Abstract
Transgenic technology has been successfully used for gene function analyses and trait improvement in cereal plants. However, its usage is limited in woody plants, especially in the difficult-to-transform but commercially viable hybrid poplar. In this work, an efficient regeneration and transformation system was established for the production of two hybrid aspen clones: Populus alba × P. berolinensis and Populus davidiana × P. bolleana. A plant transformation vector designed to express the reporter gene uidA, encoding β-glucuronidase (GUS), driven by the cauliflower mosaic virus 35S promoter, was used to detect transformation event at early stages of plant regeneration, and to optimize the parameters that may affect poplar transformation efficiency. Bacterium strain and age of leaf explant are two major factors that affect transformation efficiency. Addition of thidiazuron (TDZ) improved both regeneration and transformation efficiency. The transformation efficiency is approximately 9.3% for P. alba × P. berolinensis and 16.4% for P. davidiana × P. bolleana. Using this system, transgenic plants were usually produced in less than 1 month after co-cultivation. The growth characteristics and morphology of transgenic plants were identical to the untransformed wild type plants, and the transgenes could be inherited by vegetative propagation, as confirmed by PCR, Southern blotting, RT–PCR and β-glucuronidase staining analyses. The establishment of this system will help to facilitate the studies of gene functions in tree growth and development at a genome level, and as well as the introduction of some valuable traits in aspen breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent advances in molecular genetics and biotechnology have made plant transformation and regeneration a powerful tool for crop improvement and gene function analyses. Although transgenic plants have been obtained by Agrobacterium tumefaciens-mediated transformation in many species (Han et al. 1996; Jouanin et al. 1993; Kim et al. 1997), plant transformation and regeneration are still difficult and time-consuming in many economically important species, especially in forest tree species (Birch 1997; Han et al. 2000).
Modern methods of genetic engineering can help breeders to overcome limitations of the conventional techniques for tree improvement (Confalonieri et al. 2003). Populus is one of the major renewable resources that are highly valued by the pulp and paper industry for their fast growth and high quality fiber (Stettler et al. 1996). They are also extensively used for land reforestation and contaminated soil phytoremediation (Balatinecz et al. 2001; Rishi et al. 2001). However, due to the long generation period and callus/shoot initiation on different media, Agrobacterium-mediated poplar transformation was reported only for a limited number of model genotypes (Confalonieri et al. 2000; Dai et al. 2003; De Block 1990; Delledonne et al. 2001; Eriksson et al. 2000; Fillatti et al. 1987; Gallardo et al. 1999; Han et al. 1997, 2000; Howe et al. 1994; Hu et al. 1999; Pilate et al. 2002; Sepanen et al. 2004; Song et al. 2006; Tsai et al. 1994; Wei et al. 2006).
Recently, Yevtushenko and Misra (2010) reported the successful transformation of a hybrid poplar P. nigra L. × P. maximowiczii A. Henry (genotype NM6), with a mean transformation efficiency of 36.3%. A novel approach for in situ bud transformation of P. cathayana Rehd by Agrobacterium was also established (Yang et al. 2010). We have focused on improving salt resistance in commercially valuable hybrid poplars. P. alba × P. berolinensis and P. davidiana × P. bolleana are two salt-sensitive aspen hybrid clones widely grown in the north part of China. In the present work, we describe a rapid and efficient protocol which does not require a callus induction phases for producing transgenic hybrid poplars.
Materials and methods
Plant material and growth conditions
Two aspen hybrid clones commercially grown in the north part of China, Yinzhong (P. alba × P. berolinensis) and Shanxin yang (P. davidiana × P. bolleana), were used in this study. Generally, in vitro grown plants were sub-cultured monthly by aseptically transferring shoot apices to the fresh MS medium (Murashige and Skoog, 1962) supplemented with 0.1 mg/l NAA (MS1) in glass cultivation bottles (Table 1). Plantlets were grown in the culture room with cool white fluorescent light (~200 μmol m−2 s−1) under short day condition (12 h light/12 h dark). The temperature was kept at about 21–25°C in the daytime and 15–18°C at night.
