Abstract
Changes in endogenous phytohormone levels, DNA methylation patterns, and expression levels of related genes during induction of dormancy in two strawberry cultivars, Darselect and All Star, were studied under controlled environmental conditions. At 12°C, regardless of day length, potted, runner-derived plants of both cultivars gradually exhibited morphological traits typical of dormancy after treatment for 8 weeks. These morphological changes were accompanied by a synchronous significant decline in indole-3-acetic acid (IAA) level and increases in abscisic acid (ABA) content and global genomic DNA methylation in young leaves. Exposed at 15°C and a short-day photoperiod, the changes in morphology, phytohormone levels and DNA methylation of both cultivars were similar to those observed at 12°C. Slight but non-significant changes in IAA and ABA levels and genomic DNA methylation occurred in young leaves at both 15°C with long days and 18°C with short days. These results indicated that temperature alone was sufficient to induce strawberry to enter the typical dormant phase, and day length had no impact at 12°C. The higher temperature permissible for dormancy induction in strawberry was 15°C, but at this temperature dormancy induction was modified by day length. The expression patterns of FaPIN1, FaNCED1, FaDRM and FaROS1 were coincident with the changes in phytohormone levels and DNA methylation. Although the two tested cultivars have different temporal responses with the different degree of cold tolerance and depth of dormancy, both the endogenous phytohormone and DNA methylation were changed when induced by external environmental factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dormancy is a flexible adaptive strategy that is essential for adaptability and survival of woody and other perennial plants in stressful environments (Olsen 2010). Many biennial and perennial higher plant species undergo bud dormancy, which renders a temporal inability to initiate growth of the meristem or cells, to assist plants to survive temporarily unfavorable growing conditions (Rohde and Bhalerao 2007). Many factors are involved in dormancy regulation, which include environmental factors, such as photoperiod and temperature (Olsen 2010). Garner and Allard (1923) first demonstrated that a short photoperiod in autumn is the dormancy-inducing signal for woody plants. The importance of a short photoperiod for dormancy induction has been documented in a wide variety of tree and shrub species (Håbjørg 1978; Böhlenius et al. 2006). In addition to circadian rhythms, temperature is an important determinant of several aspects of dormancy (Olsen 2010). Some woody species of Rosaceae, such as apple (Malus domestica) and pear (Pyrus communis), respond to temperature, but not to day length, for growth cessation and dormancy induction (Nitsch 1957). In Prunus species, several experiments have demonstrated a distinct correlation between photoperiod and temperature in growth regulation and cessation (Heide 2008).
Some plant hormones, such as abscisic acid (ABA) and auxin, are involved in dormancy regulation (Swain and Singh 2005; Junttila 2007). Auxin and ABA are well known to control cell division, which might be relevant to dormancy-related processes (Horvath et al. 2008). Higher auxin levels have been detected in growing Phaseolus buds than those in dormant buds (Gocal et al. 1991), and in growing pea buds auxin-responsive genes are expressed preferentially (Stafstrom 1993). High levels of ABA were detected in Phaseolus, Elytrigia and tuberose (Polianthes tuberosa) during their dormant periods (Gocal et al. 1991; Nagar 1995; Pearce et al. 1995). Involvement of ABA in bud dormancy of Populus is also reported (Rohde et al. 2002). However, the role of ABA and auxin in dormancy induction is unclear (Junttila 2007). In addition to phytohormone, specific genes may activate plant dormancy. Accompanying the changes in phytohormone levels, the expression of genes associated with phytohormone metabolism should also differ during the induction of dormancy.
Up-regulation or down-regulation during a sequence of gene expression events occur concurrently with initialization of dormancy in response to either hormonal or environmental signals (Lang et al. 1987). Gene regulation is controlled, in part, by epigenetic mechanisms that include DNA methylation and post-translational histone modifications. Hypermethylation of CpG islands is thought to silence the expression of certain genes. Two methyltransferases are involved in establishment of the DNA methylation pattern. The de novo methylation of DNA is catalyzed by the domain-rearranged methyltransferase (DRM) family, which catalyzes cytosine methylation (Chan et al. 2005). In addition, the Repressor of Silencing 1 (ROS1) protein actively removes the 5-methylcytosine (5-mC) methyl group by a base-excision repair mechanism (Gong et al. 2002). Burn et al. (1993) reported that a reduced DNA 5-mC level occurred in both cold-treated and 5-azacytidine-treated Arabidopsis plants compared with non-vernalized plants. Additionally, DNA demethylation can occur prior to gene transcriptional activation involved in cell division and meristem growth in potato tubers exiting dormancy (Law and Suttle 2002). In dormancy induction, DNA methylation may be involved in the regulation of gene expression to synchronize plant developmental status with certain environment changes (Santamaría et al. 2009).
