Abstract
Microgametogenesis is a complex process that involves numerous well-coordinated cell activities, ending with the production of pollen grains. Pollen development has been studied at the cytological level in Arabidopsis and other plant species, where its temporal time course has been defined. However, the molecular mechanism underlying this process is still unclear, since a relative small number of genes and/or processes have been identified as essential for pollen development. We have designed a methodology to select candidate genes for functional analysis, based on transcriptomic data obtained from different stages of pollen development. From our analyses, we selected At2g22950 as a candidate gene; this gene encodes a protein belonging to the auto-regulated Ca2+-ATPase family, ACA7. Microarray data indicate that ACA7 is expressed exclusively in developing pollen grains, with the highest level of mRNA at the time of the second pollen mitosis. Our RT-PCR experiments showed that ACA7 mRNA is detected exclusively in developing flowers. Confocal microscopy experiments showed a plasma membrane localization for the recombinant GFP:ACA7 protein. We identified two different insertional mutant lines, aca7-1 and aca7-2; plants from both mutant lines displayed a normal vegetative development but showed large amounts of dead pollen grains in mature flowers assayed by Alexander’s staining. Histological analysis indicated that abnormalities are detected after the first pollen mitosis and we found a strong correlation between ACA7 mRNA accumulation and the severity of the phenotype. Our results indicate that ACA7 is a plasma membrane protein that has an important role during pollen development, possibly through regulation of Ca2+ homeostasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microgametogenesis is a complex process, involving a myriad of well-coordinated cell activities such as mitosis, cell wall biogenesis, cell-to-cell communication and cell migration, among others (for a review see McCormick 2004). This process has been very well examined at the cytological level in Arabidopsis and other species, where its temporal time course has been defined (Regan and Moffatt 1990; Sanders et al. 1999; Yamamoto et al. 2003). However, the molecular mechanisms directing pollen development are still poorly understood, as a relative small number of gametophytic mutations have been identified.
In Arabidopsis, most of the gametophytic mutants identified so far have been detected through large-scale phenotypic screening of mutant plants, performed through direct observation of pooled pollen grains stained with 4′,6-diamidino-2-phenylindol (DAPI) or Alexander’s stain (Alexander 1969; Johnson-Brousseau and McCormick 2004). However, these strategies are very laborious, time consuming and have a relatively low efficiency. For example, Bonhomme et al. (1998) detected eight genes that are important for pollen development after the analysis of 15,861 mutant lines; Howden et al. (1998) identified one mutant line with pollen defects from the analysis of 1,000 mutant lines and Lalanne et al. (2004) identified 20 mutant lines with reduced male transmission out of 3,359 analyzed mutant plants.
Considering its very particular development and function, it was thought that pollen should have had a different transcriptome in comparison to other tissues. This hypothesis was confirmed through the analysis of the Arabidopsis pollen transcriptome using the ATH1 Affymetrix DNA chip. Pina et al. (2005) purified a homogeneous population of Arabidopsis mature pollen grains through cell sorting and performed transcriptomic analysis to compare the expression profile with four different vegetative tissues (root, leaf, flower and stem). They found transcripts from 6,537 different genes in mature pollen and 11% were classified as pollen specific. Similarly, Honys and Twell (2004) prepared pollen grains from four different developmental stages: uninucleated microspores (UNM), bicellular pollen (BCP), tricellular pollen (TCP) and mature pollen grains (MPG). They found 13,977 different genes expressed during pollen development and 9.7% were not detected in other tissues, thus they were classified as pollen specific. Besides the differences in the results obtained by these two studies, a bias towards cell signaling, cell wall metabolism and vesicular trafficking was found. Interestingly, a proteomic analysis of pollen made by Holmes-Davis et al. (2005) and Noir et al. (2005) found a similar bias.
