Abstract
Natural and artificially induced mutants have provided valuable resources for plant genetic studies and crop improvement. In this study, we investigated the genetic and molecular basis of the purple sheath trait in a somaclonal mutant Z418, which was regenerated from a green sheath rice variety C418 through tissue culture. The purple sheath trait in Z418 was heritable and stable based on our 10 years of evaluation. Genetic analysis revealed that the purple sheath trait of the mutant was controlled by a single dominant gene. To map the gene, we scored 89 polymorphic SSRs markers in a F2 population of 232 plants derived from a cross between Z418 and HX-3, an indica variety with green sheath trait. The gene was initially mapped to the short arm of chromosome 6 between two SSR markers, RPM5 and RM402, with a genetic distance of 1.1 and 10.3 cM, respectively. Thirty-one SSR and indel markers located within the target region were further used to fine-map the gene to a 153-kb interval between two SSR markers (RPM8 and RPM11). The OsC1 gene, which locates within the region and encodes a MYB family transcription factor, was chosen as the candidate gene controlling the purple sheath trait in Z418. Sequencing analysis revealed that OsC1 gene and its transcript in Z418 was 34 bp longer than that in C418. The possible mechanisms for the gene mutation, the developmental and tissue-specific expression of purple anthocyanin pigmentation in Z418, were finally discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somaclonal variation induced by in vitro culture has often been observed in cultured cells and regenerated plants (Larkin and Scocroft 1981; Evans et al. 1984; Jain 2001). Such variation may present as a problem in the process of plant transformation and micropropagation of true-to-type clones, as undesirable mutations may reduce the agronomic value of the cultivars or hinder the incorporation of transgenes into plant genomes (Bao et al. 1996; Bregitzer et al. 1998; Martins et al. 2004; Toki et al. 2006). On the other hand, somaclonal variation leads to the creation of genetic variability, and can generate valuable genotypes to plant breeders. Under the continued pressure to develop sustainable food production, somaclonal mutants have provided valuable genetic resources for crop breeding (Larkin and Scocroft 1981; Evans et al. 1984; Breiman et al. 1987; Rodríguez López et al. 2004). The occurrence, principle and application of somaclonal variation have become the important topics in plant breeding, plant micropropagation as well as theoretical genetic studies.
Somaclonal variation in rice was firstly reported by Oono (1978). Since then, a large variety of somaclonal mutants have been identified, many of them show heritable variation in traits of agronomic importance, including grain quality (Shen et al. 1995; Yang et al. 1996), plant height (Sun et al. 1994; Yamagishi et al. 1996), disease resistance (Ling et al. 1985; Araijo et al. 1998; Hernalatha et al. 1999; Gao et al. 2002), abiotic stress tolerance (Adkins et al. 1995; Bertin and Bouharmont 1997; Sint Jan et al. 1997; Lutts et al. 1999) and other traits (Xie et al. 1995). In rice plants, it has been reported that some endogenous transposons, including Tos17 and mPing, can be activated by tissue culture (Hirochika et al. 1996; Jiang et al. 2003), these elements can be used as insertional mutagens to generate somaclonal mutants. Due to the economic importance and ease of regeneration, rice has become a model system to study the genetic mechanisms of somaclonal variation (Muller et al. 1990; Harada et al. 1991; Hirochika et al. 1996; Oono et al. 1999; Yang et al. 1999; Liu et al. 2004). The recent completion of rice whole genome sequence (International Rice Genome Sequencing Project 2005) and the genome annotation database (Ouyang et al. 2007) have provided unprecedented opportunities for researchers to explore the molecular mechanisms of somaclonal mutation at the genome level.
Utilization of heterosis has been a major strategy for increasing rice productivity. In China, hybrid rice cultivars can yield 10–20% more than conventional ones (Yuan 1998; Cheng et al. 2007). C418 is an elite restorer line with wide compatibility. It can overcome fertility barrier between indica and japonica subspecies and has been widely used to develop indica–japonica hybrid rice (Yang et al. 1998). However, C418 is susceptible to bacterial leaf blight disease. To improve the agronomic performance of C418 and create novel mutants for our rice breeding program, we carried out in vitro tissue culture using C418 as the parental plant. C418 has green leaf sheaths; however, among the regenerated plants, we found one somaclonal mutant (Z418) with purple colored leaf sheaths. The anthocyanin pigmentation in Z418 had a tissue- and developmental time-specific pattern. Genetic analysis demonstrated that the trait was controlled by a single dominant gene. Further mapping and sequencing analysis revealed that a Colorless1/Purple leaf (C1/Pl) like regulatory gene (OsC1) was likely responsible for the purple sheath trait in Z418. The purple sheath trait in Z418 can be used as a morphological marker for hybrid rice breeding; the somaclonal mutant also provides valuable material for future studies of anthocyanin pigmentation regulation in rice.