Strawberry is a perennial herbaceous species cultivated worldwide, and depends on timely perception of environmental signals to enter dormancy in synchrony with seasonal changes to survive cold temperatures. Robert et al. (1999) reported that the combination of photoperiod and temperature could be critical to control the initiation and termination of dormancy in strawberry plants based on vegetative growth parameters. Strawberry plants passed through a semi-dormancy phase with a decreased vegetative growth rate, which resulted in development of small leaves and short petioles (Sønsteby and Heide 2006). Both photoperiod and temperature are crucial factors in strawberry dormancy induction, but which factor plays the more important role is still unclear. In addition, the mechanisms underlying such temperature and photoperiod effects are poorly understood (Olsen 2010).
In this study, we examined the performance of two different strawberry cultivars, All Star and Darselect. All Star is a deep dormancy cultivar, which shows strong tolerance to extreme temperatures and is well suited for open field cultivation (Yang et al. 2010). Darselect is an intermediate dormancy cultivar, which shows adaptability, early flowering and good disease resistance (Yin 1999). The objective of this work was to clarify the timing and effects of phytohormone and DNA methylation on the progress of dormancy induction, simultaneously to evaluate whether photoperiod or temperature has a greater impact on strawberry dormancy induction. The levels of the phytohormone IAA and ABA, and degree of genomic DNA methylation, within young leaves were measured, and simultaneously the expression levels of several phytohormone and DNA methylation metabolism related genes were analyzed under different temperature and photoperiod conditions.
Materials and methods
Plant materials and data analysis
Runner-derived plantlets of strawberry (Fragaria × ananassa Duch.) cv. Darselect and cv. All Star were collected from a field nursery and transplanted into 10-cm diameter plastic pots on August 9, 2009. The growing medium consisted of composted garden soil, peat and vermiculite (3:1:1). The plants were grown in a glasshouse under a minimum temperature of 21°C. The plants were grown under natural light conditions. On September 1st, the potted plants were transferred to an illuminated incubator and exposed to a combination of different temperature conditions (12, 15, or 18°C, with a fluctuation range of 1°C) and a photoperiod of either 8 h (short day; SD) or 16 h (long day; LD). All treatments were applied in the illuminated incubators for 10 weeks. Relative humidity was maintained at 75% for all treatments and illumination was 300 μmol m−2 s−1 during the light period. The plants were watered with tap water as required.
The experiments were randomized, and each treatment contained three replicates with three plants in each replication. Plant growth was monitored weekly by measuring leaf area. Five-hundred milligrams of new, unfolded leaves were harvested for DNA methylation and phytohormone assays at 2-week intervals. Samples were immediately frozen in liquid nitrogen and stored at −80°C until use. Data of DNA methylation and phytohormones levels were subjected to analysis of variance (ANOVA) following standard procedures using the SPSS package. Student’s t test was used to analyse the expression of genes.
Determination of phytohormone levels
Levels of IAA and ABA were measured as previously described (Zhang et al. 2008). Briefly, 30 μl of purified leaf extract was injected with an autosampler (Waters 2707, Milford, MA, USA) into a high-performance liquid chromatograph (HPLC). Analytic conditions were as follows: a reverse phase column (Inertsil ODS-3 250 × 4.6 mm, 5 μm) was utilized at 30°C; the mobile phase was methanol with 0.6% acetic acid (dissolved in redistilled water) (45:55, v/v); the flow rate was 0.8 ml min−1. Phytohormone levels were determined by recording absorbance at 254 nm with a dual λ absorbance detector (Waters 2487, Milford, MA, USA). Data were acquired with an Empower chromatography workstation (Waters, Milford, MA, USA). Endogenous IAA and ABA were identified by their retention time compared with IAA and ABA external standards (Sigma, St. Louis, MO, USA).