From these analyses, it has been suggested that large-scale transcriptomic data could be used to select candidates for functional analysis, facilitating the discovery of gametophytic genes (McCormick 2004). We used the dataset obtained by Honys and Twell (2004) to design a methodology that, based on mRNA accumulation, allowed us to select candidate genes putatively involved in pollen development, germination and tube elongation (unpublished results from our laboratory). Using these criteria, we have identified At2g22950 as a candidate gene for functional analysis in pollen. At2g22950 encodes for ACA7, a protein belonging to the ACA family (autoinhibited Ca2+-ATPase) of calcium pumps. ACA proteins are part of the PIIB type of Ca2+-ATPases, with ten members in Arabidopsis (Kabała and Kłobus 2005). They all present a calmodulin-regulated autoinhibitory domain and show nearly 50% protein sequence identity with animal plasma membrane type (PM-type) Ca2+ pumps (Kabała and Kłobus, 2005; Boursiac and Harper 2007). Some members of this family of Ca2+ pumps have been characterized and their subcellular localization and physiological function are known (Kabala and Klobus 2005). For example, ACA8 is expressed in several Arabidopsis tissues, the protein is localized in the plasma membrane (Bonza et al. 2000) and is involved in stomatal opening and closing (Schiøtt and Palmgreen 2005). Another member of this family, ACA9, is also localized in the plasma membrane and aca9 plants have reduced fertility as pollen tubes fail to grow across the stigma (Schiøtt et al. 2004).
In this work, we performed a characterization of two allelic mutant lines in ACA7 and analyzed its subcellular distribution. Our results indicate that ACA7 encodes for a plasma membrane protein required for proper pollen development, probably through regulation of Ca2+ homeostasis.
Materials and methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col) seeds were germinated and grown hydroponically at 23ºC, under a 16-h light/8-h dark cycle (Gibeaut et al. 1997). To grow seedlings in vitro, seeds were surface sterilized for 6 min with a solution of 50% (v/v) commercial bleach. Then, seeds were rinsed thoroughly with sterile distilled water and sown on one half-concentrated Murashige and Skoog medium solidified with 0.8% (w/v) agar and supplemented with kanamycin (50 mg/L).
Selection of candidate genes for functional analyses
Microarray data from vegetative tissues (Schmid et al. 2005) and from pollen development (Honys and Twell 2004) were used to select candidate genes for functional analyses. Briefly, we used the experiment of Schmid et al. (2005) to select genes expressed only in anthers and pollen to obtain the first group of candidate genes. Then, these genes were subjected to a second round of analysis using the experiment of Honys and Twell (2004) to refine our selection. All genes that showed a signal value higher that the media for the whole experiment in uninucleated microspores (UNM) were eliminated from the analysis. Finally, we selected genes showing a peak in the signal value at tricellular pollen (TCP) or mature pollen grains (MPG). Genes accomplishing these criteria were selected and insertional mutant lines were searched in silico.
Isolation of insertional aca7 mutants
T-DNA insertion mutants for ACA7, aca7-1 (SALK_014124) and aca7-2 (SALK_143504), were identified through searching the SIGnAL T-DNA express site (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al. 2003) and seeds were obtained from the ABRC (Arabidopsis Biological Resource Center). To verify the insertion site, genomic DNA was PCR-amplified using the T-DNA left border specific primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) in combination with a gene specific primer: for aca7-1, ACA7-5F (5′-GCGATTCTTTTGCTCTTCTTTC-3′) and for aca7-2, ACA7-4R (5′-GGGTATTTTCTTCAAGCCAGCA-3′). Homozygous T-DNA insertion mutants were identified by PCR genotyping, using the primers described before to amplify the mutated alleles in combination with primers flanking the insertion site to amplify the wild type allele: for aca7-1, ACA7-5F and ACA7-5R (5′-CACCTTGTAGTCACTTGGAGA-3′) and for aca7-2, ACA7-4F (5′-TGGTCTTGTTGGTTCTGACTCT-3′) and ACA7-4R.
