Abstract
A number of genes that function in the terpenoid indole alkaloids (TIAs) biosynthesis pathway have been identified in Catharanthus roseus. Except for the geraniol 10-hydroxylase (G10H) gene, which encodes a cytochrome P450 monooxygenase, several of these genes are up-regulated by ORCA3, a jasmonate-responsive APETALA2 (AP2)-domain transcript factor. In this study, the G10H gene was transformed independently, or co-transformed with ORCA3 into C. roseus, using Agrobacterium rhizogenes MSU440. Hairy root clones expressing the G10H gene alone, or both the G10H and ORCA3 genes, were obtained. Alkaloid accumulation level analyses showed that all transgenic clones accumulated more catharanthine, with the highest accumulation level in the transgenic clone OG12 (6.5-fold higher than that of the non-expression clone). Following treatment with ABA, accumulation of catharanthine reached 1.96 mg/g DW in the transgenic clone OG12. The expression levels of TIAs biosynthesis genes in transgenic and non-transgenic clones were also investigated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an important medicinal plant, Catharanthus roseus (Madagascar periwinkle) produces a large amount of terpenoid indole alkaloids (TIAs). Among them, vinblastine and vincristine are important antitumor bisindole alkaloids. However, these two anticancer compounds are produced at a very low level in C. roseus leaves, about 5.8 μg/g for vinblastine and 0.9 μg/g (fresh weight) for vincristine, leading to their high price in the market (Favretto et al. 2001). Biotechnological methods may provide an efficient alternative for producing natural products since a number of genes involved in the TIAs’ biosynthetic pathway have been cloned (Pasquali et al. 2006). The multistep TIAs’ biosynthetic pathway is not only quite complex but also strictly regulated by some transcriptional factors (Da Costa e Silva et al. 1993; Pauw et al. 2004; van der Fits and Memelink 2000).
Due to the genetic and biochemical stability and fast growing character in a medium consisting of sucrose and simple salts, hairy root culture has been used as research material for secondary metabolites in vitro. Recently, successful efforts have been made using hairy root cultures to improve secondary metabolism compounds in Hyoscyamus niger (Zhang et al. 2004) and p-hydroxybenzoic acid (pHBA) glucose ester production in hairy roots of Beta vulgaris (Rahman et al. 2009), express foreign proteins or vaccine in tobacco (Shadwick and Doran 2007; Woods et al. 2008), and produce “unnatural” products in C. roseus (Runguphan and O’Connor 2009). Several TIAs’ biosynthesis genes have also been overexpressed in C. roseus hairy root cultures. Ectopic overexpression of anthranilate synthase holoenzyme (ASαβ) increased the amount of tryptophan, and resulted in the accumulation of several phenolic compounds (Chung et al. 2007). In the TDC-overexpressing hairy root line, serpentine accumulation was increased as much as 129% (Hughes et al. 2004). Co-transformation of ASαβ and TDC genes in C. roseus hairy root cultures remarkably increased the production of both tryptamine and alkaloids (Hong et al. 2006). Very recently the DAT gene, which is responsible for the terminal step of vindoline biosynthesis in C. roseus, was overexpressed in C. roseus hairy roots. Interestingly, overexpression of DAT did not increase vindoline production but improved the accumulation of another monoterpenoid indole alkaloid, hörhammericine (Magnotta et al. 2007).
Previously, ORCA3, a jasmonate-responsive AP2-domain regulator, enhanced the expression of several metabolite biosynthetic genes, such as CPR, TDC, and STR, and increased the accumulation of TIAs consequently in ORCA3-tagged C. roseus cell line (Van der Fits and Memelink 2000; Fig. 1). Whereas the transcription of G10H was not regulated by ORCA3 (Van der Fits and Memelink 2000). G10H encodes a cytochrome P450 monooxygenase, which hydroxylates geraniol to form 10-hydroxy-geraniol, and thus controls the first committed step in the biosynthesis of secologanin and TIAs (Collu et al. 2001). Further research revealed that the G10H promoter contains unique binding sites of several transcriptional factors, suggesting that the G10H promoter may be regulated by a different transcriptional cascade (Suttipanta et al. 2007).