Genomic DNA methylation determination
Genomic DNA was isolated using a modified CTAB method (Doyle and Doyle 1990). The concentration and purity of DNA were determined spectrophotometrically. The global DNA methylation rate was measured by reversed-phase HPLC as described by Demeulemeester et al. (1999). Approximately 40 μg DNA was hydrolyzed in 50 ml of 70% perchloric acid (100°C for 1 h) followed by adjustment of the pH to 3–5 with 1 M KOH. After centrifugation, the supernatant was filtered through a membrane (pore size 0.22 um). Filtrate samples (10 μl) were injected into the HPLC and separated with a BDS Hypersil C18 column (250 × 4.6 mm; Thermo, USA) and elution buffer (5 mM sodium heptanesulfonate, 10% methanol, pH 4.0) at a flow rate of 0.5 ml min−1 at 40°C. The absorbance of the bases was measured at 280 nm with an ultraviolet detector (Shimadzu, Tokyo, Japan). Retention times and A 280 values were calibrated with cytosine (C) and 5-methylcytosine (5-mC) standards (Sigma, St. Louis, MO, USA). The global DNA methylation rate was calculated as [concentration of 5-mC × 100]/([concentration of 5-mC] + [concentration of C]).
RNA extraction and gene isolation
Total RNA was extracted from leaves collected from the different treatments using the CTAB method (Zhang et al. 2005). DNA impurities in the isolated RNA were digested with DNaseI (TaKaRa, Beijing, China) by incubation at 37°C for 30 min followed by incubation for 10 min at 65°C to inactive DNaseI. The first-strand cDNAs were synthesized using M-MLV reverse transcriptase (Promega, Madison, WI, USA) using 1 μg total RNA as template and an oligo dT(18) as primer. These cDNAs were used as templates for amplification of ROS1 and PIN1 genes with degenerate primers (forward 5′-AGAGAAAR RACAVSAARYACAATGGA-3′ and reverse 5′-ACACAWACRWACCCTYTCCARAAGCA-3′; and forward 5′-ATGTTCAGYCTYGGKYTRTTCATGGC-3′ and reverse 5′-ATGTAGTASASMAGMGTD ATGGGCA-3′). The PCR conditions were as follows: 5 min at 94°C for initial denaturation; 32 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min 40 s; and finial extension at 72°C for 6 min. The PCR products were ligated into the Peasy-T1 vector (TransGen, Beijing, China) and sequenced. Sequences were determined by a homology search of the NCBI databases using the BLAST program. The sequences were aligned and a phylogenetic tree was constructed using Mega 4.1.
Quantitative real-time PCR analysis
Total RNA (1 μg) was used to synthesize first-strand cDNAs with M-MLV reverse transcriptase according to the manufacturer’s protocol. The cDNA was diluted fivefold, and 2 μl of the dilution was used for quantitative RT-PCR (QRT-PCR). The iQ SYBR Green Supermix (Applied Biosystems, Foster City, CA, USA) was used for amplification on an Applied Biosystems 7500 RT-PCR system. The expression levels of each sample were normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression levels. The primers for strawberry FaGAPDH and FaDRM genes were designed for real-time PCR as previously described (Chang et al. 2009; Salvatierra et al. 2010). The primers for FaROS1and FaPIN1 were designed for real-time PCR from partial sequences isolated in this work. The primers for strawberry FaNCED1 (HQ290318) genes were designed from partial sequences published in the GenBank databases. The primer sets used are listed in Table 1. The thermal cycling program was one initial cycle of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The relative gene expression data were analyzed using the 2−ΔΔCT method.
Results
Morphological changes
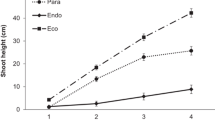
When exposed to 18°C in SD, new unfolding leaves of both All Star and Darselect showed no obvious morphological changes, and all plants showed continuous vegetable growth throughout the experimental period. In contrast, treatment at 12°C in both SD and LD induced dormancy in both cultivars; leaf area and petiole length were significantly decreased after 2 weeks and reached minimum values after treatment for 8 weeks and the plants attained the morphological traits typical of dormant strawberry plants, i.e. a more compact and restrained growth habit with small leaves on short petioles. Temperature had a highly significant effect on both cultivars at 12°C. At 15°C in SD, morphological changes were similar to those induced at 12°C (both SD and LD); both cultivars showed a significant reduction in leaf area and petiole length after treatment for 2 weeks and minimum values were attained after treatment for 8 weeks. However, at 15°C in LD, only a slight reduction in leaf size and petiole length occurred in both cultivars (Fig. 1).
