Abstract
In this study, we present a method for transient expression of the type III effector AvrGf1 from Xanthomonas citri subsp. citri strain Aw in grapefruit leaves (Citrus paradisi) via Agrobacterium tumefaciens. The coding sequence of avrGf1 was placed under the control of the constitutive CaMV 35S promoter in the binary vectors pGWB2 and pGWB5. Infiltration of grapefruit leaves with A. tumefaciens carrying these constructs triggered a hypersensitive response (HR) in grapefruit 4 days after inoculation. When transiently expressed in grapefruit leaves, two mutants, AvrGf1ΔN116 and AvrGf1ΔC83, failed to induce an HR. Moreover, using bioinformatics tools, a chloroplast transit signal was predicted at the N terminus of AvrGf1. We demonstrated chloroplast localization by using an AvrGf1::GFP fusion protein, where confocal images revealed that GFP fluorescence was accumulating in the stomatal cells that are abundant in chloroplasts. Transient expression in citrus has the potential for aiding in the development of new disease defense strategies in citrus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agrobacterium-mediated transient expression, which is based on expression of non-integrated T-DNA, provides a valuable procedure for readily assessing genetic information (Jones et al. 2009; Jones et al. 2005). Moreover, this method has led to a profusion of studies in areas such as gene-for-gene interactions, regulation and expression of multiple genes simultaneously, and gene silencing (Gurlebeck et al. 2009; Johansen and Carrington 2001; Yang et al. 2000). The Agrobacterium-mediated transient assay has been broadly applied to many plant systems (Joensuu, et al. 2010; Bhaskar et al. 2009; Cheng et al. 2009; Green, et al. 2009; Li et al. 2009; Manavella and Chan 2009; Santos-Rosa et al. 2008; Sparkes et al. 2006). Although transient expression provides certain advantages in many plant systems, there has been limited success in citrus. Agrobacterium-mediated transient expression has been especially difficult to use (Ahmad and Mirza 2005; Bespalhok et al. 2003).
Although efficient tools for delivering genes other than using A. tumefaciens are available, in general these techniques have limitations in citrus. For example, virus-based transient-expression vectors are routinely used for tree crops, such as citrus, but they are especially difficult to transform and the approach suffers from the time-consuming process of generating stable vectors in the plant (Folimonov et al. 2007; Porta and Lomonossoff 1996). Duan et al. (1999) also reported a non-uniform gene transfer to cells using A. tumefaciens and particle bombardment as transient methods. Although generating transgenic plants has become more commonly used in citrus, transient expression of ectopic genes continues to have major drawbacks. The development of an efficient, robust, and relatively simple Agrobacterium-mediated transient protocol for citrus will undoubtedly expedite investigation of gene function in this host plant.
One of the most devastating diseases that limits citrus production worldwide is citrus canker, which is caused by Xanthomonas citri subsp. citri (X. citri—Brunings and Gabriel 2003). X. citri relies on highly specialized delivery machinery, named the type three secretion system (T3SS), to introduce virulence factors or type III effector (T3-effector) proteins inside the host cell, which suppress defense responses in the host (Büttner and Bonas 2010). Durable resistance has been a goal of plant breeding programs for decades although with limited success. Many conventional breeding and molecular-based approaches have been applied to citrus although most are very time consuming due to the regeneration process. By using transient expression, it is plausible to gain insights into the potential value of resistance-associated genes in a relatively short period. Therefore, the aim of this work was to establish an easy and efficient method for Agrobacterium-mediated transient expression and, additionally, apply the procedure to elucidate the mechanism of AvrGf1 function in grapefruit leaves. The T3-effector protein, AvrGf1, from X. citri subsp. citri strain AW (Xcc-Aw) triggers a localized apoptosis-like defense reaction commonly referred to as the hypersensitive reaction (HR) in grapefruit leaves (Rybak et al. 2009). This response is triggered by the direct or indirect recognition of the T3-effector protein by one or more cognate resistance (R) proteins in the host (Chisholm et al. 2006; Bonas and Van den Ackerveken 1999).
Materials and methods
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are provided in the Supplementary Table S1.