Biosynthesis of TIAs in C. roseus. Solid arrows indicate single-step enzymatic conversions, whereas broken arrows indicate multiple-step reactions. AS anthranilate synthase, TDC tryptophan decarboxylase, G10H geraniol 10-hydroxylase, CPR cytochrome P450-reductase, SLS secologanin synthase, STR strictosidine synthase, SGD strictosidine β-d-glucosidase, D4H desacetoxyvindoline 4-hydroxylase, and DAT deacetylvindoline 4-O-acetyl transferase. Genes up-regulated by ORCA3 are underlined. (revised from van der Fits and Memelink 2000)
Although several alkaloids have been successfully produced in C. roseus hairy roots, no study on improving catharanthine, one of the most important medical alkaloids, has been reported. In this study, a construct harboring the G10H gene driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter, or a combined construct harboring both the G10H driven by the CaMV 35S promoter and the ORCA3 gene driven by the stress-inducible rd29A promoter from Arabidopsis (Yamaguchi-Shinozaki and Shinozaki 1994), was introduced into C. roseus hairy roots. The accumulation of four alkaloids, vindoline, catharanthine, vinblastine, vincristine, and the transcriptional levels of several TIA biosynthesis genes are investigated in the engineered hairy roots.
Materials and methods
Construction of plant expression vectors
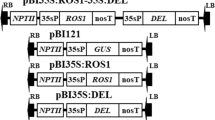
Plasmids pGEMT-G10H and pGEM-T easy-ORCA3, carrying C. roseus G10H and ORCA3 genes, respectively, were kindly provided by Professor J. Memelink. The G10H cDNA was isolated from pGEMT-G10H and inserted into the NcoI and PmaCI sites in pCAMBIA1301 (http://www.cambia.org) to generate the plant expression vector pG10H (Fig. 2a). To construct the pOG vector which carries both the G10H and the ORCA3 genes, the stress/ABA-inducible rd29A promoter was cloned from Arabidopsis using the rd29A forward (5′-CCGGTACCCGACTCAAA ACAAACTTACG-3′) and reverse (5′-C GCCGCGGAATCAAACCCTTTA TTCCTG-3′) primers. The 824 bp PCR product was cloned into pUC19 for sequence confirmation. This intermediate plasmid was named pUC19-Prd29A. The ORCA3 gene was excised from pGEM-T easy-ORCA3 and cloned into pUC19-Prd29A. Then, the Prd29A-ORCA3 fragment was inserted just upstream of the OCS terminator in p1301P (via KpnI and PstI), a modified pCAMBIA1301 vector with a CaMV 35S promoter and an OCS terminator introduced into the multicloning sites. The whole expression cassette (Prd29A-ORCA3-OCS) was transferred (as a KpnI-HindIII fragment) onto pCAMBIA1301 to form a new intermediate plasmid pORCA3. Finally, the plant expression vector pOG was generated by replacing the β-glucuronidase (GUS) gene with the G10H fragment excised from the plasmid pG10H. The plant expression vectors (pG10H and pOG) were transformed into A. rhizogenes MSU440 as described previously (Wise et al. 2006).