RT-PCR
Total RNA was isolated from different tissues using TRIzol reagent (Invitrogen) and cDNA was obtained using the SuperScript III First-Strand Synthesis System (Invitrogen). ACA7 cDNAwas amplified using primers ACA7-UTR3 (5′-CCTTAGCCTATTTAACTCAA-3′) and ACA7-3R (5′-TGGTCTTGTTGGTTCTGACTCT-3′). At5g46630, encoding a clathrin adaptor complex subunit, was used as control (Czechowski et al. 2005) using the primers Cla1F (5′-GAAACATGGTGGATCCAT-3′) and Cla1R (5′-CTCAACAACAAATTTGAATC-3′).
Generation of GFP:ACA7 construction and analysis of subcellular localization
The full length ACA7 open reading frame (3,042 bp) was amplified using primers aca7-9F (5′-CACCATGGAGAGTTACCTCAACTC-3′) and aca7-4R (5′-GGGTATTTTCTTCAAGCCAGCA-3′), cloned into pENTR/D/TOPO (Invitrogen) and sequenced. The resulting plasmid (pENTR:ACA7) was recombined to pK7WGF2 (Karimi et al. 2002) using the LR Clonase enzyme mix (Invitrogen) to generate a construction expressing the GFP:ACA7 fusion protein under the control of the 35S promoter. Competent Agrobacterium tumefaciens cells (GV3101) were transformed and positive clones were selected. We used a positive Agrobacterium clone to transiently transform epidermal cells of tobacco plants (Petite Havana SR1). Tobacco leaves were analyzed by LSCM using an Olympus FluoView1000 spectral confocal microscope. For co-localization analysis, a membrane marker (PIP2A:RFP) was co-transformed and cells expressing both fluorescent proteins were analyzed.
Microscopic analyses
Alexander staining of pollen was performed as described before (León et al. 2007). For histological analysis of pollen development, flowers at different developmental stages were fixed overnight in 3% (v/v) glutaraldehyde, 0.1 M sodium cacodylate (pH 7.2), dehydrated in acetone series to 100% and embedded in Embed 812 resin according to the recommendations of the manufacturer (EMS). Anther transverse sections (2 μm) were stained with 1% toluidine blue. Alexander stained pollen and anther cross sections were viewed using an Olympus IX81 microscope and bright-field photographs were taken using a MicroPublisher 3.3 RTV digital camera.
Results
Selection of At2g22950 as candidate gene
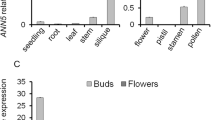
Using available microarray data we designed an approach that allowed us to select candidate genes for functional analysis, based on their expression pattern throughout pollen development. After this, we searched for mutant lines in all candidate genes selected for further analysis of pollen development, germination and tube elongation. At2g22950 was selected as candidate gene, since mRNAs are detected only in developing flowers, anthers and pollen according to microarray data and RT-PCR analysis (Fig. 1a, b). On the other hand, according to Honys and Twell (2004), ACA7 expression is restricted to bicellular pollen (BCP) and tricellular pollen (TCP) stages, showing no or low expression in uninucleated microspores (UNM) and in mature pollen grains (MPG), respectively (Fig. 1c).