Vector and Agrobacterium strains
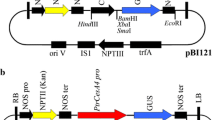
The binary vector, pCAMBIA2301 (Huang et al. 2009), which contains a β-glucuronidase (GUS)-encoding gene (uidA), driven by the cauliflower mosaic virus 35S promoter, and a selection marker gene (nptII) (Fig. 1), was introduced into A. tumefaciens strain LBA4404, GV3101 and EHA105, respectively, as described previously (Holsters et al. 1978). Growth of pCAMBIA2301-containing Agrobacterium culture was carried out as described previously (Ho et al. 1998).
Schematic representation of T-DNA region in the plasmid vector pCAMBIA2301. LB T-DNA left border repeat, 35S-t CaMV35S terminator, npt-II neomycin phosphotransferase II gene, 35S-p CaMV35S promoter, MCS multiple cloning sites, uidA β-glucuronidase gene, NOS-t NOS terminator, RB T-DNA right border repeat
GUS expression analysis
To optimize the transformation procedure, factors that may affect the regeneration and transformation frequencies, including Agrobacterium strain, explant source, physiological condition for regeneration, and phytohormone type and concentration, were examined by GUS transient expression assay. For each treatment, 120 leaf explants were used and each experiment was repeated at least three times. Juvenile, adult and senescent leaf explants (2–3 cm2) collected from in vitro 21-day-old poplar plants of Yinzhong (P. alba × P. berolinensis) grown on MS1, were co-cultivated on MS2 (Table 1) for 2 days with different A. tumefaciens strains (LBA4404, GV3101 and EHA105) harboring pCAMBIA2301 under different conditions. The explants were then transferred to A. tumefaciens inhibition medium MS3 (Table 1) for GUS expression analysis. Histochemical GUS staining was conducted as described previously (Gallagher 1992). After infection and co-cultivation, leaf explants were incubated overnight at 37°C in a reagent mix containing 2 mM 5-bromo-4- chloro-3-indolyl-β-d-glucuronide (X-Gluc), 0.1 M sodium phosphate buffer (pH 7.0), 0.5 mM each potassium ferri- and ferrocyanide, 10 mM EDTA (pH 7.0), and 0.1% Triton X-100. After staining, the tissues were cleared of chlorophyll using 95% ethanol. GUS transformation efficiency was expressed as the percentage of leaf explants showing GUS activity relative to the total number of co-cultivated leaf explants. For GUS detection in transgenic plants, whole leaves from the regenerated kanamycin-resistant plants were used.
Plant transformation
Leaf explants excised from 3-week-old plantlets were inoculated by swirling in the Agrobacterium culture for 8–10 min. The inoculated explants were placed horizontally and co-cultivated on MS2 (Table 1) at 24 ± 2°C in the dark for 2 days. The co-cultivated explants were washed four times in sterilized distilled water for 5 min each, briefly blotted dry on a sterilized filter paper, and cultured on MS3 (Table 1) at 24 ± 2°C, 16 h light/8 h dark cycle with supplemental lighting (Philips, 600 W) of 200 μEm−2 s−1, for 7 days. Then, the explants were transferred onto MS4 (Table 1), followed by sub-culturing on fresh MS4 every 14 days until the transgenic shoots were formed. This usually takes about 21–30 days after co-cultivation. Transgenic shoots (1 cm in length) were excised from the explants and rooted on MS5 (Table 1). Transgenic plants were transplanted into soil and maintained in a greenhouse. Transformation frequency was defined as the percentage of co-cultivated leaf explants that produced transgenic shoots. For each leaf explant, only one shoot was selected and further analyzed.
Statistical analysis
For statistical analyses, the Student’s t test was used to generate every P value. The alpha level was 0.01. The tests were one-tailed. The data were normalized and all samples were normally distributed with homogeneity of variance. Differences were considered to be statistically significant when the confidence intervals showed no overlap for the mean values with an error of 0.01.