Phytohormonal changes
Treatment with 18°C in SD induced no significant changes in IAA and ABA levels during the experimental period. When treated at 15°C in LD, a slow decrease in IAA level and increase in ABA content were recorded throughout the experiment. However, with exposure to 15°C in SD the IAA level decreased significantly after treatment for 2 weeks and remained at a low level for the next 4 weeks. The same pattern of IAA content was observed under 12°C, regardless of photoperiod (LD or SD). In contrast, the levels of ABA increased significantly from the fourth week to the sixth week of treatment in both cultivars (Fig. 2). The changes in IAA levels coincided with plant morphological changes, whereas changes in ABA levels lagged 2 weeks behind the changes in IAA levels.
Contents of ABA and IAA in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. IAA and ABA levels were quantified using high-performance liquid chromatography. Each data point represents the average of three independent samples; error bars indicate the standard deviation
DNA methylation changes
With exposure to 15°C in SD, or 12°C under SD and LD, global DNA methylation in All Star increased significantly after 4 weeks of induction and peaked after induction for 6 weeks. In contrast, the DNA methylation level in Darselect began to increase significantly after 2 weeks of treatment and peaked after 4 weeks’ treatment (Fig. 3). In plants of both cultivars treated at 15°C in LD, fluctuations in global DNA methylation levels were barely detected and only increased slightly after treatment for 8 weeks. Increased DNA methylation lagged behind changes in IAA level, but was synchronized with changes in ABA level.
Percentage global genomic DNA methylation in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. Global genomic DNA methylation was quantified using high-performance liquid chromatography. Each data point represents the average of three independent plants; error bars indicate standard deviation
Characterization of FaROS1and FaPIN1 genes of strawberry
Two gene fragments were amplified from the cDNAs with the degenerate primers. One fragment comprised 1,485 bp and encoded 495 amino acids (Fig. 4). Comparison of the amino acid sequence of FaROS1 (JN004136) with other protein sequences lodged in GenBank databases indicated that FaROS1 shared 68.35% identity with Arabidopsis thaliana (AtROS1, AAP37178), 71.66% identity with Nicotiana tabacum (NtROS1, BAF52855) and 67.88% identity with Oryza sativa (OsROS1, EFC70183). Based on the amino acid alignment, a phylogenetic tree was constructed. At amino acid level FaROS1, AtROS1, NtROS1 and OsROS1 were clustered together (Fig. 5).
Alignment of amino acid sequences of AtROS1 (Arabidopsis thaliana), NtROS1 (Nicotiana tabacum) and FaROS1 (Fragaria ananassa) proteins. Black shading indicates amino acid positions identical in all three proteins, blue shading indicates residues identical in two proteins. The conserved HhH-GPD domain and distinct FES motif are underlined
Phylogenetic relationships of FaROS1 (Fragaria ananassa) and related proteins from other plants. The studied proteins comprised the following accessions: Arabidopsis thaliana (AAP37178), Nicotiana tabacum (BAF52855), Ricinus communis (XP 002528585), Oryza sativa (EFC70183), Vitis vinifera (CBI17085), Triticum aestivum (CBH32632)
The second fragment amplified comprising 299 bp was deposited in the GenBank database (accession number JN419023) and shared high identity with other PIN1 proteins lodged in Genbank isolated from Arabidopsis thaliana (AAD04376), Pisum sativum (AAO38045) and Triticum aestivum (AAS19858), reaching about 84.85, 89.90, and 91.92%. Thus, this fragment was confirmed to represent a PIN1 gene.
Quantitative real-time PCR
With exposure to 12°C under either LD or SD, expression of FaDRM in All Star began to increase significantly after 4 weeks and peaked after 6 weeks. In Darselect the FaDRM expression level began to increase significantly after 2 weeks of treatment and reached a maximum after 4 weeks. When plants were exposed to 15°C under SD, the changes in DRM expression was moderate compared with those at 12°C. No obvious changes in FaDRM expression level occurred at 15°C in LD except for a slight increase after treatment for 6 weeks (Fig. 6).