ACA7 expression profile in different tissues and developmental stages. a Expression profile in Arabidopsis during development and in different tissues. Digital northern analysis was performed using the AtGenExpress server at http://jsp.weigelworld.org/expviz/expviz.jsp using the microarray data generated by Schmid et al. (2005). R roots, S seedlings, L leaf, W whole plants, A apex, F flowers, FO floral organs, Sd developing seeds. b RT-PCR detection of ACA7 mRNA in different tissues. RNA samples from roots (R), steams (S), leaves (L) and closed (CF) or open flowers (OF) were obtained using TRIzol and PCR of cDNA samples was preformed as described in “Materials and methods”. Amplification of At5g46630, encoding a clathrin adaptor complex subunit, was used as control (Czechowski et al. 2005). c Expression profile throughout pollen development. Digital northern analysis was performed using the microarray data generated by Honys and Twell (2004). Each point represents the media of two technical replicates, except for MPG. UNM uninucleated microspores, BCP bicellular pollen, TCP tricellular pollen, MPG mature pollen grains
At2g22950 encodes for a putative auto-regulated Ca2+-ATPase located in the plasma membrane
At2g22950 is annotated as ACA7, a member of the Arabidopsis family of calmodulin auto-regulated Ca2+-ATPases. Sequence analysis indicated that ACA7 shares a 93% of protein identity with ACA2 (At4g37640), its closest paralog. Expression analysis using microarray indicated that both genes show different expression patterns, being ACA2 widely expressed.
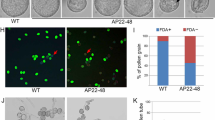
We performed protein sequence analysis using the InterPro tool at http://www.ebi.ac.uk/interpro/index.html; this analysis indicated that ACA7 contains conserved domains among ACA family members, including a N-terminal calmodulin binding domain, a cation transporter domain and E1-E2 ATPase domain, suggesting that it codes for a functional Ca2+ ATPase. in silico analysis of subcellular localization using the SUBA tool (http://suba.plantenergy.uwa.edu.au/) indicated that ACA7 is probably a plasma membrane protein, based on the presence of 10–12 putative transmembrane domains. To probe this, the full-length ACA7 cDNA (3,042 bp) was translationally fused to GFP; the construction was introduced in Agrobacterium and infiltrated in tobacco leaves (Petite Havana SR1). Fluorescence was detected in abaxial epithelial cells 48 h after by confocal microscopy (Fig. 2a). To establish the subcellular localization of ACA7, we performed co-infiltration experiments using a construction expressing PIP2A:RFP, a plasma membrane protein (Nelson et al. 2007), as membrane marker (Fig. 2b). Confocal microscopy analysis of co-infiltrated cells demonstrated a good correlation between green and red signals, suggesting that ACA7 is a membrane protein (Fig. 2c). Taking together, these analyses suggest that ACA7 encodes a pollen-expressed plasma membrane Ca2+-ATPase.
Subcellular localization of GFP:ACA7. Epidermal tobacco leaves (Petite Havana SR1) were used to determine ACA7 subcellular localization. a Localization of the GFP:ACA7 fusion protein. b Localization of the plasma membrane marker, PIP2A:RFP. c Merge of a and b. d Bright field image of the analyzed epidermal cell. Bar 40 μm
aca7 plants produce dead pollen grains
To investigate the physiological role of ACA7, two insertional mutant lines were obtained (aca7-1 and aca7-2, Fig. 3a). The exact insertion point was experimentally determined; for aca7-1 the insertion was mapped to the first intron, 280 bp downstream the start codon. For aca7-2, the insertion was mapped to the last exon, 105 bp upstream the stop codon. Homozygous mutant plants were identified by PCR and mature pollen grains were analyzed using Alexander’s stain. This technique is used to discriminate between live pollen grains, which are rounded and have a purple cytoplasm and a green exine layer, from dead pollen grains, that are completely green and flattened. Anthers from both mutant lines showed high amounts of dead pollen grains (Fig. 3c, d) compared to anthers from wild type plants (Fig. 3b).