Media and growth conditions
Escherichia coli DH5α was used as the cloning host throughout this study and cultured in Luria broth (LB) medium (Sambrook et al. 1989) at 37°C. For solid medium, 1.5% BD Bacto agar was added. A. tumefaciens strains were grown in YEP medium (An 1987) and MSO medium without plant growth regulators (Murashige and Skoog 1962). Antibiotics were used at the following concentrations: ampicillin (Amp), 100 μg ml−1; kanamycin (Kn), 50 μg ml−1; rifamycin SV (Rif), 100 μg ml−1; spectinomycin (Spc), 100 μg ml−1 and chloramphenicol (Cm), 50 μg ml−1.
Plant material
Six-year-old Duncan grapefruit (Citrus paradisi) plants were grown from seeds in 15 cm plastic pots with Metromix 300 (W. R. Grace & Co., Cambridge, MA, USA) in glasshouses at temperatures ranging from 25 to 30°C. Before inoculation, the plants were pruned and fertilized to produce uniformly aged shoots. A scale to standardize the age of citrus leaves was adopted in this study and consisted of young leaves (2-week-old first leaves developed after pruning back the branches), intermediate-aged leaves (3–5-week-old leaves that developed after the pruning), and mature leaves (more than 5-week-old leaves following pruning).
Recombinant DNA techniques
All standard DNA manipulations were performed as described (Sambrook et al. 1989). Restriction enzymes and T4 DNA ligase (Promega, Madison, WI, USA) were used according to the manufacturer’s instruction, and Phusion high-fidelity polymerase was used following the recommendations of the manufacturer (New England Biolabs, Beverly, Mass.). Competent E. coli DH5α cells were transformed by vector constructs as described (Sambrook et al. 1989). The selected plasmids were introduced into A. tumefaciens strain GV3101 (Van Larebeke et al. 1974) by electroporation.
Generation of full-length and deletion mutants of AvrGf1
To generate the construct encoding Xcc-Aw avrGf1 gene, we amplified the entire coding sequence of avrGf1 gene from pL799 clone plasmid DNA by polymerase chain reaction (PCR) using custom oligonucleotide primers. Next, the PCR product was cloned into the vector pENTR-D (Invitrogen) and transferred via LR-recombination (Invitrogen) into the binary vectors, pGWB2 and pGWB5 (Nakagawa et al. 2007—Fig. 1a) creating pGavrGf1_2 and pGavrGf1_5, respectively. We also generated four independent deletions of the avrGf1 open reading frame (ORF) by designed custom oligonucleotide primers to amplify 519 and 416 amino acids (a.a.), deleting 13 a.a. and 116 a.a. from the N terminus of avrGf1, respectively, creating p2∆N13 and p2∆N116 constructs, in pGWB2 plasmid, and p5∆N13 and p5∆N116 constructs in pGWB5 plasmid. The same procedure was applied to create p2∆C7, p2∆C83, p5∆C7 and p5∆C83 constructs, in which 7 and 83 a.a. were deleted from the C terminus, respectively. In all four deletions, the stop codon was deleted to allow an in-frame fusion with the green fluorescence protein (GFP) reporter gene in pGWB5 plasmid, or with the terminator—NosT in the pGWB2 plasmid. In both plasmids, pGWB2 and pGWB5, the expression of AvrGf1 was driven by the constitutive cauliflower mosaic virus 35S (35S) promoter. The constructs were then transformed into competent cells of E. coli DH5α and electroporated into competent cells of A. tumefaciens strain GV3101 generating the strain designations, 31::Gf1_2 and 31::Gf1_5, which carry the entire avrGf1 coding sequence, and the avrGf1 deletion mutants designated AvrGf1∆N13, AvrGf1∆N116, AvrGf1∆C7 and AvrGf1∆C83 (Fig. 1b).