Plant expression constructs and molecular analyses of transgenic hairy root lines. a Schematic map of pG10H and pOG constructs used to transform C. roseus. PCR analyses for the presence of rolB and rolC genes (b), G10H gene (c) and ORCA3 gene (d) in different hairy root lines. M λ/EcoT14 I digested DNA marker; P positive control; N negative control; M3, M5 control hairy root lines; GH2, GH3 hairy root lines transformed with pG10H; OG10 OG11, OG12 and OG13 hairy root lines transformed with pOG. e Expression analyses of G10H and ORCA3 in transgenic lines and control hairy root lines under different treatments. RT-PCR assays were performed and the RPS9 gene was analyzed to serve as a quantitative internal control. CK control; ABA hairy roots treated with 100 μmol/l ABA for 24 h; Cold, hairy roots cultured at 4°C for 24 h
Plant transformation and hairy root cultivation
Plant materials (C. roseus L. G. Don) were grown in the greenhouse under a 16 h light/8 h dark photoperiod (~1,450 μmol m−2 s−1), with 50 ± 10% relative humidity. The temperatures were set to 26°C at daytime and 21°C at night. The Agrobacterium rhizogenes strain MSU440 harboring pG10H or pOG was used for C. roseus transformation as described previously (Hughes et al. 2002). Hairy roots formed at the infected site in about 3 weeks after inoculation. The hairy roots were excised from the plant when they were 2–4 cm in length, and transferred onto hormone-free, half-strength B5 (1/2 B5) solid medium supplemented with 30 g/l sucrose, 200 mg/l timentin (GlaxoSmithKline, China), and 10 mg/l hygromycin (Gamborg et al. 1968). All hairy root cultures were grown on petri dishes in the dark at 26°C. The rapidly growing hygromycin-resistant lines were used to establish hairy root lines. Root tips 2–4 cm in length of well-established hairy root lines were transferred to fresh solid media without hygromycin every 6 weeks. Wild-type hairy root lines formed after infection with the A. rhizogenes strain MSU440 were established to be used as control.
PCR analysis
Genomic DNA was extracted from control and transformed hairy roots with 400 μl of homogenizing buffer (250 mM NaCl, 25 mM EDTA, 0.5% SDS, 200 mM Tris–HCl, pH 7.4). The supernatant of a 10,000g centrifugation was mixed with 300 μl of isopropanol. DNA was collected at the interface, washed, and re-suspended in double-distilled water. The quality and the concentration of the extracted DNA were confirmed using a 0.8% (w/v) agarose gel. To detect the presence of Agrobacterium rol (B, C), G10H and ORCA3 genes in the hairy root cultures, PCR analyses were performed. The primers for rol B and rol C gene detection are as follows: rolB forward (5′-CGAGGGGATCCGATT TGCTTT-3′), rolB reverse (5′-GACGCCCTCCTCGCCTTCCT-3′), rolC forward (5′-TC GCCATGCCTCACCAACTCAC-3′), and rolC reverse (5′-CCTTGATCGAGCCGGGT GAGAA-3′). Primers for G10H and ORCA3 gene detection are 35S-F (5′-TTCGCAAGACCCTTCCTC-3′) and G10H-R (5′-CCGCTTCTCCGCTCTGGCTA-3′), and Prd29A-F and ORCA3-R (5′-CGTCGTAGAAGGCTCCGCAGG-3′), respectively. For PCR reaction, an aliquot of 10 ng of DNA was combined with 2.5 μl of 10× buffer, 1.5 μl of 15 mM MgCl2, 1 U of Taq polymerase (Sangon, Shanghai, China), 0.5 μl of 2.5 mM dNTP and 2 μl of 10 mM each primer. PCR programs were set as following: 3 min at 94°C, then 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min and holding at 4°C. Five microliters of PCR products were visualized on 0.8% (w/v) agarose gel.
Abscisic acid (ABA) or cold treatment
Hairy roots from each line were divided into three groups: one was for cold treatment, the second for ABA treatment, and the third was used as control. About 300 mg (fresh weight, FW) hairy roots of each line were transferred into a 250 ml culture bottle containing 30 ml 1/2 B5 liquid media. For cold treatment, cultures were kept at 4°C for 24 h in the dark, and the control group was cultured at 26°C for 24 h in the dark without shaking. For ABA treatment, hairy root cultures were grown under the same conditions as control, but the liquid media were supplement with 100 μmol/l ABA (Sigma).