Identification of aca7 insertional mutants and pollen phenotype. a Identification of insertional mutant lines in ACA7. Two independent insertional mutant lines, aca7-1 (SALK_014124) and aca7-2 (SALK_143504) were identified as described in “Materials and methods” and homozygous mutant plants were identified. Arrows indicate the position of the primers used for PCR genotyping. Black boxes represent exons and the lines between them, introns. Mature anthers from wild type (b), aca7-1/- (c) and aca7-2/- (d) were mounted in a drop of Alexander’s stain and viewed using a bright field microscope. Live pollen grains are round and purple, whereas dead pollen grains are green and flattened. Bar 40 μm
Seeds from aca7-1/- plants were sown and we repeated the phenotypic analysis. Unexpectedly, we detected two populations of plants: one showed high amounts of dead pollen grains (ranging between 50 and 65%) and the other displayed a lower amount of dead pollen grains (ranging between 15 and 25%, Fig. 4a). Flower RNA was prepared from both populations and ACA7 mRNA was detected by RT-PCR. Interestingly, in plants displaying a strong phenotype (SP) a very weak signal was detected, whereas in plants displaying a mild phenotype (MP) a clear product was detected, similar to wild type plants (Fig. 4b). This strong correlation between the severity of the phenotype and the abundance of ACA7 mRNA suggests that pollen phenotype is linked to ACA7.
Correlation between ACA7 expression and pollen phenotype. a Quantification of dead pollen grains in aca7-1/- plants showing a strong phenotype (SP) or mild phenotype (MP) compared to wild type plants (WT). b Flower RNA was prepared from plants showing a strong phenotype (SP), mild phenotype (MP) or wild type plants (WT) and RT-PCR analysis of ACA7 expression was performed. Amplification of At5g46630 cDNA, encoding a clathrin adaptor complex subunit, was used as control (Czechowski et al. 2005). L, DNA ladder
The first pollen mitosis is altered in mutant plants
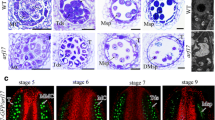
To determine the stage of pollen development in which defects are first noticeable, we performed histological analysis. Transverse sections (2 μm) of flowers from mutant plants (aca7-1/-, from the group displaying a strong phenotype) in different developmental stages were obtained and stained with blue toluidine. At tetrad stage (Fig. 5a), a homogeneous population was present inside the anthers of mutant plants, indicating that no cytological alterations have occurred yet. The same was observed at uninucleated microspores stage; developing pollen grains showed a discernible exine layer, a large central vacuole and a peripheral nucleus (Fig. 5b). At the time of the first mitosis, two pollen populations were present in the anthers of mutant plants. One of the populations went through the first mitosis to produce bicellular pollen grains, composed by a large vegetative cell and a small, peripheral generative cell. This population showed a normal morphology. However, the other pollen population displayed a distorted morphology: the cytoplasm was detached from the cell wall and the large vacuole was still present, whereas the generative cell was not (Fig. 5c). These abnormalities became more noticeable in advanced stages of pollen development (Fig. 5d, e), leading to abnormal pollen grains deprived of cytoplasm and structurally collapsed at the time of pollen release (Fig. 5f). Notably, a normal development and degeneration of the tapetum layer was seen throughout pollen development.
Histological analysis of aca7-1/- anthers. Anther transverse sections representing different developmental stages of pollen were analyzed: a tetrads, b uninucleated microspores, c bicellular pollen grains, d tricellular pollen grains, e and f mature pollen grains in closed and open flowers, respectively. Bar 40 μm
Discussion
Use of transcriptomic data to select candidate genes for functional analysis
Transcriptomic profiling has been used to select candidate genes for functional analysis in plants (Costaglioli et al. 2005; Suh et al. 2005; Soler et al. 2007; Breeze et al. 2011). The underlying hypothesis is that genes involved in a particular process, as developmental transitions or organ biogenesis, must be expressed in order to accomplish its biological role. Certainly, transcription could not be used as a single parameter to predict the relevance of a gene in a particular process. Mechanisms such as mRNA stability, translation regulation and post-translational modifications could strongly modify the final biological activity of a gene, regardless of its transcriptional status. However, transcription plays a key regulatory control and thus, mRNA abundance could be a good starting point to identify candidate genes involved in developmental transitions.