Materials and methods
Obtaining the somaclonal mutant Z418 by tissue culture
Young spikes (3–5 cm) of a green sheath cultivar C418 (japonica) were inoculated on a nutrient medium comprising Murashige and Skoog (MS) basal medium supplemented with 2 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), 5% sucrose and 7 g/L agarose. Explants were cultured at 25°C in darkness. Four weeks later, induced calli were transferred to a shoot regeneration medium comprising MS medium with 2 mg/L 6-benzylaminopurine (6-BA), 1 mg/L 6-furfurylamino-purine (KT), 3% sucrose and 7 g/L agarose. Cultures were incubated at 25°C under constant white light. The regenerated plants were grown in soil in the greenhouse at the Jiangsu Academy of Agricultural Sciences (JAAS) to harvest seeds. During 1999–2008, seeds from the regenerated plants and their progenies were planted in the experimental field at JAAS for phenotype evaluation. The purple leaf-sheath trait was examined at both seedling and adult stages.
Crossing experiments
To determine the inheritance pattern and genetic basis of the purple sheath trait in Z418, we crossed Z418 with C418 and two indica varieties (93-11 and HX-3) with green leaf sheath. Z418 were used as the pollen parent in all crosses. F1 plants were self-pollinated to produce F2 seeds. F1 and F2 plants were grown in a greenhouse or growth chambers at Michigan State University, with the temperatures at 30°C/25°C (day/night) and a photoperiod at 12:12 (L:D). Since normally the leaf sheath of Z418 begin to turn purple at 6-leaf stage, we evaluated phenotypes of the F2 plants after that stage (at seedling or tiller stage).
Screening and development of SSR markers
A total of 256 simple sequence repeats (SSRs) covering all the 12 rice chromosomes were screened to identify polymorphic makers between the mutant Z418 (japonica) and a green sheath variety HX-3 (indica). These SSR markers were developed by Temnykh et al. (2001) and McCouch et al. (2002), and the sequences are available at http://www.gramene.org. Genomic DNA was extracted from freshly harvested young leaves of Z418 and HX-3 using CTAB method. PCR amplification was performed in a 25 μl volume containing 20 ng of genomic DNA, 1× PCR buffer, 1.5 mM of MgCl2, 0.2 mM of each dNTP, 0.2 μM each of forward and reverse primers and 1.0 unit Taq DNA polymerase. Thermal cycling consisted of an initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 45 s; 55°C or 60°C (depending on the melting temperature of the primers) for 45 s; 72°C for 50 s, and a final extension at 72°C for 5 min. Microsatellite alleles were separated by running the reactions on 3% agarose gels containing 0.5 μg/ml ethidium bromide, and/or on 6% denaturing polyacrylamide gels as described by Gao et al. (2009). To obtain more polymorphic molecular markers for genetic linkage mapping, a total of 29 new SSR and indel (insertion/deletion) markers were developed based on the orthologous sequence differences between Nipponbare (japonica) and 93-11 (indica). These newly developed markers are listed in Supplementary Table 1.
Genetic linkage analysis
Two hundred and thirty-two F2 plants derived from the F1 hybrids of Z418 and HX-3 were used as the mapping population for genetic linkage analysis. Genomic DNA was extracted from the leaves of parental and F2 plants using the CTAB method. Twenty nanogram DNA from each plant was used for the amplification of polymorphic markers in a 25 μl PCR reaction. Linkage analysis and map construction were performed using Mapmaker 3.0 software (Lander et al. 1987). The maximum-likelihood map order for markers was determined with a log of odds (LOD) score threshold of 3.0. The map distances were calculated in centiMorgan (cM) using Kosambi function.
Southern hybridization
Genomic DNA (6 μg each) from Z418, C418, Nipponbare and HX-3 was digested by XbaI or HindIII(New England, Ipswich, MA)at 37°C for 8 h. The digested DNA fragments were separated by electrophoresis on a 1.0% (w/v) agarose gel at 70 V for 5 h, then transferred onto a Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ). A 567-bp fragment (AP005652: 67511–68077) of OsC1 gene was used as probe for the hybridization reaction. The probe was amplified from Z184 genomic DNA (forward primer: 5′-ATCGCTCAGTCTCACACCG-3′; reverse primer: 5′-CGACAGTACCAAATAGCTCC-3′), and labeled with digoxigenin-labeling mix (Roche, Penzberg, Germany). Hybridization was performed using a digoxigenin detection system (Roche, Mannheim, Germany) according to the manufacture’s instructions.