Comparative FaDRM gene expression in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. Real-time PCR was performed on total RNA isolated from new leaves. Each date represents the average of three independent plants; error bars indicate the standard deviation. Asterisks above the error bars indicate a significant difference between the treatment and control (0 week) using Student’s t test (*p < 0.05, **p < 0.01)
The expression pattern of FaROS1 was identical in plants of both cultivars exposed to 15°C in SD or 12°C in both LD and SD. During the initial 2 weeks, no notable changes in FaROS1 transcript levels were detected, but a significant decline occurred after 4 weeks of treatment, and the low expression level was maintained until 8 weeks of treatment. Subsequently, FaROS1 transcript levels returned to normal after 10 weeks of treatment. No significant differences in FaROS1 expression levels were observed in plants treated at 15°C in LD (Fig. 7).
Comparative FaROSI gene expression in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. Real-time PCR was performed on total RNA isolated from new leaves. Each date represents the average of three independent plants; error bars indicate the standard deviation. Asterisks above the error bars indicate a significant difference between the treatment and control (0 week) using Student’s t test (*p < 0.05, **p < 0.01)
When plants were exposed to 15°C in SD or 12°C in LD and SD, the expression of FaPIN1 gene was down-regulated significantly after beginning the treatment, and the expression was maintained at low level throughout the process of induction in both cultivars, while the expression of FaNCED1 gene was up-regulated significantly at 4 weeks of treatment, and maintained at relatively high level during the treatment. However, on exposing the plants at 15°C in LD, the FaPIN1 gene expression levels decreased significantly after 6 weeks of treatment in All Star and 8 weeks in Darselect (Fig. 8), while the expression of FaNCED1 gene changed insignificantly until 6 weeks of treatment in both cultivars (Fig. 9).
Comparative FaPIN1 gene expression in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. Real-time PCR was performed on total RNA isolated from new leaves. Each date represents the average of three independent plants; error bars indicate the standard deviation. Asterisks above the error bars indicate a significant difference between the treatment and control (0 week) using Student’s t test (*p < 0.01)
Comparative FaNCED1 gene expression in young leaves of the strawberry cultivars All Star and Darselect under different temperature and photoperiod regimes as indicated. Real-time PCR was performed on total RNA isolated from new leaves. Each date represents the average of three independent plants; error bars indicate the standard deviation. Asterisks above the error bars indicate a significant difference between the treatment and control (0 week) using Student’s t test (*p < 0.01)
Discussion
Our experiments with two different strawberry cultivars showed that a temperature of 18°C, even under the SD condition, did not induce dormancy. Plants grew continuously throughout the treatment period, regardless of the photoperiod, in a similar manner to Malus and Pyrus plants exposed to 21°C (Nitsch 1957; Heide and Prestrud 2005). In contrast, with exposure to temperature as low as 12°C, the temperature alone was sufficient to induce the strawberry plants into a typical dormant state, regardless of day length. Only SD induced dormancy of plants exposed to 15°C (Fig. 1). Thus, 15°C represented an intermediate temperature under which an interaction between photoperiod and temperature existed in the regulation of dormancy. These results provided circumstantial evidence for temperature ‘gating’ of phytochrome-controlled photoperiodic processes as suggested by Halliday et al. (2003). At a temperature less than 12°C, temperature was the predominant and highly efficient factor in dormancy induction of strawberry.
A previous study of leafy spurge (Euphorbia esula) indicated that during the dormancy transition, some genes involved in auxin responses showed highly differential expression and expression of genes involved in auxin metabolism increased (Horvath et al. 2008). Our study clarified that, at 12°C, the decrease in IAA levels was detected after 2 weeks and reached a minimum level after 4 weeks. The changes in IAA levels coincided with morphological changes (Fig. 2), which might indicate that auxin was involved in the control of dormancy-related processes, and the low auxin content in the leaf constrained vegetative growth and resulted in the compact growth habit. Transcript levels of FaPIN1 gene dramatically decreased and with low level of the expression throughout the course of dormancy induction (Fig. 8). The PIN gene family encodes a class of auxin efflux carriers that mediate polar auxin transport (Blakeslee et al. 2005). The PIN1 protein is a crucial component of auxin transport models (Santos et al. 2010). Down-regulation of FaPIN1 in the leaf might weaken the capability for basipetal transport of IAA, which would lead to inhibition of root growth by IAA deficiency in roots (Woodward and Bartel 2005). Furthermore, the changes in IAA levels preceded the changes in ABA levels and DNA methylation during dormancy induction. Thus, IAA responded sooner to temperature during dormancy induction in strawberry. However in potato buds, the IAA level showed the opposite trend, increasing steadily during dormancy and then declining after the termination of dormancy (Sorce et al. 2000).