Plant genomic DNA isolation and PCR analysis
Genomic DNA was isolated from leaves (0.5 g) of transgenic and control plant as described previously (Kang et al. 2010). To ensure that samples were free from Agrobacterium contamination, each sample was taken from shoots growing on rooting medium with 25 mg/l kanamycin for 4 weeks. For PCR analyses of uidA, gene-specific primers (forward 5′-GCCGGAATCCATCGCAGCGTA-3′ and reverse 5′-CCCGCTTCGAAACCAATGCCT-3′) were used to amplify a 602-bp fragment.
Southern blot analysis of transgenic plants
For Southern blot analysis, four micrograms of genomic DNA isolated from the leaves of transformed and control poplar plants was digested with EcoRI, electrophoresed on 0.8% agarose gels, and transferred onto Hybond N+ nylon membranes by vacuum blotting system. The 602-bp fragment of uidA-coding region was used as a hybridization probe. Standard procedures for Southern blot analysis and probe labeling were conducted with DIG DNA Labeling and Detection Kit1 (Roche, Germany) following the manufacturer’s instruction.
Reverse transcriptase-PCR analyses
Total RNA was isolated from leaves of 3-week-old poplar plants with the TRIZOL Reagent (Invitrogen, Shanghai, China) following the manufacturer’s instruction. After treated with DNase I (Promega), 2 μg of total RNA was subjected to reverse transcription reaction using the reverse transcriptase ReverTra Ace (TOYOBO, Japan) at 42°C for 1 h. Reverse transcriptase-mediated PCR (RT–PCR) was performed using uidA gene specific primers to amplify a PCR product of 602 bp. Expression level of PtEF1β was also determined with forward (5′-GACAAGAAGGCAGCGGAGGAGA G-3′) and reverse (5′-CAATGAGGGAATCCACTGACACAA G-3′) primers (to serve as a quantitative control).
Kanamycin-resistance stability evaluation
Leaf explants excised from PCR, RT–PCR and GUS staining confirmed plantlets (3-week-old) were directly cultured on MS4 (Table 1) at 24 ± 2°C with cool white fluorescent light (~200 μmol m−2 s−1) on a 12 h light/12 h dark photoperiod. Regenerated kanamycin-resistant shoots (1 cm in length) were excised from the explants and rooted on MS5 (Table 1). Transgenic plants with well-developed root systems were obtained usually after 2–3 weeks on MS5.
Results
Optimization of the transformation procedure
The commercial aspen hybrid clone Yinzhong (P. alba × P. berolinensis) was used for the preliminary GUS expression experiments. After infection with A. tumefaciens EHA 105 harboring pCAMBIA2301 (Fig. 1), GUS expression were detected in 3 days on MS3 (Figs. 2a, 3d), with the best staining 6 days after transfer (Fig. 2a). Agrobacterium strain is one of the major factors affecting transformation efficiency. EHA105 produced the highest transformation frequency while GV3101 showed little or not virulence (Fig. 2b). Therefore, EHA105 harboring pCAMBIA2301 was chosen for the following examination of other factors. Addition of acetosyringone (Fig. 2c), age of leaf (Fig. 2d), growth stage of bacterium (Fig. 2e) and dark period during co-cultivation (Fig. 2f) all significantly affect the expression of uidA gene. The best results in terms of GUS expression were obtained using EHA105 pCAMBIA2301 A. tumefaciens strain, dipping juvenile leaf explants into a bacterial suspension (OD600 = 0.6) for 8–10 min, and co-cultivating them in dark conditions on MS2 medium supplemented with 50 mg/l acetosyringone.