Reverse transcription-polymerase chain reaction (RT-PCR)
To investigate the expression profiles of the candidate gene OsC1, we collected leaves at 4 and 6 leaf stages, sheaths at 5 and 6 leaf stages from both Z418 and C418 for RT-PCR analysis (additional sheath tissues were collected for Z418 at 7-leaf stage). Total RNA was isolated using the TRIZOL Reagent (Invitrogen, Carlsbad, CA). Four microgram total RNA from each sample was converted into single-strand cDNA with reverse transcriptase (Invitrogen, Carlsbad, CA). Reactions were diluted 4- to 5-fold, and 2 μl of the diluted cDNA were used as templates for PCR amplifications. RT-PCR primers were designed based on the full cDNA sequence of OsC1 gene (GenBank accession number: Y15219. Forward primer: 5′-ATCGCTCAGTCTCACACCG-3′, reverse primer: 5′-CGACGAACTAATGTCACG CAC-3′). The rice actin gene was amplified in parallel with the target gene as an internal standard for quantitative comparison of mRNA levels, using actinF (5′-CAAGGCCAATCGTGAGAA-3′) and actinR (5′-AGCAATGCCAGGGAACATAGT-3′) primers.
Comparison of cDNA and genomic DNA sequences of the OsC1 gene between Z418 and the wild-type C418
The same RT-PCR primer pair described above was used to amplify cDNA and the corresponding genomic DNA sequences of the OsC1 gene in both Z418 and C418. PCR products were recovered and purified from 1% agarose gel using the QIA quick Gel Extraction kit (Qiagen, Chatsworth, CA). PCR products were cloned using TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Sequencing reactions were carried out by the Genomics Technology Supporting Facility at Michigan State University. Multiple randomly selected recombinant clones were sequenced for each sample to verify the result.
Phylogenetic analysis of MYB-related genes
R2R3 conserved domains of 121 MYB-related genes, which include 91 MYB genes from O. sativa and 30 MYB-related sequences from other species, were used for the phylogenetic analysis. The 91 O. sativa MYB genes used in this study are described in Supplementary Table 2; other 30 MYB sequences from wild rice species as well as other plants and animals are listed in Supplementary Table 3. Multiple pairwise alignments of the 121 conserved R2R3 sequences were generated using ClustalW program (Larkin et al. 2007) under the default settings. The phylogenetic tree was constructed using neighbor-joining method in MEGA4 (Tamura et al. 2007). The analysis was based on 1,000 bootstrap replicates with complete deletion parameters using the nucleotide: maximum composite likelihood model.
Results
Inheritance and expression pattern of purple pigmentation in the leaf sheath of Z418
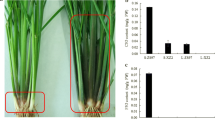
In 1998, young spikes of C418 were cultured to induce somaclonal mutation. Among the 51 regenerated plants, a mutant Z418 displayed purple leaf sheath and good agronomic characters. To evaluate the genetic stability of the purple sheath trait, we planted Z418 and its self-fertilized offspring for 10 successive years (1998–2008). During the period of time, no phenotypic segregation was observed in the plants of the later generations, indicating that the purple sheath trait in Z418 is heritable, and Z418 is homozygous for the trait. The purple pigmentation in Z418 shows a tissue-specific and developmental stage-specific pattern. Z418 has green leaf sheath before 5-leaf stage (Fig. 1a); its sheaths begin to turn purple at around 6-leaf stage (Fig. 1b), and the same color remains thereafter (Fig. 1c). Z418 also displays purple color in stigma (Fig. 1f, g), but not in its spikes and seeds (Fig. 1h–j). In contrast, the original plant C418 has green sheaths and stigmata throughout its life cycle.
The purple sheath in Z418 is controlled by a single dominant gene
All F1 plants from crosses between Z418 and the three green sheath cultivars uniformly exhibited purple color in leaf sheath, indicating dominant inheritance of the trait. In F2 populations, plants from the three crosses segregated for purple and green sheath traits, and the segregation showed a good fit to the 3:1 ratio (Table 1). All these results demonstrate that the purple sheath trait in Z418 is controlled by a single dominant gene.
Molecular mapping of the purple sheath gene in Z418
Among the 256 SSR markers screened for polymorphism between the parental varieties Z418 and HX-3, 89 (34.8%) of the SSR loci were polymorphic. The polymorphic rate of SSR markers on different chromosomes ranged from 19.4 to 60%, with the highest rate on chromosome 9 (60.0%), and the lowest on chromosome 2 (19.4%).
A total of 232 F2 plants derived from the cross Z418/HX-3, including 171 purple sheath and 61 green sheath plants, were used as the mapping population. Bulked segregating analysis was used to map the purple sheath gene in Z418. First, DNAs from 20 green sheath and 20 purple sheath individuals were bulked into two DNA pools and then genotyped with 89 polymorphic SSR markers, 2 SSRs on chromosome 6, RM204 and RM3, did not fit 1:2:1 (P1:F1:P2) segregation pattern, suggesting that the target gene is located on chromosome 6. Subsequently, all 232 F2 individuals were genotyped with 6 polymorphic SSR markers on chromosome 6. Our linkage analysis initially mapped the purple sheath gene on the short arm of rice chromosome 6 between two SSR markers, RPM5 and RM402, with a genetic distance of 1.1 and 10.3 cM, respectively (Fig. 2).