The levels of ABA and DNA methylation changed significantly at 4 weeks of dormancy induction in strawberry (Fig. 2). Our results indicated that compounds that participate in the dormancy process underwent drastic changes in contents during the first 4 weeks of dormancy induction. Our results were consistent with those of previous studies in which ABA concentration was shown to increase significantly in poplar apical buds after 3–4 weeks of dormancy induction (Ruttink and Arend 2007). Expression of FaNCED1 was stimulated during the dormancy induction. In the ABA biosynthetic pathway, the 9-cis-epoxycarotenoid dioxygenase (NCED) is the key enzyme in ABA biosynthesis in higher plants (Iuchi et al. 2001). These results indicate that the expression of FaNCED1 gene plays a role in the regulation of ABA level during dormancy. Thus, there may be a close correlation occurs between dormancy and the levels of FaNCED1 transcript. However, in the present study, the ABA levels remained relatively high even after 8 weeks of dormancy induction, by which time the strawberry plants had entered a dormant state, which was at variance with the conclusion of Ruttink and Arend (2007) that the dormant phase was achieved when the ABA level was low and very few changes in gene expression occur.
In our research, when strawberries were induced to enter dormancy, global DNA methylation levels in young leaves were increased. FaDRM transcript levels dramatically increased between the second and fourth week, whereas expression of FaROS1 remained at a low level from the fourth to the eighth week (Fig. 7). The up-regulation of FaDRM coupled with maintenance of the low expression level of FaROS1 contributed to the increased global DNA methylation level. Previous studies showed that global DNA methylation increased in Castanea sativa during bud set and, in contrast, decreased during bud burst (Santamaría et al. 2009). The increased methylation of genomic DNA, especially within the promoter regions of housekeeping genes, down-regulates expression of downstream genes (Law and Suttle 2002). In the present study, FaGAPDH showed constant expression levels throughout the experimental period. Expression of the housekeeping gene GAPDH was unchanged, because of the total DNA hypermethylation during dormancy induction in strawberry.
During dormancy induction in strawberry, the changes in ABA content and DNA methylation patterns occurred 2 weeks later in All Star compared with those in Darselect, while the ABA level was higher in All Star than in Darselect (Fig. 2), The results might somehow be due to All Star as a deep dormancy cultivar with the higher tolerance to cold (Yang et al. 2010). When dormancy in strawberry plants were induced by environmental factors, the two tested cultivars have different responses in time with the different degree of cold tolerance and depth of dormancy. However, the inside responding materials, endogenous phytohormone and DNA methylation, were homologous in both cultivars during the induction of dormancy. The experimental results lead to the suggestion that dormancy induction was mostly caused by intrinsic signals, somehow regardless of the cultivars.
Our experimental results clearly indicated that dormancy is a gradual process, which was induced by the interaction of both temperature and photoperiod factors, which trigger changes in the levels of phytohormones such as IAA and ABA and in the expression of the related genes. During the induction of strawberry dormancy, DNA methylation increased, which lagged behind changes in IAA levels but was synchronized with changes in ABA levels (Fig. 3). Considering the report that ABA plays a pivotal role in seed development and dormancy by direct induction of histone ubiquitination, acetylation and methylation-dependent chromatin (Chinnusamy et al. 2008), it might be speculated that ABA and IAA regulate DNA methylation by an unknown mechanism in plant dormancy. DNA hypermethylation, in combination with slow-down metabolic, is likely to repress the growth-associated gene expression during induction of dormancy. The present study quantified global genomic DNA methylation; further research is required to investigate the methylation of specific genes involved in dormancy regulation combined with the study of responses to exogenous phytohormones in different cultivars showing different dormancy induction characteristics.
