A (1988) Development of an embryogenic suspension culture of soybean (Glycine max Merrill.). Plant Cell Tiss Organ Cult 15:125–136
Fki L, Masmoudi R, Drira N, Rival A (2003) An optimised protocol for plant regeneration from embryogenic suspension cultures of date palm, Phoenix dactylifera L., cv Deglet Nour. Plant Cell Rep 21:517–524
Fraga H, Vieira L, Caprestano C et al (2012) 5-Azacytidine combined with 2,4-D improves somatic embryogenesis of Acca sellowiana (O. Berg) Burret by means of changes in global DNA methylation levels. Plant Cell Rep. doi:10.1007/s00299-012-1327-8
Fu C, Li L, Wu W et al (2012) Assessment of genetic and epigenetic variation during long-term Taxus cell culture. Plant Cell Rep 31:1321–1331
Gehrke CW, McCune RA, Gama-Sosa MA et al (1984) Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr 301:199–219
Grafi G (2004) How cells dedifferentiate: a lesson from plants. Dev Biol 268:1–6
Grafi G, Chalifa-Caspi V, Nagar T et al (2011) Plant response to stress meets dedifferentiation. Planta 233:433–438
Grandbastien M-A (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3:181–187
He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Arabidopsis Tissues. PLoS Genet 8:e1002911
Hirochika H (1993) Activation of tobacco retrotransposons during tissue culture. EMBO J 12:2521–2528
Huang H, Han S, Wang Y et al (2012) Variations in leaf morphology and DNA methylation following in vitro culture of Malus xiaojinensis. Plant Cell Tiss Organ Cult. doi:10.1007/s11240-012-0179-9
Ito H, Gaubert H, Bucher E et al (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115–119
Jacob Y, Martienssen RA (2011) Chromatin reprogramming: gender equality during Arabidopsis germline differentiation. Curr Biol 21:R20–R22
Jaligot E, Rival A, Beulé T et al (2000) Somaclonal variation in oil palm (Elaeis guineensis Jacq.): the DNA methylation hypothesis. Plant Cell Rep 19:684–690
Jaligot E, Beulé T, Rival A (2002) Methylation-sensitive RFLPs: characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor Appl Genet 104:1263–1269
Jaligot E, Beulé T, Baurens FC et al (2004) Search for methylation-sensitive amplification polymorphisms associated with the “mantled” variant phenotype in oil palm (Elaeis guineensis Jacq.). Genome 47:224–228
Jaligot E, Adler S, Debladis E et al (2011) Epigenetic imbalance and the floral developmental abnormality of the in vitro-regenerated oil palm Elaeis guineensis. Ann Bot 108:1463–1475
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188
Kramut P, Te-chato S (2010) Effect of culture media, plant growth regulators and carbon sources on establishment of somatic embryo in suspension culture of oil palm. J Agri Technol 6:159–170
Krogan NT, Long JA (2009) Why so repressed? Turning off transcription during plant growth and development. Curr Opin Plant Biol 12:628–636
Kubis SE, Castilho AMM, Vershinin AV, Heslop-Harrison JS (2003) Retroelements, transposons and methylation status in the genome of oil palm (Elaeis guineensis) and the relationship to somaclonal variation. Plant Mol Biol 52:69–79
Li W, Liu H, Cheng ZJ et al (2011) DNA Methylation and Histone Modifications Regulate De Novo Shoot Regeneration in Arabidopsis by Modulating WUSCHEL Expression and Auxin Signaling. PLoS Genet 7:e1002243
Lippman Z, Gendrel A-V, Black M et al (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471–476
LoSchiavo F, Pitto L, Giuliano G et al (1989) DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones and hypomethylating drugs. Theor Appl Genet 77:325–331
Ma J, He Y, Hu Z et al (2012) Characterization and expression analysis of AcSERK2, a somatic embryogenesis and stress resistance related gene in pineapple. Gene 500:115–123
Matthes M, Singh R, Cheah SC, Karp A (2001) Variation in oil palm (Elaeis guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor Appl Genet 102:971–979
McCue AD, Nuthikattu S, Reeder SH, Slotkin RK (2012) Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA. PLoS Genet 8:e1002474
Miguel C, Marum L (2011) An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot 62:3713–3725
Mirouze M, Paszkowski J (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14:267–274
Monteuuis O, Doulbeau S, Verdeil J-L (2008) DNA methylation in different origin clonal offspring from a mature Sequoiadendron giganteum genotype. Trees 22:779–784
Morcillo F, Gagneur C, Adam H et al (2006) Somaclonal variation in micropropagated oil palm. Characterization of two novel genes with enhanced expression in epigenetically abnormal cell lines and in response to auxin. Tree Physiol 26:585–594
Neelakandan A, Wang K (2012) Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep 31:597–620
Ngezahayo F, Xu C, Wang H et al (2009) Tissue culture-induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol 9:91