Fine mapping of the target gene
To refine the localization of the purple sheath gene in Z418, we selected 319 F2 green sheath plants derived from Z418/HX-3 cross, and screened for recombinants among these plants using RM204 and RM402 markers. We identified 82 and 65 recombinants based on the genotypes at RM204 and RM402 locus, respectively. These 147 recombinant plants were further used as the material for the fine mapping of the target gene.
Thirty-one SSR markers located between RM204 and RM402 were further genotyped and screened for polymorphism between HX-3 and Z418. These markers include two (RM11 and RM253) of the SSRs reported early and 29 additional SSR or indel markers newly developed in this study (Supplementary Table 1). Among these markers, RM253, RPM8 and RPM11 were polymorphic between the parents. These 3 markers were subsequently used to genotype the 147 F2 recombinant plants which were polymorphic either at RM204 or RM402 locus (described above). Among the 82 F2 recombinants polymorphic at RM204 locus, 4 plants were also polymorphic at RPM8 locus. On the other hand, all the 65 plants polymorphic at RM402 showed homogenous genotype at RPM8. The results indicate that RM204 and RPM8 are located on the same side of the candidate gene. Among the 65 plants polymorphic at RM402 locus, 6 and 3 individuals were also polymorphic at RM253 and RPM11 locus, respectively. Therefore, along with RM402, these two markers were located on the other side of the target gene. As the result of our linkage analysis, the target gene was finally anchored to a 153-kb interval between RPM8 and RPM11 markers (Fig. 3). Based on the Rice Genome Annotation Project Data (http://rice.plantbiology.msu.edu), this region contains a total of 18 non-transposon-related genes (Supplementary Table 4). An expressed gene (Os06g10350) encoding a MYB family transcription factor was chosen as the candidate gene controlling the purple leaf sheath in Z418. Further Blastn searches revealed that this candidate gene has been reported as the rice homolog (OsC1) of maize C1 gene (Reddy et al. 1998, Saitoh et al. 2004), which is a basic regulatory gene required for pigmentation pathway in maize.
Fine mapping of the purple sheath gene. The location of the target gene was narrowed down to a region of approximately 153 kb (spanning 3 BACs: AP004991, AP005652, AP004754) between two SSR markers, RPM8 and RPM11. Eighteen non-transposon-related genes located within the region were shown. The candidate gene Os06g10350 encodes a MYB family transcription factor OsC1. The numbers of recombinant were indicated near the SSR markers
Expression analysis and sequence comparison of OsC1 gene
RT-PCR analysis showed that the OsC1 gene expressed in both leaf and sheath tissues at all the developmental stages tested (leaves at 4 and 6-leaf stages, sheathes at 5 and 6-leaf stages) in both Z418 and C418. However, while all RT-PCR products from C418 appeared as a single band on the 1% agarose gel, all RT-PCR products from Z418 apparently showed two bands (Fig. 4). Further sequencing and alignment analysis showed that the bigger transcript of OsC1 gene in Z418 (GenBank accession number: HQ379703) was 34 bp longer than that in C418 (GenBank accession number: HQ379701). The smaller-sized transcript detected from Z418 contained a 161-bp deletion within the third exon of the OsC1 gene (GenBank accession number: HQ379704), possibly resulted from alternative splicing of OsC1 mRNA. To verify the homozygosity of the OsC1 gene in Z418, three recombinant clones of the PCR amplified gene product were randomly selected for sequencing. These three clones contain a uniform gene sequence, which has a 34-bp indel at the position between 973 and 1006 (GenBank accession number: HQ379705) when compared with the sequence of the same gene from C418 (GenBank accession number: HQ379702). Additionally, nucleotide difference was found in the small flanking region (5 and 6 bp in length) of the indel (Supplementary Fig. 1). The cDNA sequence of OsC1 from Z418 is identical to that has been reported by Saitoh et al. (2004), GenBank accession number: Y15219).
RT-PCR analysis on the expression of OsC1 gene in leaf and sheath tissues collected at different developmental stages of Z418 and C418. 1. Leaf of C418, 4-leaf stage; 2. sheath of C418, 4-leaf stage; 3-4. leaf and sheath of C418, 6-leaf stage; 5. Leaf of Z418, 4-leaf stage; 6. sheath of Z418, 4-leaf stage; 7-8. leaf and sheath of Z418, 6-leaf stage. The rice actin gene was amplified as an internal standard for quantitative comparison of mRNA levels
Amino acid sequences analysis showed that the OsC1 gene in Z418 encodes a 272-amino acid sequence, which is 13 amino acids longer than that in C418 due to the 34-bp indel in the gene sequence (Fig. 5). The amino acid sequence of OsC1 in Z418 shows high similarity to the orthologous gene from Nipponbare and two wild rice species, O. rufipogon and O. glaberrima. Further comparison indicates that OsC1 protein in Z418 shares the conserved R2R3 MYB domain in the N-terminal region with other 8 MYB-related proteins, suggesting the functional importance of these motifs. In contrast, the C-terminal region of these proteins is highly variable in sequence and in size. Surprisingly, a high sequence divergence in the C-terminal region is also present between Z418 and its wild-type C418, which is probably caused by the frameshift of the amino acid sequence due to the indel in the gene (Fig. 5).