Abbreviations
- ABA:
-
Abscisic acid
- IAA:
-
Indole-3-acetic acid
- SD:
-
Short day
- LD:
-
Long day
- DRM:
-
Domain-rearranged methyltransferase
- NCED:
-
9-cis-Epoxycarotenoid dioxygenase
- ROS1:
-
Repressor of silencing 1
- HPLC:
-
High-performance liquid chromatograph
- QRT-PCR:
-
Quantitative real-time PCR
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
References
Blakeslee JJ, Peer WA, Murphy AS (2005) Auxin transport. Curr Opin Plant Biol 8:494–500
Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043. doi:10.1126/science.1126038
Burn JE, Bagnall DJ, Metzger JD, Dennis ES, Peacock WJ (1993) DNA methylation, vernalization, and the initiation of flowering. Proc Nat Acad Sci USA 90(1):287–291
Chan SW, Henderson I, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360. doi:10.1038/nrg1601
Chang LL, Zhang ZH, Han BM, Li H, Dai HY, He P, Tian HZ (2009) Isolation of DNA-methyltransferase genes from strawberry (Fragaria × ananassa Duch.) and their expression in relation to micropropagation. Plant Cell Rep 28:1373–1384. doi:10.1007/s00299-009-0737-8
Chinnusamy V, Gong ZZ, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol l50(10):1187–1195. doi:10.1111/j.1744-7909.2008.00727.x
Demeulemeester MAC, Van Stallen N, De Proft MP (1999) Degree of DNA methylation in chicory (Cichorium intybus L.): influence of plant age and vernalization. Plant Sci 142:101–108. doi:10.1016/S0168-9452(99)00010-2
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Garner WW, Allard HA (1923) Further studies in photoperiodism, the response of the plant to relative length of day and night. J Agric Res 23:871–920
Gocal GFW, Pharis RP, Yeung EC, Pearce D (1991) Changes after decapitation of indole-3-acetic acid and abscisic acid in the larger axillary bud of Phaseolus vulgaris L. cv Tender Green . Plant Physiol 95:344–350. doi:10.1104/pp.95.2.344
Gong ZZ, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 3:803–814. doi:10.1016/S0092-8674(02)01133-9
Håbjørg A (1978) Photoperiodic ecotypes in Scandinavian trees and shrubs. Meld Nor Landbrukshøgsk 57(33):1–20
Halliday KJ, Salter MG, Thingnaes E, Whitelam CG (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33:875–885. doi:10.1046/j.1365-313X.2003.01674.x
Heide OM (2008) Interaction of photoperiod and temperature in the control of growth and dormancy of Prunus species. Sci Hortic 115:309–314. doi:10.1016/j.scienta.2007.10.005
Heide OM, Prestrud AK (2005) Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol 25:109–114. doi:10.1093/treephys/25.1.109
Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson1 JV (2008) Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536. doi:10.1186/1471-2164-9-536
Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333. doi:10.1046/j.1365-313x.2001.01096.x
Junttila O (2007) Regulation of annual shoot growth cycle in northern tree species. In: Taulavuori E, Taulavuori K (eds) Physiology of northern plants under changing environment. Res Signpost, Kerala, pp 177–210
Lang GA, Early JD, Martin GC, Darnell RL (1987) Endodormancy, paradormancy and ecodormancy: physiological terminology and classification for dormancy research. Hortscience 22:371–377
Law RD, Suttle JC (2002) Transient decreases in methylation at 5′-CCGG-3′ sequences in potato (Solanum tuberosum L.) meristem DNA during progression of tubers through dormancy precede the resumption of sprout growth. Plant Mol Biol 51:437–447. doi:10.1023/A:1022002304479
Nagar PK (1995) Change in absicisic-acid, phenols and indoleacetic-acid in bulbs of tuberose (polianthes-tuberosa L) during dormancy and spring. Sci Hortic 63(1–2):77–82
Nitsch JP (1957) Photoperiodism in woody plants. Proc Am Soc Hortic Sci 70:526–544. doi:10.1146/annurev.pp.07.060156.001203
Olsen JE (2010) Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Mol Biol 73:37–47. doi:10.1007/s11103-010-9620-9