Factors that affect the transient expression of uidA gene in leaf segments of Yinzhong (Populus alba × P. berolinensis). a Post-cultivation periods with Agrobacterium strain EHA105 harboring pCAMBIA2301. b Agrobacterium strains. c Acetosyringone (AS) concentrations. d Ages of leaves. e Growth stages of Agrobacterium bacteria. f Dark periods during co-cultivation. Results are presented as means and standard errors from three independent experiments. **Significant difference in comparison to the controls at P < 0.01 (Student’s t test)
Regeneration of transgenic Yinzhong (Populus alba × P. berolinensis) plants. a An explant on MS4 for 2 weeks after infection. b An explant on MS4 for 4 weeks after infection. c Regenerated shoots excised from explants and cultured on MS5 for shoot elongation. d GUS staining of explants 3 days on MS3 after co-cultivation with Agrobacterium strain EHA105 harboring pCAMBIA2301. e GUS staining of an uninfected explant 3 days on MS3 after co-cultivation (negative control). f GUS staining of an explant 2 weeks on MS4 after co-cultivation. g GUS staining of an uninfected explant 2 weeks on MS4 after co-cultivation (negative control). h A regenerated shoot rooted on MS1 for 2 weeks. i GUS staining of a transgenic shoot. j GUS staining of a transgenic root. k GUS staining of a wild type root (bar 1 cm)
Stable transformation experiments
Based on the preliminary experiments, a total of 1,136 infected leaf segments of Yinzhong (P. alba × P. berolinensis) were transferred onto MS4 after 7 days pre-cultivation on MS3 (Table 1). Kanamycin-resistant shoots emerged in 2 weeks (Fig. 3a) and were ready for rooting in 4 weeks (Fig. 3b). For whole plant regeneration, regenerated kanamycin-resistant shoots were separated from the leaf explants when they were about 1 cm long, and transferred onto MS5 (Table 1) to induce roots (Fig. 3c). After 2 weeks, most of them had rooted (Fig. 3h). The rooting percentages were approximately 95%. In total, 106 independent kanamycin-resistant lines were obtained with a transformation frequency of 9.3%. Rooted shoots were then propagated on MS5.
To determinate the stability of kanamycin-resistance in regenerated shoots, leaf explants of wild type grown on MS1 and 3-week-old regenerated plants grown on MS5 were subjected to a second round regeneration on MS4. After 40 days, shoots were regenerated from the leaf explants of regenerated kanamycin-resistant shoot, but all the wild type leaf explants died (data now shown). The expression of uidA gene of the kanamycin-resistant shoots propagated on MS5 was further confirmed by GUS expression analysis (Fig. 3i–k).
The efficiency of the above-described transformation system was tested also in P. davidiana × P. bolleana (Shanxin yang) and a detailed regeneration protocol is shown in Fig. 4. Among the 420 co-cultivated leaf explants, 69 of them produced kanamycin-resistant shoots in 3 weeks (Fig. 4d, f). As expected, control leaf explant did not regenerate shoots (Fig. 4e). Expression of the uidA gene was detected by histochemical GUS analysis in transgenic shoots (Fig. 4d). The kanamycin-resistant shoots rooted on MS5 in 2 weeks (Fig. 4g, h; plant in the right) while wild-type shoots did not produce roots (Fig. 4g, h; plant in the left). For both varieties, regenerated plants were propagated on MS5 (Fig. 4i) and grown in soil in the greenhouse for further confirmation of GUS expression (Fig. 4j).
A flowchart of the Agrobacterium-mediated transformation system for Shanxin yang (Populus davidiana × P. bolleana). a 3-week-old plants grown on MS1 ready for leaf explant preparation. b Leaf explants were co-cultivated with EHA105 harboring pCAMBIA2301 on MS2 for 2 days. c Leaf explants were cultured on MS3 for 7 days to inhibit the growth of EHA105. d GUS staining of an explant 3 weeks on MS4 after co-cultivation. Transgenic (solid arrow) and non-transgenic (empty arrow) shoots are shown. e Uninfected explants on MS4 3 weeks after co-cultivation (negative control). f Infected explants on MS4 3 weeks after co-cultivation. g Root induction of regenerated shoots on MS5. Left non-transgenic plant, right transgenic plant. h Bottom views of g. i Transgenic shoots propagated on MS5. j 7-week-old transgenic plants grown in a greenhouse (bar 2 cm)
Confirmation of transgenic plants by PCR, Southern blot, RT–PCR, and GUS histochemical analyses
To confirm that the uidA gene was inserted into the populus genome, uidA gene was PCR-amplified using specific primer pair, yielding the expected band size of 602 bp (Fig. 5a, b). Southern blot analyses of selected transgenic plants confirmed that most transgenic plants had 1–2 copies of the transgene (Fig. 5a, b). Transcripts of uidA gene were detected in all the PCR-positive lines (Fig. 6a, b). GUS staining further confirmed the expression of uidA gene in transgenic plants (Fig. 6c).