Amino acid sequence comparison of OsC1 from Z418, C418 and Nipponbare (rice cultivars) and other MYB protein from different plant species, including MYB-OR from O. rufipogan, MYB-OG from O. glaberrima, C1 and Pl from maize, PAP1 and TT2 from arabidopsis, AN2 from petunia and IpMYB1 from japanese morning glory. Highlighted are the conserved amino acid residues. R2 and R3 repeats of MYB-DNA binding motif were marked. The arrow indicates the location of the lost amino acid residue in Nipponbare
Detection of OsC1 and other MYB-related genes in the rice genome
To examine the copy number of OsC1 gene in the genome of Z418 and other rice plants, a fragment of OsC1 gene was used as the probe to hybrid with either EcoRI or HindIII digested total DNA from Z418 and plants of three rice varieties with green leaf sheath. In all instances a strong hybridization band was detected, which most likely represented the OsC1 gene (Fig. 6). Meanwhile, multiple weak hybridization signals were also detected on the blots, which might represent other MYB-related genes sharing low homology with OsC1.
Southern blot analysis of OsC1 gene from Z418 and 3 rice cultivars with green leaf sheath. Genomic DNA from Z418, C418, Nipponbare and HX-3 was digested by XbaI and HindIII and separated in 1.0% (w/v) agarose gel, then transferred onto a Hybond N+ membrane. A 567-bp digoxigenin-labeled DNA probe, which was prepared by PCR amplification of the coding region of OsC1 gene from genomic DNA of Z184, was used to hybrid with the digested DNA fragments
To investigate the distribution of MYB-related genes in rice genome, R2R3 domain sequences of OsC1 gene in rice and C1 gene in maize were used as queries to search against Genbank and the MSU Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu). A total of 91 MYB-related genes were identified. These genes are distributed across all the 12 rice chromosomes; all but 2 of them contain the conserved and intact R2R3 domains (Supplementary Table 2). These MYB-related genes appear to be clustered on the rice chromosomes. For example, the chromosome 1 contains 19 MYB-related genes, while the chromosome 10 only has 2 MYB genes and both are located on the long arm.
To provide insight into the evolutionary relationship of rice OsC1 gene and other MYB-related genes, the conserved R2R3 protein sequences of 89 MYB-related genes from Oryza sativa ssp. japonica, and 30 MYB-related genes from other species were used to build a phylogenetic tree (Fig. 7). All MYB-related sequences from different animals were grouped into a major family, while all those from plant species were grouped into another big family consisting of five subfamilies. The OsC1 gene of Z418 and two wild rice species, the C1 gene of maize were grouped into to the same clade (II-5), suggesting the relatively recent divergence of these genes.
Discussion
The hypothesis that OsC1 is the homolog of maize C1 gene in rice (OsC1) was first proposed by Saitoh et al. (2004) based on a comparative map between maize and rice. In the same study, it was found that rice lines with colored and colorless apiculus had different alleles of OsC1 gene. Two colorless lines had a 10-bp deletion in the OsC1 gene compared with the colored rice line T65, suggesting a loss-of-function mutation in their OsC1 allele due to the deletion. In our current study, the gene regulating the purple leaf-sheath trait in the somaclonal mutant Z418 is mapped to a 153-kb interval on the short arm of chromosome 6, where OsC1 is located. Sequence analysis reveals that Z418 had the same OsC1 sequence as that in T65, while the original plant C418 and other 3 green sheath varieties have a 34-bp deletion in OsC1 gene compared to Z418. Therefore, our data strongly support the previous hypothesis that OsC1 is the rice homolog of Maize C1, which is the basic regulatory factor required for anthocyanin pigmentation in rice plants. Z418 and C418 have a functional and non-functional (due to the 34-bp deletion) alleles for OsC1 gene, respectively. How did the gain-of-function mutation occur in the OsC1 gene of the original plant (C418) during tissue culture? Further investigation into this question would increase our understanding of the genetic basis for somaclonal variation in rice plants.
Possible mechanisms for the mutation in the OsC1 gene of C418
The genetic mechanisms of somaclonal variation in plants are still not well understood. A variety of genetic changes have been reported to be associated with culture-induced phenotypic variation in plants, including polyploidy, aneuploidy, chromosome rearrangement (Larkin and Scocroft 1981; Lee and Phillips 1988; Jain 2001), point mutation (Dennis et al. 1987; Yang et al. 1996), DNA deletion (Harada et al. 1991), epigenetic modifications (Muller et al. 1990; Kubis et al. 2003), and transposon insertion (Hirochika et al. 1996; Courtial et al. 2001; Jiang et al. 2003; Liu et al. 2004). Our previous data suggests that multiple molecular mechanisms are responsible for somaclonal variation in rice (Gao et al. 2009). In this study, it is unclear how tissue culture has induced a 34-bp insertion in the OsC1 gene of Z418. Here, we propose three possible mechanisms for future investigations.