Pearce DW, Taylor JS, Robertson JM, Harker KN, Daly EJ (1995) Changes in abscisic acid and indole-3-acetic acid in axillary buds of Elytrigia repens released from apical dominance. Physiol Plant 94:110–116
Robert F, Risser G, PeÂtel G (1999) Photoperiod and temperature effect on growth of strawberry plant (Fragaria × ananassa Duch.): development of a morphological test to assess the dormancy induction. Sci Hortic 82:217–226. doi:10.1016/S0304-4238(99)00054-0
Rohde A, Bhalerao R (2007) Dormancy in the perennial context. Trends in Plant Sci 12:217–223. doi:10.1105/tpc.003186
Rohde A, Prinsen E, De Rycke R, Engler G, van Montagu M, Boerjan W (2002) PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in Poplar. Plant Cell 14:1885–1901. doi:10.1105/tpc.003186
Ruttink T, Arend M (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19:2370–2390. doi:10.1105/tpc.107.052811
Salvatierra A, Pimentel P, Moya-Leon MA, Caligari PDS, Herrera R (2010) Comparison of transcriptional profiles of flavonoid genes and anthocyanin contents during fruit development of two botanical forms of Fragaria chiloensis ssp. chiloensis. Phytochemistry 71:1839–1847. doi:10.1016/j.phytochem.2010.08.005
Santamaría ME, Hasbún R, Valera MJ, Meijón M, Valledor L, Rodríguez JL, Toorop PE, Cañal MJ, Rodríguez R (2009) Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J Plant Physiol 166:1360–1369. doi:10.1016/j.jplph.2009.02.014
Santos F, Teale W, Fleck C, Volpers M, Ruperti B, Palme K (2010) Modelling polar auxin transport in developmental patterning. Plant Biol. 12:s1 3–14. doi:10.1111/j.1438-8677.2010.00388.x
Sønsteby A, Heide OM (2006) Dormancy relations and flowering of the strawberry cultivars Korona and Elsanta as influenced by photoperiod and temperature. Sci Hortic 110:57–67. doi:10.1016/j.scienta.2006.06.012
Sorce C, Lorenzi R, Ceccarelli N, Ranalli P (2000) Changes in free and conjugated IAA during dormancy and sprouting of potato tubers. Aust J Plant Physiol 27(4):371–377. doi:10.1071/PP99150
Stafstrom JP (1993) Axillary bud development in pea: apical dominance, growth cycles, hormonal regulation and plant architecture. In: Amasino RM (ed) Cellular communication in plants. Plenum Press, New York, pp 75–86
Swain SM, Singh DP (2005) Tall tales from sly dwarfs: novel functions of gibberellins in plant development. Trends Plant Sci 10:1360–1385. doi:10.1016/j.tplants.2005.01.007#
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735. doi:10.1093/aob/mci083
Yang FX, Jin F, Yan X (2010) Comprehensive evaluation of different strawberry varieties’ tolerance to coldness. J. Fruit Sci 27(3):368–372. doi:1009-9980(2010)03-368-05
Yin KL (1999) The new French strawberry variety “Darselect”. South China Fruits 28(5):41–42
Zhang YG, Cheng JH, Han ZH, Xu XF, Li TZ (2005) Comparison of methods for total RNA isolation from Malus xiaojinensis and cDNA amplified using LD-PCR. China Biotechnol Bull 4:50–53 (ISSN:1002-5464.0.2005-04-015)
Zhang XZ, Zhao YB, Wang GP, Chang RF, Li CM, Shu HR (2008) Dynamics of endogenous cytokinins during phase trogen change in Malus domestica Borkh. Acta Hort 774:29–33
Acknowledgments
This project was supported by the National Key Technologies R&D Program of China (grant nos. 2011BAD12B02 and 2006BAD07B06-04), and the Key Laboratory of the Beijing Municipality of Stress Physiology and Molecular Biology for Fruit Trees.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Q. Zhao.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Wang, Y., Zhang, X. et al. Dynamics of phytohormone and DNA methylation patterns changes during dormancy induction in strawberry (Fragaria × ananassa Duch.). Plant Cell Rep 31, 155–165 (2012). https://doi.org/10.1007/s00299-011-1149-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1149-0