PCR and Southern blot analyses of transgenic plants. a Yinzhong plants. b Shanxin yang plants. A 602-bp PCR product was detected in most transgenic lines. M λ-EcoT14 I digest DNA markers, P PCR product with pCAMBIA2301 plasmid DNA as template, W PCR product with double distilled water as template, WT PCR product with wild type plant genomic DNA as template, L1-16 and L1-8 PCR products with genomic DNA from regenerated Yinzhong and Shanxin yang as template, respectively. For Southern blot analysis of transgenic plants, genomic DNA was digested with EcoRI, electrophoresed, and probed with a DIG-labeled 602-bp PCR product of uidA gene as above. The number of bands reflects the number of transgene insertions. Molecular weight DNA markers are shown on the left. Lane 1 wild type plant (WT), lanes 2–10 regenerated poplar lines. Lane 4 (L3) and Lane 6 (L5) in b presented non-transformed plants
RT–PCR and histochemical GUS staining analyses of transgenic plants. a, b RT–PCR analysis of wild type and transgenic lines of Yinzhong (a) and Shanxin yang (b). Total RNA was isolated from wild type (WT) and different transgenic lines (L1–L16). RT–PCR was performed with uidA-specific primers (30 cycles) or PtEF1β-specific primers (25 cycles). The PtEF1β gene was used as an internal control. c Histochemical GUS staining of wild type and transgenic (L1–L8) leaves of Shanxin yang
Discussion
A major problem in the production of transgenic populus is regeneration of shoots from transformed cells. To date, although poplars have been transformed for research purposes far more than any other species of forest tree, many commercially important genotypes and species still remain difficult to transform (Han et al. 1996, 2000). Several laboratories have reported highly efficient transformation systems for poplars (Fladung et al. 1997; Leple et al. 1992; Tsai et al. 1994; Tzifra et al. 1996, 1997), however, most of the work are restricted to a few model hybrids and species of section Leuce (Han et al. 2000; Song et al. 2006), and the transformation efficiencies are low, ranging mostly from 2 to 13%. Recently, an efficient transformation system was established for a commercial hybrid poplar P. nigra L. × P. maximowiczii A. Henry (Yevtushenko and Misra 2010). Here, we report the establishment of a transformation system for another two hybrid clones commercially grown in China, P. alba × P. berolinensis and P. davidiana × P. bolleana.
A large number of factors that affect the efficiency of DNA delivery into the targeted cells and the regeneration of transgenic plants are involved in Agrobacterium transformation, including Agrobacterium strain, explant source, genotype, physiological condition for regeneration, and phytohormone type and combination. We observed that bacterium strain and the age of leaf explant are the two major factors that affect the transformation efficiency for these two hybrid clones. Addition of TDZ improved the regeneration and transformation frequency (data now shown). The 1 week pre-cultivation on MS3 is also a key step to improve transformation efficiency. Unlike the system reported for Populus trichocarpa Genotype Nisqually-1 (Song et al. 2006), in which the entire process from co-cultivation to whole plant regeneration takes about 5–8 months, shoots can be directly regenerated without callus induction in this system. Figure 7 illustrates the stepwise process using leaf discs as explants. As set out in Fig. 7, the entire process from co-cultivation to whole plant regeneration takes less than 2 months.
Stepwise protocol for transforming Yinzhong (Populus alba × P. berolinensis) and Shanxin yang (Populus davidiana × P. bolleana) using leaf disc as explants. Medium MS1 to MS5 are shown in Table 1
Asexual transformation methods provide a means for introducing new traits that are difficult to obtain via traditional breeding, and allow modification of valuable clones without the genetic recombination that occurs during sexual reproduction (Han et al. 2000). With the availability of a rapid and efficient transformation-regeneration system from leaf explants, it is now feasible to introduce genes for desirable traits, such as disease and abiotic stress resistance, into these two economically important aspen hybrid clones.