Schematic of AvrGf1 protein and the deletion mutants. a Organization of the binary vectors, pGavrGf1_2 and pGavrGf1_5. b AvrGf1 deletion mutations were cloned into pGWB2 and pGWB5 to create truncated AvrGf1 proteins: AvrGf1∆N13 (13 amino acids were deleted after the start codon) and AvrGf1∆N116 (116 amino acids were deleted after the start codon) mutants generated in the N terminus; AvrGf1∆C7 (7 amino acids were deleted plus the stop codon) and AvrGf1∆C83 (83 amino acids were deleted plus the stop codon) mutants generated in the C terminus
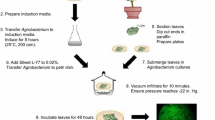
Preparation of A. tumefaciens cells for transient expression in citrus leaves
A. tumefaciens cells were prepared as follows: (a) A. tumefaciens strain GV3101 (GV3101) or transformed derivatives were cultured on YEP agar supplemented with appropriate antibiotics; (b) the strains were incubated at 28°C overnight; (c) a small loopful of each strain was then inoculated into 30 ml YEP liquid medium containing appropriate antibiotics; (d) each culture was incubated at 28°C with shaking at 150 rev m−1 overnight until an OD (Abs = 600 nm) of 0.7–0.9 was reached; (e) the OD was then adjusted to 0.3 (Abs = 600 nm); (f) 3.0 ml of each culture was pelleted at 1,150 g for 5 min and resuspended in 0.9 ml MS basal medium (Murashige and Skoog 1962), supplemented with 30 g l−1 sucrose, 50 mg l−1 myo-inositol, and 10 ml l−1 vitamins (glycine 40 mg l−1, nicotinic acid 100 mg l−1, pyridoxine HCl 200 mg l−1, thiamine 200 mg l−1); (g) 1 μl of 10 mol l−1 acetosyringone was added; (h) each culture was then incubated at 28°C, shaking at 200 rev m−1 for 1 h; and (i) the bacterial suspensions were infiltrated with a 27 gauge hypodermic needle and syringe into grapefruit or Nicotiana benthamiana leaves. Infiltrated plants were incubated in a growth room at a constant temperature of 28°C.
Plant inoculations
Bacterial cultures of Xanthomonas and Agrobacterium for plant inoculations were grown for 18 h at 28°C on nutrient agar (NA) and YEP solid medium, respectively, amended with appropriate antibiotics. Bacterial cells for Xanthomonas were harvested and suspended in sterile tap water, then adjusted to an optical density (OD600) of 0.3 (3 × 108 colony-forming units (cfu) ml−1) with a Spectronic 20 spectrophotometer (Spectonic-UNICAM, Rochester, NY, USA). Each bacterial suspension was infiltrated using a 1 ml syringe and 27 gauge hypodermic needle into the abaxial leaf surface by infiltrating an area of ~0.5 cm2 in each side of the leaf or infiltrating the entire leaf (Klement 1963). For each strain, three leaves were infiltrated and served as replications. Each strain was tested three separate times under the same conditions. A. tumefaciens strains were infiltrated in the same manner as the X. citri inoculum except that the cell preparation differed for the Agrobacterium-mediated transient assay as described below.
Hypersensitive response and GFP assay in transiently transformed grapefruit leaves
For every construct designed here, three independent experiments were carried out to evaluate the expression of avrGf1 and mutants, using three individual citrus leaves per strain. For the hypersensitive response (HR) assay, grapefruit leaves transiently transformed with 31::Gf1_2, 31::Gf1_5, AvrGf1∆N13, AvrGf1∆N116, AvrGf1∆C7 and AvrGf1∆C83 strains were incubated at 28°C and the HR was assessed up to 5 days after infiltration (DAI). Transient grapefruit leaves transformed with 31::Gf1_5 and AG::2202 strains were assessed for GFP. Approximately, 0.5 cm2 of the infiltrated area was placed on a glass microscope slide, covered with water and then a coverslip. The GFP fluorescence was observed with a Leica TCS SP5 confocal laser-scanning microscope (CLSM) with 100x oil objective lens.
Protein structural analysis and localization
Predictions of protein localization were performed using neural-network-based predictors for subcellular localization of proteins PCLR (Schein et al. 2001), ChloroP (Emanuelsson et al. 1999) and LOCtree (Nair and Rost 2005). Protein localization was conducted in young grapefruit leaves collected 6 days after infiltration (DAI). GFP fluorescence was observed as mentioned above.