First, the mutation in OsC1 gene may result from homologous recombination mediated by a transposon. In this study, we found 91 MYB-related genes in the genome of Nipponbare (Oryza sativa L. ssp. japonica), and among them 6 genes were on the same chromosome where OsC1 gene is located (chromosome 6). Therefore, it is possible that Z418 has obtained the 34-bp sequence from another MYB gene via homologous recombination during tissue culture. It has been well documented in maize that transposon Ac can induce homologous recombination of pericarp color1 (P1) gene (Athma and Peterson 1991). Transposons contribute 35% of the rice genome (International Rice Genome Sequencing Project 2005), and several rice transposons have been reported to be active (Hirochika et al. 1996; Jiang et al. 2003; Fujino et al. 2005; Tsugane et al. 2006). It is likely that a transposon in rice was activated by tissue culture, and the activated transposon mediated the homologous recombination between genes of MYB family. A second possibility is that the insertion was caused by a transposition event. Although in many cases mutation in regulatory genes was caused by deletions (Nesi et al. 2001; Sweeney et al. 2006; Furukawa et al. 2006), it is also reported to be associated with insertions of transposons. For example, the insertion of a 7-bp nucleotide sequence has caused the mutation of the regulatory gene InWDR1 in Japanese morning glory (Morita et al. 2006). Also, there are evidences that transposons can capture gene fragments (Jiang et al. 2004). Since transposon insertion has been considered as an important cause of genetic mutations, we cannot rule out the possibility that it may play a role in the mutation of OsC1 gene. As our last hypothesis, the 34-bp sequence insertion might belong to extra-genomic DNA sequences (Lolle et al. 2005) and has been integrated into the OsC1 locus during tissue culture.
Developmental time- and tissue-specific pattern of purple pigmentation in Z418
Spatial and temporal expression of regulatory genes has been reported to be associated with the anthocyanin deposition in plant tissues (Procissi et al. 1997). For instance, the expression of the transparent test 2 (TT2) gene in Arabidopsis, which is responsible for proanthocyanidin accumulation in the endothelium, is restricted to the seed at early stage of embryogenesis (Nesi et al. 2001). In maize, different alleles of the B gene cause tissue- and developmental time-specific patterns of anthocyanin synthesis in plant and seed tissues, and the level of B gene expression correlates with the accumulation of anthocyanins in different tissues (Radicella et al. 1992). Transcriptional control has been suggested to responsible for the expression of the regulatory genes in specific tissues (Damiani and Wessler 1993; Mol et al. 1998; Selinger and Chandler 1999; Zhang and Peterson 2005). In addition, expression of regulatory genes can also subject to post-transcriptional control (Pairoba and Walbot 2003) or epigenetic modification (Cocciolone et al. 2001; Rhee et al. 2010).
The developmental time- and tissue-specific pattern of purple coloration in Z418 suggests that a complex mechanism is involved in the regulation of the pigmentation pathway. Some recent data has indicated that besides OsC1 gene, one or more genes may involved in the regulation of the purple pigmentation in rice plants. Yue et al. (2006) have mapped three main-effect QTLs and a pair of epistatic QTL for the purple leaf sheath coloration in rice plants. In another study, Wang et al. (2009) identified a major dominant gene PSH1 (t) controlling the purple leaf sheath in a RIL line, and mapped it to a 23.5-kb interval on chromosome 1. Further cloning and functional analysis of these QTLs and candidate genes will enhance our understanding of regulation in pigmentation pathway, and how OsC1 gene has interacted with other regulatory gene(s) to control the time- and tissue-specific pigmentation in rice.