Previously, McCormick (2004) suggests that transcriptomic data could be useful to select candidate genes to perform functional analysis during pollen development. We started with a very simple hypothesis: in the group of genes that are only expressed during pollen development and that showed a peak in their mRNA abundance at tricellular stage or mature pollen grains, there is an over representation of genes that are important for pollen development. We decided to use these criteria to narrow down the population of candidate genes and maximize the effectiveness of our screening. For instance, genes that showed high levels of expression at UNM could reflect the activity of the promoter in the pre-meiotic stage (Honys and Twell 2004), and thus they were not considered for our analysis. To date, roughly 10% of the analyzed mutant lines (representing 154 non redundant genes) show defects in pollen development, germination and/or tube elongation (unpublished results from our laboratory).
At2g22950 encodes for ACA7, a putative Ca2+ pump located in the plasma membrane
Sequence analysis indicated that At2g22950 encodes for ACA7, a member of the PIIB type of auto-regulated Ca2+ ATPases family in Arabidopsis. This family encompasses ten members and only few have been characterized at the functional level (Kabała and Kłobus 2005; Boursiac and Harper 2007). Protein sequence analysis showed that ACA7 presented the characteristic domains conserved across the ACA family, including the N-terminal calmodulin sensor domain that regulates the activity of this family of Ca2+ pumps (Boursiac and Harper 2007). Despite the presence of this molecular signatures, suggesting that ACA7 encodes for a calcium pump, a functional confirmation of the activity in vivo using a yeast model would be necessary in order to confirm its molecular role.
On the other hand, our subcellular localization experiment indicated that ACA7 is a plasma membrane protein. Other two members of the ACA family, ACA8 and ACA9, have been experimentally confirmed as plasma membrane proteins (Bonza et al. 2000; Schiøtt et al. 2004) and a third member, ACA10, is possibly also a membrane protein; nevertheless, the formal experimental confirmation is pending (George et al. 2008). On the other hand, ACA2, the closest paralog of ACA7 in Arabidopsis, has been shown to be an endomembrane system protein (Harper et al. 1998). But it is important to consider that even when sequence similarity could suggest a similar protein distribution, is not always the case. For example, Dixon et al. (2009) studied the subcellular distribution of three glutathione transferases of the theta class (GSTTs) from Arabidopsis. The three genes arose probably by gene duplication (Cannon et al. 2004) and shown extensive protein sequence identity and similar enzymatic activity (Cannon et al. 2004; Dixon et al. 2009). However, whereas GSTT1 and 2 are localized in peroxisomes, GSTT3 was directed to the nucleus (Dixon et al. 2009). The significance of the subcellular localization of ACA7 may be related to calcium homeostasis. Ca2+ ions are important second messengers, hence the cytoplasmic concentration is kept tightly controlled at low levels. When calcium is released from intracellular compartments, as endoplasmic reticulum, cytoplasmic calcium levels must be restored by the action of Ca2+ pumps. We hypothesized that ACA7 participates in maintaining calcium homeostasis during pollen development and thus, it regulates the action of calcium binding proteins and other signaling pathways that use Ca2+ as second messenger.