Abbreviations
- AS:
-
Acetosyringone
- 6-BA:
-
6-Benzyladenine
- GUS:
-
β-Glucuronidase
- MS:
-
Murashige and Skoog (1962)
- NAA:
-
Naphthalene-acetic acid
- nptII:
-
Neomycin phosphotransferase gene
- TDZ:
-
Thidiazuron
References
Balatinecz JJ, Kretschmann DE, Leclercq A (2001) Achievements in the utilization of poplar wood-guideposts for the future. For Chron 77:265–269
Birch RG (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Physiol Plant Mol Biol 48:297–326
Confalonieri M, Belenghi B, Balestrazzi A, Negri S, Facciotto G, Schenone G, Delledonne M (2000) Transformation of elite white poplar (Populus alba L.) cv. ‘Villafranca’ and evaluation of herbicide resistance. Plant Cell Rep 19:978–982
Confalonieri M, Balestrazzi A, Bisoffi S, Carbonera D (2003) In vitro culture and genetic engineering of Populus spp.: synergy for forest tree improvement. Plant Cell Tissue Org 72:109–138
Dai WH, Cheng ZM, Sargent W (2003) Plant regeneration and Agrobacterium-mediated transformation of two elite aspen hybrid clones from in vitro leaf tissues. In Vitro Cell Dev Biol Plant 39:6–11
De Block M (1990) Factors influencing the tissue culture and the Agrobacterium tumefaciens-mediated transformation of hybrid aspen and poplar clones. Plant Physiol 93:1110–1116
Delledonne M, Allegro G, Belenghi B, Balestrazzi A, Picco F, Levine A, Zelasco S, Calligari P, Confalonieri M (2001) Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol Breed 7:35–42
Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18:784–788
Fillatti JJ, Sellmer J, McCown B, Haissig B, Comai L (1987) Agrobacterium-mediated transformation and regeneration of Populus. Mol Gen Genet 206:192–199
Fladung M, Kumar S, Ahuja MR (1997) Genetic transformation of Populus genotypes with different chimaeric gene constructs: transformation efficiency and molecular analysis. Transgene Res 6:111–121
Gallagher SR (1992) GUS protocols: using the GUS gene as a reporter of gene expression. Academic Press, San Diego
Gallardo F, Fu JM, Francisco RC, Garcia-Gutierrez A, Francisco MC, Kirby EG (1999) Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta 210:19–26
Han KH, Gordon MP, Strauss SH (1996) Cellular and molecular biology of Agrobacterium-mediated transformation of plants and its application to genetic transformation of Populus. In: Bradshaw HD Jr, Stettler RF, Heilman PE, Hinckley TM (eds) Biology of Populus and its implication for management and conservation. National Research Council, Ottawa, pp 201–222
Han KH, Ma CP, Strauss SH (1997) Matrix attachment regions (MARs) enhance transformation frequency and transgene expression in poplar. Transgenic Res 6:415–420
Han KH, Meilan R, Ma CP, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19:315–320
Ho KC, Chang SH, Tsay JY, Tsai CJ, Chiang VL, Chen ZZ (1998) Agrobacterium tumefaciens mediated transformation of Eucalyptus camaldulensis and production of transgenic plants. Plant Cell Rep 17:675–680
Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187
Howe GT, Goldfarb B, Strauss SH (1994) Agrobacterium-mediated transformation of hybrid poplar suspension cultures and regeneration of transformed plants. Plant Cell Tissue Org 36:59–71
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17:808–812
Huang YH, Mei M, Zeng JW, Zhou BR, Xia R, Yi GJ (2009) Study on expression character of gas gene contained in two different plasmids on plant cell and Agrobacterium tumefaciens. Genomics Appl Biol 28:34–38
Jouanin L, Brasileiro ACM, Leple JC, Pilate G, Cornu D (1993) Genetic transformation: a short review of methods and their applications, results and perspectives for forest trees. Ann Sci For 50:325–336