RT-PCR analysis of gene expression
Total RNA was isolated from the hairy roots of each line grown under different conditions (control, ABA or cold treatment) using the RNAiso reagent (TaKaRa, Japan), and then digested with RNase-Free DNase (Promega). cDNA was prepared from 2 μg total RNA per reaction using ReverTra Ace (TOYOBO, Japan). Expression levels of G10H and ORCA3 in transgenic hairy root lines were analyzed using primers G10H-F (5′-CCATCGCCATTGCCGTTCAT-3′) and G10H-R, and ORCA3-F (5′-G GCGGCCGGAGGAGGTTGTTC-3′) and ORCA3-R, respectively. The expression level of RPS9 (40S ribosomal protein S9) was investigated with primers RPS9-F (5′-AGCGCCGCCTTCAAACCCTT-3′) and RPS9-R (5′-GCCAGGACGACCACCACCGA-3′) to serve as a quantitative internal control.
Alkaloid extraction and HPLC analysis
Lyophilized hairy roots, 100 mg (dry weight, DW) of each hairy root line, were weighted and used for the alkaloid extraction as described previously (Tikhomiroff and Jolicoeur 2002). For HPLC analysis, individual stock solutions of standard samples, catharanthine, vindoline (Aladdin-Reagent, China), vinblastine, and vincristine (Sigma), were prepared at a concentration of 5 mg/ml in methanol, and stored at −20°C. The stock solutions were diluted in methanol at different concentrations (0.5–20 μg/ml for vindoline, vinblastine and vincristine, and 10–200 μg/ml for catharanthine). The HPLC analysis was performed using an Agilent 1100 Series HPLC System. A Zorbax Eclipse XDB-C18 (150 mm × 4.6 μm) column, coupled with a 1 cm × 4.3 mm ODS guard column, was used at a column temperature of 35°C. The injection volume was 10 μl. The mobile phase and eluent profile were performed as described earlier (Tikhomiroff and Jolicoeur 2002). The flow-rate was 0.9 ml/min and the DAD detection wavelength was 220 nm. The peaks’ purity was determined using the Agilent ChemStation A09.01 software. Quantification analysis was repeated for three replicates of each line in parallel, and the means and standard deviations were calculated.
RT-PCR analyses
The transcripts of several marker genes involved in C. roseus TIAs’ biosynthesis such as, ASα, CPR, SLS, TDC, STR, D4H and DAT, were analyzed using the following primers: ASα-F: 5′-GGCGGCGAAGCATGGGAACT-3′, ASα-R: 5′-CCTCGGTCCGCTGG GG TTTC-3′; CPR-F: 5′-TCGGCCCTGGTACTGGACTAGC-3′, CPR-R: 5′-TGGCATCACCGCAGACGTAAAC-3′; SLS-F: 5′-CTGCCAGCTTTTGCCATATGC-3′, SLS-R: 5′-CCAGCCTTCAAAGCCTTCATTC-3′; TDC-F: 5′-TCGGCATCTCACCTCAAGTTCT-3′, TDC-R: 5′-TCGGGACATATACAGGCGCTT-3′; STR-F: 5′-TTCGCCTACGCATCTCCCTTCT-3′, STR-R: 5′-CCGCCGGGAACATGTAGCTCTT-3′; D4H-F: 5′-TGGCGGCCTTCAAATTCTTCTT-3′, D4H-R: 5′-TCCGTACAATCTTGGCGAAACC-3′; DAT-F: 5′-CGGAGGTTTGGCGGTTGCTT-3′ and DAT-R: 5′-CCGTGGCACATCGACTGAGAAA-3′. The expression levels of G10H and ORCA3 were also analyzed at the same time. The RPS9 served as a quantitative internal control.
Statistical analysis
All data in this work were obtained from at least three independent experiments with three replicates each. For data analyzed with the Student’s t test, the difference between treatments was considered as significant when P < 0.05, 0.01 or 0.001 in a two-tailed analysis.
Results and discussion
Generation of transgenic hairy root lines
The G10H gene alone or integrated with ORCA3 gene was introduced into C. roseus. Hairy root lines resistant to hygromycin were selected. A total of 25 hygromycin-resistant hairy root lines transformed with pG10H (GH) and 38 hygromycin-resistant hairy root lines transformed with pOG (OG) were obtained, of which 12 GH and 9 OG lines survived the subsequent selection process (three to five rounds of selection). Two GH lines (GH2 and GH3) and three OG lines (OG10, OG12 and OG13), which showed fast growth were chosen for further examinations. As control, two wild-type hairy root lines (M3 and M5) were also generated following inoculation with MSU440, which contains neither pG10H nor pOG.