References
Adkins SW, Kunanuvatchaidach R, Godwin ID (1995) Somaclonal variation in rice—drought tolerance and other agronomic characters. Aust J Bot 43:201–209
Araijo LG, Prabhu AS, Freire AB (1998) Variation for rice blast resistance in early somaclonal generations derived from immature panicles. Pesqui Agropecu Bras. 33:1349–1359
Athma P, Peterson T (1991) Ac induces homologous recombination at the maize P locus. Genetics. 128:163–173
Bao PH, Granata S, Castiglione S, Wang G, Giordani C, Cuzzoni E, Damiani G, Bandi C, Datta SK, Datta K, Potrykus I, Callegarin A, Sala F (1996) Evidence for genomic changes in transgenic rice (Oryza sativa L.) recovered from protoplasts. Transgenic Res 5:97–103
Bertin P, Bouharmont J (1997) Use of somaclonal variation and in vitro selection for chilling tolerance improvement in rice. Euphycita. 96:135–142
Bregitzer P, Halbert SE, Lemaux PG (1998) Somaclonal variation in the progeny of transgenic barley. Theor Appl Genet 96:421–425
Breiman A, Rotem-Abarbanel D, Karp A, Shaskin H (1987) Heritable somaclonal variation in wild barley (Hordeum spontaneum). Theor Appl Genet 74:1432–2242
Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY (2007) Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot 100:959–966
Cocciolone SM, Chopra S, Flint-Garcia SA, McMullen MD, Peterson T (2001) Specific patterns of a maize Myb transcription factor are epigenetically regulated. Plant J. 27:467–478
Courtial B, Feuerbach F, Eberhard S, Rohmer L, Chiapello H, Camilleri C, Lucas H (2001) Tnt1 transposition events are induced by in vitro transformation of Arabidopsis thaliana and transposed copies integrate into genes. Mol Genet Genomics. 265:32–42
Damiani RD Jr, Wessler SR (1993) An upstream open reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc Natl Acad Sci USA 90:8244–8248
Dennis ES, Brettell RS, Peacock WJ (1987) A tissue culture induced Adh1 null mutant of maize results from a single base change. Mol Gen Genet 210:181–183
Evans DA, Sharp WR, Medina-Filho HP (1984) Somaclonal and gametoclonal variation. Am J Bot 71:759–774
Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Gen Genet 273:150–157
Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Takamure I, Kadowak K (2006) The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49:91–102
Gao DY, Xu ZG, Chen ZY, Sun LH, Sun QM, Lu F, Hu BS, Liu YF (2002) Identification of a resistance gene to bacterial blight (Xanthomonas oryzae pv. oryzae) in a somaclonal mutant HX-3 of indica rice. Yi Chuan Xue Bao 29:138–143
Gao DY, Vallejo VA, He B, Gai YC, Sun LH (2009) Detection of DNA changes in somaclonal mutants of rice using SSR markers and transposon display. Plant Cell Tiss Organ Cult 98:187–196
Harada T, Sato T, Asaka D, Matsukawa I (1991) Large-scale deletions of rice plastid DNA in anther culture. Theor Appl Genet 81:157–161
Hernalatha RG, Jebaraj S, , Raja JAJ, Raguchander T, Ramanathan A, Sarniyappan R, Balasubrarnanian P (1999) Employing a crude toxin preparation from Sarocladium oryzae as a molecular sieve to select sheath rot-resistant somaclones of rice. J Plant Biochem Biotechnol 8:75–80
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93:7783–7788
Jain SM (2001) Tissue culture-derived variation in crop improvement. Euphytica 118:153–166
Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431:567–569
Kubis SE, Castilho AM, Vershinin AV, Heslop-Harrison JS (2003) Retroelements, transposons and methylation status in the genome of oil palm (Elaeis guineensis) and the relationship to somaclonal variation. Plant Mol Biol 52:69–79
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Larkin PJ, Scocroft WR (1981) Somaclonal variation: a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:197–214
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Ann Rev. Plant Physiol Plant Mol Biol 39:413–437
Ling DH, Vidhyaseharan P, Borromeo ES, Zapata FJ, Mew TW (1985) In vitro screening of rice germplasm for resistance to brown spot disease using phytotoxin. Theor Appl Genet 71:133–135
Liu ZL, Han FP, Tan M, Shan XH, Dong YZ, Wang XZ, Fedak G, Hao S, Liu B (2004) Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theor Appl Genet 109:200–209
Lolle SJ, Victor JL, Young JM, Pruitt RE (2005) Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434:505–509
Lutts S, Bouharmont J, Kinet JM (1999) Physiological characterisation of salt-resistant rice (Oryza sativa) somaclones. Aust J Bot 47:835–849
Martins M, Sarmento D, Oliveira MM (2004) Genetic stability of micropropagated almond plantlets as assessed by RAPD and ISSR markers. Plant Cell Rep 23:492–496
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47:457–470
Muller E, Brown PTH, Harke S, Lorz H (1990) DNA variation in tissue-culture-derived rice plants. Theor Appl Genet 80:673–679
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Oono K (1978) Test tube breeding of rice by tissue culture. Trop Agric Res Ser 11:109–123
Oono D, Niizeki M, Senda M, Ishikawa R, Akada S, Harada T (1999) An analysis of somaclonal variation in progenies regenerated from rice calli. Rice Genet Newslett 16:81–83
Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35:D883–D887