ACA7 is required for pollen development
ACA8 and ACA9 are plasma membrane pumps that transport Ca2+ ions out to the apoplast, and thus are important to maintain Ca2+ homeostasis in plant cells. ACA8 is expressed mainly in vegetative tissues, particularly during the transition between seedling and juvenile phase, and their transcripts are absent in pollen (Bonza et al. 2000; George et al. 2008). aca8 mutant plants have no apparent phenotypic defects when compared to wild type plants. However, ACA8 overexpression from a 35S promoter rescued the phenotypic defects of aca10 mutant plants, which showed some abnormalities during the transition from juvenile to adult growth stage. This suggests that ACA8 and ACA10 are partially functional redundant, which is a good argument to suggest that ACA10 is also a plasma membrane protein. On the other hand, ACA9 is a plasma membrane protein involved in pollen tube elongation (Schiøtt et al. 2004). aca9 plants show defects in pollen tube elongation and have a reduced fertility as pollen tubes fail to grow across the stigma. Microarray data from different stages of pollen development, indicated that ACA9 is expressed preferentially after the tricellular stage, and their transcripts were highly abundant in mature pollen grains. It is known that mature pollen grains store many mRNAs (Mascarenhas et al. 1984; McCormick 2004) and has been demonstrated that pollen germination is almost independent of transcription but not translation, suggesting that pre-synthesized mRNAs are important for pollen germination (Capková et al. 1988; Honys and Twell 2004). Thus, many proteins important for pollen germination and tube elongation are translated from pre-existing mRNAs, stored during the last stages of pollen development. This seems to be the scenario for ACA9, based on its transcript accumulation profile and the physiological evidence. On the other hand, ACA7 showed a different transcription profile: it is expressed early during development and it is almost absent in mature pollen grains (Fig. 1). This transcriptional profile is in agreement with our hypothesis that ACA7 is important for pollen development. Our histological data indicated that pollen grains from aca7 plants showed the earliest noticeable defects at the time of the first pollen mitosis, coincident with the detection of ACA7 mRNA in microarray experiments (Figs. 1, 5). Furthermore, the existence of two populations of plants showing a mild and strong phenotype, allowed us to determine the existence of a correlation between the abundance of ACA7 mRNA and the phenotype severity (Fig. 5). It is known that T-DNAs inserted into intronic regions could be removed through splicing of the intron that contains the T-DNA (Funck et al. 2010; Chehab et al. 2011). Moreover, splicing efficiency of the intron could be modified in some individuals (Chehab et al. 2011). Considering these evidences, we suggest that differential splicing efficiency of the intron harboring the T-DNA insertion, could lead to the generation of two populations of plants, displaying quantitatively different phenotypes directly related to ACA7 mRNA abundance.
Ca2+ and pollen development
Ca2+ plays an important role as a second messenger and its concentration is modulated by channels that increase the cytoplasmic levels of Ca2+, and efflux pumps that restore Ca2+ concentration to low levels. Ca2+ is considered as a very important player for pollen germination and tube elongation (Brewbaker and Kwack 1963; Malho et al. 1995; Pierson et al. 1996; Taylor and Hepler 1997) but its role during pollen development has received less attention. Chen et al. (2008) studied Ca2+ dynamics during pollen development in Torenia fournieri using the method of potassium antimonate precipitation. They found Ca2+ precipitates in the nucleus and cytoplasm of early uninucleated microspores at initial stages of pollen development. However, intracellular detection of Ca2+ was progressively reduced throughout development, concomitantly with an increase in Ca2+ detection in the cell wall (Chen et al. 2008). These data suggest that Ca2+ is transported outside developing pollen grains after UNM stage in Torenia. Is appealing to propose that in Arabidopsis exists a similar Ca2+ dynamics during pollen development, and ACA7 may play a central role transporting Ca2+ from the cytoplasm to the cell wall. Unregulated Ca2+ levels in the cytoplasm could interfere with protein function and/or signal transduction pathways that use Ca2+ as second messenger, causing the phenotypic alterations described during pollen development in aca7 plants.
Taking together, our results suggest that ACA7 is a plasma membrane protein involved in transporting Ca2+ outside developing pollen grains, and this activity is important to support normal pollen development, particularly the progression to uninucleated microspores to bicellular pollen grains.