Molecular analysis of genetically engineered hairy roots
PCR analysis of Agrobacterium rol genes revealed that both control and transgenic hairy roots contained the rolB and rolC genes (Fig. 2b). To eliminate the interference from C. roseus endogenous genes, both forward primers for the integration confirmation of G10H and ORCA3 into the transformed hairy root genome land on the promoter sequences. PCR analyses confirmed the integration of G10H and/or ORCA3 in all the tested transgenic lines (Fig. 2c, d).
To investigate the expression of the introduced G10H and ORCA3 genes, total RNA was isolated from the generated hairy root lines following different treatments. We observed that G10H, which is driven by the constitutive CaMV 35S promoter, was overexpressed in all transgenic root lines (Fig. 2e); the expression of G10H was affected by ABA or cold treatment. As shown in Fig. 2e, the expression level of G10H increased in transgenic lines GH2 and GH3 following ABA or cold treatment. The transcriptional factor gene ORCA3, which is promoted by the stress-inducible promoter Prd29A, was also overexpressed in all the examined transgenic hairy root lines, especially following ABA or cold treatment (Fig. 2e). However, the transcripts of ORCA3 were only detectable following cold treatment in the control hairy root line (M3), implying that the rd29A promoter from Arabidopsis was sensitive to environmental stress in the C. roseus hairy roots. ABA treatment increased ORCA3 expression in OG13 whereas it decreased the expression slightly in OG10. In all hairy root lines, including M3, ORCA3 was expressed at a comparatively higher level following cold treatment, suggesting that the transcripts of endogenous ORCA3 gene could be induced by low temperature.
As a jasmonate-responsive transcriptional factor, transcription of ORCA3 can be activated by jasmonic acid, a plant hormone, which acts as an intermediate signal response to stress, and overexpression of ORCA3 led to increased accumulation of terpenoid indole alkaloids (Van der Fits and Memelink 2000). The Arabidopsis rd29A gene can be induced by ABA and abiotic stress such as low temperature, dehydration, and high salinity (Yamaguchi-Shinozaki and Shinozaki 1994). Thus the Rd29A promoter has been successfully used to promote inducible overexpression of different transcript factors in tobacco (Yamaguchi-Shinozaki and Shinozaki 1993), potato (Behnam et al. 2007; Pino et al. 2007) and peanut (Bhatnagar-Mathur et al. 2007). When compared with other constitutive promoters, it has fewer negative phenotype effects on transgenic plants (Pino et al. 2007). In the preliminary experiments, we transformed the GUS reporter gene driven by the rd29A promoter into C. roseus hairy roots, and confirmed the inducible expression of the GUS gene following ABA or cold treatment by histochemical dye staining (data not shown). The above results showed that the rd29A promoter can drive the overexpression of ORCA3 following ABA or cold treatment. The different expression patterns of ORCA3 in the OG lines, cultured under different conditions, (CK, ABA or cold) may be due to the effects of insertion sites where the heterologous gene was integrated or other unknown reasons.