Kang BG, Ye X, Osburn LD, Stewart CN Jr, Cheng ZM (2010) Transgenic hybrid aspen overexpressing the Atwbc19 gene encoding an ATP-binding cassette transporter confers resistance to four aminoglycoside antibiotics. Plant Cell Rep 29:643–650
Kim MS, Klopfenstein NB, Chun YW (1997) Agrobacterium-mediated transformation of Populus species. In: Klopfenstein NB, Chun YW, Kim MS, Ahuja MR (eds) Micropropagation, genetic engineering, and molecular biology of Populus. Gen Tech Rep RM-GTR-297, US Dep Agric-For Serv, Fort Collins, Colo, pp 51–59
Leple JC, Brasileiro ACM, Michel MF, Delmotte F, Jouanin L (1992) Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep 11:137–141
Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C et al (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20:607–612
Rishi AS, Nelson ND, Goyal A (2001) Genetic modification for improvement of Populus. Physiol Mol Biol Plants 7:7–21
Sepanen SK, Syrjala L, Weissenberg KV, Teeri TH, Paajanen L, Pappinen A (2004) Antifungal activity of stilbenes in in vitro bioassays and in transgenic Populus expressing a gene encoding pinosylvin synthase. Plant Cell Rep 22:584–593
Song JY, Lu SF, Chen ZZ, Lourenco R, Chiang VL (2006) Genetic transformation of populus trichocarpa genotype Nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol 47:1582–1589
Stettler RF, Bradshaw HD Jr, Heilman PE, Hinckley TM (eds) (1996) Biology of Populus and its implication for management and conservation. National Research Council Canada, Ottawa
Tsai CJ, Podila GK, Chiang VL (1994) Agrobacterium-mediated transformation of quaking aspen (Populus tremuloides) and regeneration of transgenic plants. Plant Cell Rep 14:94–97
Tzfira T, Ben-Meir H, Vainstein A, Altman A (1996) Highly efficient transformation and regeneration of aspen plants through shoot-bud formation in root culture. Plant Cell Rep 15:566–571
Tzfira T, Jensen CS, Wang W, Zuker A, Vincour B, Altman A et al (1997) Transgenic Populus tremula: a step-bystep protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15:219–235
Wei H, Meilan R, Brunner AM, Skinner JS, Ma CP, Strauss SH (2006) Transgenic sterility in Populus: expression properties of the poplar PTLF, Agrobacterium NOS and two minimal 35S promoters in vegetative tissues. Tree Physiol 26:401–410
Yang L, Sun Y, Xie L, Liang A (2010) A novel approach for in situ bud transformation of Populus by Agrobacterium. Scand J For Res 25:3–9
Yevtushenko DP, Misra S (2010) Efficient Agrobacterium-mediated transformation of commercial hybrid poplar Populus nigra L. × P. maximowiczii A. Henry. Plant Cell Rep 29:211–221
Acknowledgments
We thank Prof. Gui-Feng Liu (Northeast Forestry University, China) for providing the Shanxin yang (P. davidiana × P. bolleana) clone. This work was supported by the following grants: the National Natural Science Foundation of China (30872044; 30800880; 3100012; 31000288), the National Basic Research Program of China (2010CB126600), the National mega project of GMO crops (2008ZX08001-003; 2008ZX08004-002; 2008ZX08010-004); Shanghai Science & Technology Committee: 10DZ2271800); and Shanghai Key Laboratory of Bio-Energy Crops.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Jouanin.
Rights and permissions
About this article
Cite this article
Wang, H., Wang, C., Liu, H. et al. An efficient Agrobacterium-mediated transformation and regeneration system for leaf explants of two elite aspen hybrid clones Populus alba × P. berolinensis and Populus davidiana × P. bolleana . Plant Cell Rep 30, 2037–2044 (2011). https://doi.org/10.1007/s00299-011-1111-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1111-1