Results
Optimization of leaf age for transient expression in grapefruit
To test the efficiency of the protocol and the constructs, we used AvrGf1 (31::Gf1_5) as a marker to monitor the efficacy of transient expression, and leaves in all growth stages were assessed. The HR was initially visible on 2-week-old (young) grapefruit leaves 3 DAI and became more pronounced at 4 DAI in an infiltrated area of ~0.5 cm2 (Fig. 2a). In contrast to the young leaves, infiltrated mature leaves did not display any visible cell death in any of the experiments or replicates conducted here (Fig. 2b). Wild-type A. tumefaciens strain GV3101, GV3101 carrying the empty vector—pGWB6 (31::WB6), and A. tumefaciens strain AGL-1 expressing pCAMBIA2201 (AG::2201) were similarly tested and used as HR negative controls. When infiltrated in young grapefruit leaves, none of these strains triggered an HR-like symptom 4 DAI (Fig. 2c).
Expression of AvrGf1 in grapefruit leaves using Agrobacterium-mediated transient expression. a Infiltration of Agrobacterium cultures, 31::Gf1_2 and 31::GF1_5, in 2-week-old grapefruit leaf. b Agrobacterium-mediated transient expression in mature leaf. c Phenotype of grapefruit leaves inoculated with A. tumefaciens strain GV3101 (GV3101) and A. tumefaciens carrying an empty vector (AG::2201 and 31::WB6). d AvrGf1 mutants transiently infiltrated in grapefruit leaves. NΔ116 and NΔ13, represents 116 and 13 amino acids deleted from the N-terminal, respectively; CΔ7 and CΔ83, represents 7 and 83 amino acids deleted from the C-terminal, respectively, and AvrGf1 is the expression of the avrGf1 full length. *Leaves were photographed at 4 DAI. Similar results were observed in at least three independent experiments
AvrGf1 requires full N- and C-terminal for hypersensitive response in grapefruit
We analyzed young grapefruit leaves transiently transformed with avrGf1-truncated mutants over a 6-day period following infiltration. First, we analyzed young leaves expressing truncated versions of the N- and C-terminal domains. The mutant AvrGf1ΔN13, carrying a deletion in the first 13 a.a. in the N terminus, and AvrGf1ΔC7, from which 7 a.a. were deleted from the C terminus, did not impair HR elicitation, showing a strong HR 4 DAI (Fig. 2d). We investigated the effects of deleting more amino acids in both N- and C-terminal of AvrGf1. Transient analysis of young grapefruit leaves transformed with either AvrGf1ΔN116 or AvrGf1ΔC83 mutants, where 116 a.a. from the N terminus and 83 from the C terminus were deleted, respectively, nullified the HR (Fig. 2d).
Evaluation of transient expression in grapefruit by visualization of GFP
Leaves were infiltrated with 31::Gf1_5 ranging in size from ~0.5 cm2 of the leaf area to the entire leaf. When the entire leaf was infiltrated, the leaf abscised from the tree between 2 and 3 DAI and the GFP was not visualized in these leaves. Optimal GFP expression was visualized at 6 DAI and using an infiltrated area of ~0.5 cm2 in diameter. Analysis of the images revealed that the GFP fluorescence derived from AvrGf1::GFP strongly accumulated in the stomatal guard cells and less intensely in the surrounding cells (Fig. 3a).
AvrGf1::GFP accumulates in the stomata cells and targets the chloroplasts. Confocal images of abaxial epidermal leaf cells expressing GFP fusion protein 6 days after inoculation. a GFP expression in grapefruit leaves transiently transformed with 35S::GFP construct (first row), and non-infiltrated grapefruit leaf scanning by GFP and Red channels (second row) b AvrGf1::GFP targets the chloroplasts in grapefruit cells. The white arrows indicate the accumulation of GFP in the area of “degraded” chloroplasts
AvrGf1 targets the plant-cell chloroplast
Subcellular localization of AvrGf1 protein transiently expressed in grapefruit leaf was examined. The analysis of the AvrGf1 amino acid sequence by the predictors indicated the presence of a chloroplast transit signal at the N terminus in the first 87 a.a. After infiltration, the abaxial epidermis of the grapefruit leaves was viewed by confocal laser-scanning microscope (CLSM). Infiltrated grapefruit leaves viewed by CLSM clearly showed GFP expression in the stomatal cells (Fig. 3b). Comparing the non-infiltrated leaf with the infiltrated, we observed that the chloroplast was intact in the non-infiltrated leaf (Fig. 3b), while the chloroplasts in the infiltrated leaf were not visible. The lack of intact chloroplasts in infiltrated cells and the strong GFP signal in the stomata, which are abundant for chloroplasts, provides evidence that AvrGf1 indeed targets chloroplasts for destruction.