Pairoba CF, Walbot V (2003) Post-transcriptional regulation of expression of the Bronze2 gene of Zea mays L. Plant Mol Biol 53:75–86
Procissi A, Dolfini S, Ronchi A, Tonelli C (1997) Light-dependent spatial and temporal expression of regulatory genes in developing maize seeds. Plant Cell 9:1547–1557
Project International Rice Genome Sequencing (2005) The map-based sequence of the rice genome. Nature 436:793–800
Radicella JP, Brown D, Tolar LA, Chandler VL (1992) Allelic diversity of the maize B regulatory gene: different leader and promoter sequences of two B alleles determine distinct tissue specificities of anthocyanin production. Genes Dev 6:2152–2164
Reddy VS, Scheffler BE, Wienand U, Wessler SR, Reddy AR (1998) Cloning and characterization of the rice homologue of the maize C1 anthocyanin regulatory gene. Plant Mol Biol 36:497–498
Rhee Y, Sekhon RS, Chopra S, Kaeppler S (2010) Tissue culture-induced novel epialleles of a Myb transcription factor encoded by pericarp color1 in maize. Genetics (online)
Rodríguez López CM, Wetten AC, Wilkinson MJ (2004) Detection and quantification of in vitro-culture induced chimerism using simple sequence repeat (SSR) analysis in Theobroma cacao (L.). Theor Appl Genet 110:157–166
Saitoh K, Onishi K, Mikami I, Thidar K, Sano Y (2004) Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics. 168:997–1007
Selinger DA, Chandler V (1999) Major recent and independent changes in levels and patterns of expression have occurred at the b gene, a regulatory locus in maize. Proc Natl Acad Sci USA 96:15007–15012
Shen YW, Cai QH, Gao MW, Liang ZQ (1995) Isolation and genetic characterization of somaclonal mutants with large-sized grain in rice. Cereal Res Commun 23:235–241
Sint Jan V, Van Costa, de Macedo C, Kinet JM, Bouharmont J (1997) Selection of Al-resistance plants from a sensitive rice cultivar, using somaclonal variation, in vitro and hydroponic cultures. Euphytica 97:303–310
Sun LH, Wang YF, Jiang N, Li HB (1994) A recessive tall culm somatic mutant with wide compatibility in rice (Oryza sativa L.). Acta Genet Sin 21:67–73
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix–loop–helix protein conditioning red pericarp in rice. Plant Cell. 18:283–294
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:152–1441
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47:969–976
Tsugane K, Maekawa M, Takagi K, Takahara H, Qian Q, Eun CH, Iida S (2006) An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J. 45:46–57
Wang WY, Ding HF, Li GX, Jiang MS, Li RF, Liu X, Zhang Y, Yao FY (2009) Delimitation of the PSH1(t) gene for rice purple leaf sheath to a 23.5 kb DNA fragment. Genome 52:268–274
Xie QJ, Rush MC, Linscombe SD (1995) Inheritance of homozygous variation in rice. Crop Sci 36:1491–1495
Yamagishi M, Shimada T, Niizeki H, Otani M, Koda T, Handa T, Takamura Y, Shimizu E, Higashi M, Nakamura K, Arakawa K, Matsumoto N (1996) Gametoclonal variation in anther culture-derived rice plants: mutation breeding of new elite lines showing short culm and early heading. J Genet Breed 50:269–275
Yang CD, Zhuang JY, Zhao CZ, Qian HR, Wu LB, Zheng KL (1996) Studies on the differences between tissue culture variety Heizhenmi and its donor by conventional and RFLP analysis. Acta Agron Sin. 22:688–692
Yang ZY, Zhang ZX, Zhao YC, Gao Y (1998) Development of a japonica restorer line with wide compatibility C418 and its characters. Hybrid Rice 13:31–34
Yang H, Tabei Y, Kamad H, Kayano T, Takaiwa F (1999) Detection of somaclonal variation in cultured rice cells using digoxigenin based random amplified polymorphic DNA. Plant Cell Rep 18:520–526
Yuan LP (1998) Hybrid rice breeding in china. In: Virmani SS, Siddiq EA, Muralidharan K (eds) Advances in hybrid rice technology. International Rice Research Institute, Los Banos, pp 27–33
Yue B, Cui KH, Yu SB, Xue WY, Luo LJ, Xing YZ (2006) Molecular marker-assisted dissection of quantitative trait loci for seven morphological traits in Rice (Oryza sativa L.). Euphytica 150:131–139
Zhang F, Peterson T (2005) Comparisons of maize pericarp color1 alleles reveal paralogous gene recombination and an organ-specific enhancer region. Plant Cell 903–914
Acknowledgments
We are grateful to Dr. Ning Jiang at Michigan State University for her kind support on this study and to two anonymous reviewers for their valuable and critical comments. We also thank Dr. Chunhua Zhang for her valuable comments on the manuscript. This research was partly funded by a grant from National Natural Science Foundation of China (30471066) and the Scholarship for Overseas Training Program from the government of Jiangsu province in China to D.G.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W. Harwood.
A contribution to the Special Issue: Plant Biotechnology in Support of the Millennium Development Goals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, D., He, B., Zhou, Y. et al. Genetic and molecular analysis of a purple sheath somaclonal mutant in japonica rice. Plant Cell Rep 30, 901–911 (2011). https://doi.org/10.1007/s00299-011-1004-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1004-3