Ooi S-E, Choo C-N, Ishak Z, Ong-Abdullah M (2012) A candidate auxin-responsive expression marker gene, EgIAA9, for somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). Plant Cell Tiss Org Cult 110:201–212
Ozeki Y, Davies E, Takeda J (1997) Somatic variation during long term subculturing of plant cells caused by insertion of a transposable element in a phenylalanine ammonia-lyase (PAL) gene. Mol Gen Genet 254:407–416
Pannetier C, Arthuis P, Lievoux D (1981) Néoformation de jeunes plantes d’Elaeis guineensis à partir de cals primaires obtenus sur fragments foliaires cultivés in vitro. Oléagineux 36:119–122
Peredo EL, Arroyo-García R, Revilla MÁ (2009) Epigenetic changes detected in micropropagated hop plants. J Plant Physiol 166:1101–1111
Peschke VM, Phillips RL, Gengenbach BG (1987) Discovery of transposable element activity among progeny of tissue culture—derived maize plants. Science 238:804–807
Reuveni O, Israeli Y (1990) Measures to reduce somaclonal variation in in vitro propagated bananas. Acta Horticulturae 275:307–314
Richards EJ (2011) Natural epigenetic variation in plant species: a view from the field. Curr Opin Plant Biol 14:204–209
Rigal M, Mathieu O (2011) A “mille-feuille” of silencing: epigenetic control of transposable elements. Biochim Biophys Acta 1809:452–458
Rival A, Jaligot E (2011) Epigenetics and plant breeding. CAB reviews: perspectives in agriculture, veterinary science, nutrition and natural resources 6:048
Rival A, Beulé T, Barre P et al (1997) Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis Jacq) tissue cultures and seed-derived plants. Plant Cell Rep 16:884–887
Rodriguez APM, Wetzstein HY (1998) A morphological and histological comparison of the initiation and development of pecan (Carya illinoinensis) somatic embryogenic cultures induced with naphthaleneacetic acid or 2,4-dichlorophenoxyacetic acid. Protoplasma 204:71–83
Roowi SH, Ho C-L, Alwee SSRS et al (2010) Isolation and Characterization of Differentially Expressed Transcripts from the Suspension Cells of Oil Palm (Elaeis guineensis Jacq.) in Response to Different Concentration of Auxins. Mol Biotechnol 46:1–19
Schoëpke C, Taylor N, Carcamo R et al (1996) Regeneration of transgenic cassava plants (Manihot esculenta Crantz) from microbombarded embryogenic suspension cultures. Nat Biotechnol 14:731–735
Smulders M, de Klerk G (2011) Epigenetics in plant tissue culture. Plant Growth Regul 63:137–146
Smýkal P, Valledor L, Rodríguez R, Griga M (2007) Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Rep 26:1985–1998
Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9:465–476
Tang X, Lim M-H, Pelletier J et al (2012) Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J Exp Bot 63:1391–1404
Tanurdzic M, Vaughn MW, Jiang H et al (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6:2880–2895
Vasil V, Redway F, Vasil IK (1990) Regeneration of plants from embryogenic suspension culture protoplasts of wheat (Triticum aestivum L.). Nat Biotechnol 8:429–434
Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A (2010) Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 185:1108–1118
Wang Q-M, Wang L (2012) An evolutionary view of plant tissue culture: somaclonal variation and selection. Plant Cell Rep 31:1535–1547
Yaish MW, Colasanti J, Rothstein SJ (2011) The role of epigenetic processes in controlling flowering time in plants exposed to stress. J Exp Bot 62:3727–3735
Yang J, Bi H-P, Fan W-J et al (2011) Efficient embryogenic suspension culturing and rapid transformation of a range of elite genotypes of sweet potato (Ipomoea batatas [L.] Lam.). Plant Sci 181:701–711
Yao Y, Bilichak A, Golubov A, Kovalchuk I (2012) ddm1 plants are sensitive to methyl methane sulfonate and NaCl stresses and are deficient in DNA repair. Plant Cell Rep 31:1549–1561
Zeng F, Zhang X, Jin S et al (2007) Chromatin reorganization and endogenous auxin/cytokinin dynamic activity during somatic embryogenesis of cultured cotton cell. Plant Cell Tiss Organ Cult 90:63–70
Zhang C–C, Yuan W-Y, Zhang Q-F (2012) RPL1, a Gene Involved in Epigenetic Processes Regulates Phenotypic Plasticity in Rice. Mol Plant 5:482–493
Acknowledgments
This work was supported by the French National Research Agency (ANR) through the Vitropalm Emergence Project (ANR-06-EMPB-036-01), and by PalmElit SAS. The authors are thankful to the dedicated staff of CRAPP-INRAB Research Station in Pobè, Benin for the kind supply of selected plant material.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Guiderdoni.
Rights and permissions
About this article
Cite this article
Rival, A., Ilbert, P., Labeyrie, A. et al. Variations in genomic DNA methylation during the long-term in vitro proliferation of oil palm embryogenic suspension cultures. Plant Cell Rep 32, 359–368 (2013). https://doi.org/10.1007/s00299-012-1369-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-012-1369-y