Discussion
In this study, we present a straightforward Agrobacterium-mediated transient expression method in attached grapefruit leaves, which had not been achieved in previous studies. To develop this method we used the Xcc-Aw T3-effector avrGf1 gene, which triggers HR in grapefruit, as a reporter gene. We also explored the requirements of this HR reaction elicited by AvrGf1 in grapefruit leaves using the transient expression method developed here. Our data demonstrated that when only a few amino acids were deleted from the AvrGf1N- and C-domains, the mutants AvrGf1ΔN13 and AvrGf1ΔC7 induced strong cell death 4 DAI. However, when we increased the number of amino acids deleted in each domain, the mutants AvrGf1ΔN116 and AvrGf1ΔC83 did not trigger HR symptoms, even 10 DAI, suggesting that both domains, N- and C-terminal, are directly involved with HR elicitation in grapefruit leaves.
Analysis of ectopic expression of AvrGf1 through Agrobacterium-transient transformation showed that AvrGf1 induces an apoptosis-like response when expressed in grapefruit cells in the absence of any additional bacterial factor. However, our findings show that transient expression in mature leaves was impaired. Thus, successful application of this method in citrus leaves is strongly related to the use of young leaves. This observation is supported by previous studies, where young leaves have been shown to be more responsive to Agrobacterium-mediated transient transformation (Bhaskar et al. 2009; Li et al. 2009; Folimonov et al. 2007).
Xanthomonas citri subsp. citri strain Aw delivers AvrGf1 via the type III secretion system causing development of strong tissue necrosis in the period of 2–4 DAI (Rybak et al. 2009). Although Duan et al. (1999) successfully demonstrated the phenotype induced by the pthA gene, the symptoms induced by transient expression of pthA were considerably reduced compared with X. citri. In our method, we consistently expressed AvrGf1 in grapefruit, with the HR triggered being indistinguishable from that induced by X. citri—Aw (Rybak et al. 2009).
Based on protein sequence analysis, we determined that the N terminus region of AvrGf1 resembles a chloroplast-targeting signal. Although visualization of the chloroplast in the stomatal cells was simple and straightforward in non-infiltrated leaves, the visualization of intact chloroplasts in infiltrated leaf tissue was difficult due to the HR initiated by the recognition of AvrGf1. The intense fluorescence observed in the guard cells along with the predicted chloroplast localization signal identified by the neural-network, PCLR, ChloroP and LOCtree, strongly support the hypothesis that AvrGf1 carries the chloroplast localization signal and targets them in grapefruit cells. Likewise, several other type III effectors have also been predicted to have N terminus regions carrying chloroplast-target signals (Jelenska et al. 2007; Guttman et al. 2002).
Pseudomonas HopL1 effector, which carries an N terminus chloroplast-targeting signal, is a virulence factor shown to suppress host defenses (Jelenska et al. 2007). Thus, we also predicted that the N terminus should be essential for HR elicitation. Indeed, the N terminus plays an essential role in HR elicitation as shown by the mutant AvrGf1ΔN116 not inducing an HR. In agreement with previous studies demonstrating that the C terminus domain is involved in HR induction (Catanzariti et al. 2010; Abramovitch et al. 2003), deletion of 83 a.a. from AvrGf1 coding sequence, also disrupted the elicitation of HR and GFP expression in grapefruit.
In conclusion, we have demonstrated an Agrobacterium-mediated transient expression method for citrus leaves and its applicability in citrus for characterization of gene function. This technique has been intensively used in our laboratory to evaluate different protein activity in grapefruit leaves through HR assay and β-Glucuronidase activity by fluorometric measurement (unpublished data). The method developed here is a more rapid and efficient alternative to particle bombardment, protoplast transformation, or virus-based vectors for the transient expression of foreign proteins in citrus plants. Moreover, we provide solid evidence that AvrGf1 targets the chloroplast in host cells, and we highlight the importance of the N- and C-termini in this incompatible reaction.