References
Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44:117–122
Alonso JM, Stepanova AN, Leisse TJ et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Bonhomme S, Horlow C, Vezon D, de Laissardière S, Guyon A, Férault M, Marchand M, Bechtold N, Pelletier G (1998) T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol Gen Genet 260:444–452
Bonza MC, Morandini P, Luoni L, Geisler M, Palmgren MG, De Michelis MI (2000) At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol 123:1495–1506
Boursiac Y, Harper JF (2007) The origin and function of calmodulin regulated Ca2+ pumps in plants. J Bioenerg Biomembr 39:409–414
Breeze E, Harrison E, McHattie S et al (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23:873–894
Brewbaker J, Kwack B (1963) The essential role of calcium ion in pollen germination and pollen tube growth. Am J Bot 50:859–865
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Čapková V, Hrabětová E, Tupý J (1988) Protein synthesis in pollen tubes: preferential formation of new species independent of transcription. Sex Plant Reprod 1:150–155
Chehab EW, Kim S, Savchenko T, Kliebenstein D, Dehesh K, Braam J (2011) Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol 156:770–778
Chen SH, Liao JP, Lup MZ, Kirchoff B (2008) Calcium distribution and function during anther development of Toreneiafournieri (Linderniaceae). Ann Bot Fennici 45:195–203
Costaglioli P, Joubès J, Garcia C et al (2005) Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. BBA Mol Cell Biol L 1734:247–258
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dixon DP, Hawkins T, Hussey PJ, Edwards R (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60:1207–1218
Funck D, Eckard S, Müller G (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10:70
George L, Romanowsky SM, Harper JF, Sharrock RA (2008) The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol 146:716–728
Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115:317–319
Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H (1998) A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem 273:1099–1106
Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5:4864–4884
Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5:R85
Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, Twell D (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149:621–631
Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39:761–775
Kabała K, Kłobus G (2005) Plant Ca2+-ATPases. Acta Physiol Plant 27:559–574
Karimi M, Inzé D, Depicker A (2002) Gateway(™) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Lalanne E, Michaelidis C, Moore JM et al (2004) Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167:1975–1986
León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143:1534–1546
Malho R, Read ND, Trewavas AJ, Pais MS (1995) Calcium channel activity during pollen tube growth and reorientation. Plant Cell 7:1173–1184
Mascarenhas NT, Bashe D, Eisenberg A, Willing RP, Xiao CM, Mascarenhas JP (1984) Messenger RNAs in corn pollen and protein synthesis during germination and pollen tube growth. Theor Appl Genet 68:323–326
McCormick S (2004) Control of male gametophyte development. Plant Cell 16:S142–S153
Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J51:1126–1136
Noir S, Bräutigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337:1257–1266
Pierson ES, Miller DD, Callaham DA, van Aken J, Hackett G, Hepler PK (1996) Tip-localized calcium entry fluctuates during pollen tube growth. Dev Biol 174:160–173
Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control and gene expression regulation. Plant Physiol 138:744–756
Regan SM, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2:877–889
Sanders PM, Bui AQ, Weterings K et al (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11:297–322
Schiøtt M, Palmgreen M (2005) Two plant Ca2+ pumps expressed in stomatal guard cells show opposite expression patterns during cold stress. Physiol Plantarum 124:278–283
Schiøtt M, Romanowsky SM, Bækgaard L, Jakobsen MK, Palmgren MG, Harper JF (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA 101:9502–9507
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37:501–506
Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M (2007) A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiol 144:419–431
Suh MC, Samuels AL, Jetter R et al (2005) Cuticular lipid composition, surface structure and gene expression in Arabidopsis stem epidermis. Plant Physiol 139:1649–1665
Taylor LP, Hepler PK (1997) Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol 48:461–491
Yamamoto Y, Nishimura M, Hara-Nishimura I, Noguchi T (2003) Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol 44:1192–1201
Acknowledgments
The authors are greatly indebted to Ariel Orellana and Reinaldo Campos for their constant encouragement and support. This work was supported by FONDECYT grant number 11080037 from the Chilean Government and Universidad Andres Bello VRID UNAB-DI-23-10/R to GL. NL is supported by a CONICYT fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Toriyama.
Rights and permissions
About this article
Cite this article
Lucca, N., León, G. Arabidopsis ACA7, encoding a putative auto-regulated Ca2+-ATPase, is required for normal pollen development. Plant Cell Rep 31, 651–659 (2012). https://doi.org/10.1007/s00299-011-1182-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1182-z