References
Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22:60–69
Ahmad M, Mirza B (2005) An efficient protocol for transient transformation of intact fruit and transgene expression in Citrus. Plant Molecular Biology Reporter 23:419a–419k
An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153:292–305
Bespalhok JCF, Kobayashi AK, Pereira LFP, Galvão RM, Vieira LGE (2003) Transient gene expression of β-glucuronidase in citrus thin epicotyl transversal sections using particle bombardment. Braz Arch Biol Technol 46:1–6
Bhaskar PB, Venkateshwaran M, Wu L, Ane JM, Jiang J (2009) Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS One 6:e5812
Bonas U, Van den Ackerveken G (1999) Gene-for-gene interactions: bacterial avirulence proteins specify plant disease resistance. Curr Opin Microbiol 2:94–98
Brunings AM, Gabriel DW (2003) Xanthomonas citri: breaking the surface. Mol Plant Pathol 4:141–157
Büttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 2:107–133
Catanzariti A, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ et al (2010) The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact 23:49–57
Cheng YQ, Yang J, Xu FP, An LJ, Liu JF, Chen ZW (2009) Transient expression of minimum linear gene cassettes in onion epidermal cells via direct transformation. Appl Biochem Biotechnol 159:739–749
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Duan YP, Castañeda A, Zhao G, Erdos G, Gabriel DW (1999) Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol Plant Microbe Interact 12:556–560
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Folimonov AS, Folimonova SY, Bar-Joseph M, Dawson WO (2007) A stable RNA virus-based vector for citrus trees. Virology 368:205–216
Green BJ, Fujiki M, Mett V, Kaczmarczyk J, Shamloul M, Musiychuk K (2009) Transient protein expression in three Pisum sativum (green pea) varieties. Biotechnol J 4:230–237
Gurlebeck D, Jahn S, Gurlebeck N, Szczesny R, Szurek B, Hahn S (2009) Visualization of novel virulence activities of the Xanthomonas type III effectors AvrBs1, AvrBs3 and AvrBs4. Mol Plant Pathol 10:175–188
Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT (2002) A functional screen for the type III (hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722–1726
Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr Biol 17:499–508
Joensuu JJ, Conley AJ, Lienemann M, Brandle JE, Linder MB, Menassa R (2010) Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol 152:622–633
Johansen LK, Carrington JC (2001) Silencing on the spot induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126:930–938
Jones HD, Doherty A, Wu H (2005) Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods 1:5
Jones HD, Doherty A, Sparks CA (2009) Transient transformation of plants. Methods Mol Biol 513:131–152
Klement Z (1963) Rapid detection of the pathogenicity of phytopathogenic pseudomonads. Nature 199:299–300
Li JF, Park E, von Arnim AG, Nebenfuhr A (2009) The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5:6
Manavella PA, Chan RL (2009) Transient transformation of sunflower leaf discs via an Agrobacterium-mediated method: applications for gene expression and silencing studies. Nat Protoc 4:1699–1707
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nair R, Rost B (2005) Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol 348:85–100
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41
Porta C, Lomonossoff GP (1996) Use of viral replicons for the expression of genes in plants. Mol Biotechnol 5:209–221
Rybak M, Minsavage GV, Stall RE, Jones JB (2009) Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol Plant Microbe Interact 2:249–262
Sambrook J, Fristsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, NY
Santos-Rosa M, Poutaraud A, Merdinoglu D, Mestre P (2008) Development of a transient expression system in grapevine via agro-infiltration. Plant Cell Rep 27:1053–1063
Schein AI, Kissinger JC, Ungar LH (2001) Chloroplast transit peptide prediction: a peek inside the black box. Nucleic Acids Res 29:E82
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1:2019–2025
Van Larebeke N, Engler G, Holsters M, Van den Elsacker S, Zaenen I, Schilperoort RA, Schell J (1974) Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252:169–170
Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22:543–551
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Merkle.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Figueiredo, J.F.L., Römer, P., Lahaye, T. et al. Agrobacterium-mediated transient expression in citrus leaves: a rapid tool for gene expression and functional gene assay. Plant Cell Rep 30, 1339–1345 (2011). https://doi.org/10.1007/s00299-011-1045-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1045-7