Analysis of alkaloid concentration
The profiles of four kinds of alkaloids (catharanthine, vindoline, vinblastine, and vincristine) in both control and transgenic hairy root lines were determined by HPLC. Identification of alkaloids from the hairy roots’ extract was established by comparison of the retention time and the UV spectra with those of authentic standards (Fig. 3). The purity of the peaks was determined using the Agilent ChemStation A09.01 software to make sure that a peak contained only one compound. Our results demonstrated that although vindoline, vinblastine, and vincristine were not detectable in either control or transgenic root lines, catharanthine was detected in all hairy root lines. Compared with the control hairy root lines (M3 and M5), the accumulation of catharanthine increased in all transgenic lines (Fig. 4a). Under normal conditions, the amount of catharanthine was 0.019 and 0.029% on a dry weight basis in the wild-type lines M3 and M5, but reached 0.063–0.107% in GH lines and 0.075–0.123% in OG lines. The range of the catharanthine amount was not significantly different between GH lines and OG lines, though the expression level of ORCA3 in the OG lines was higher than in the GH lines under the control conditions (Fig. 2e), implies that the G10H gene plays a more important role in catharanthine synthesis. The transgenic line OG12 accumulated catharanthine at about 0.123% on a dry weight basis under normal conditions, which is about 6.5-fold of that of M3, and 4.2-fold of that of M5. ABA or cold treatment slightly increased the catharanthine accumulation in M3 and M5 root lines, but had a different influence on different transgenic hairy root lines. As for the GH lines, neither ABA nor cold treatment has an effect on the catharanthine amount. In the OG lines, ABA treatment significantly increased the catharanthine amount in OG12 and OG13, but had no effect on OG10, whereas under cold treatment the increased catharanthine amount was observed only in OG12 but not in OG10 and OG13. The highest amount of catharanthine was observed in OG12 (0.196% on a dry weight basis) following treatment with ABA (Fig. 4a).
HPLC analyses of TIAs accumulation in representative hairy root line OG12. a The pure standards. 1 vincristine, 2 vindoline, 3 catharanthine, 4 vinblastine. b Control. c 100 μmol/l ABA treatment for 24 h. d 4°C treatment for 24 h. The abscissas represent the retention time (min) and the y-axes represent the absorbance (mAU) at 220 nm. The injection volume is 10 μl
Catharanthine contents (a) and RT-PCR analyses of the TIAs’ biosynthesis genes (b) in hairy root lines after different treatments. Asterisks indicate statistically significant differences of transgenic lines compared with the control line M3 as determined with Student’s t test (*0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001)
An earlier study has demonstrated that although the production of tryptamine and tryptophan was increased in C. roseus cell suspension cultures overexpressing ORCA3, production of TIAs was increased only when loganin was supplemented in the medium (Van der Fits and Memelink 2000). This was due to the fact that G10H was not up-regulated under ORCA3 overexpression (Fig. 1). Our observation that catharanthine production was increased in all transgenic hairy root lines implies that G10H gene plays an important role in TIA biosynthesis. Very recently, ORCA3 was overexpressed in C. roseus hairy roots, but no significant increases in TIA metabolites were observed, even after the hairy roots were fed with loganin, tryptophan or loganin and tryptophan (Peebles et al. 2009). A possible explanation of this inconsistency is that only one hairy root line was analyzed in the article. Compared with the transgenic lines OG10 and OG13, G10H-overexpressing lines (GH2 and GH3) produced equivalent or even more catharanthine (except OG12). Furthermore, the amounts of catharanthine in different transgenic lines were not consistent with the expression levels of G10H and/or ORCA3 (Fig. 2e), suggesting the complexity of TIA biosynthesis.
Expression analyses of TIA biosynthetic genes
In the ORCA3-tagged C. roseus cell suspension line, the expressions of ASα, TDC, CPR, STR, and D4H increased, whereas SLS was not analyzed (Van der Fits and Memelink 2000). To understand the relationship between catharanthine accumulation and TIA biosynthesis-related gene expression in the hairy root lines, total RNA was extracted from both transgenic and control hairy root lines cultured under different conditions, and RT-PCR analyses were performed (Fig. 4b).The same amplification cycles were carried out for the same gene under different treatments, and the amount of catharanthine in hairy root lines corresponding to the treatments was determined (Fig. 4a). Compared with the lines M3, M5, GH2, and GH3, STR was strongly expressed in OG10, OG12, and OG13 under normal conditions (CK) or following cold treatment. The SLS expression was mildly higher in OG10 and OG12 under normal conditions. The expression of the other analyzed genes (ASα, CPR, and TDC) showed no significant differences between transgenic lines and control lines, though slight expression variations existed. The expression of D4H and DAT, two downstream genes of TIA in the biosynthesis pathway, were hardly detectable in most hairy root lines. In this study, only STR- and SLS-enhanced expression was observed in ORCA3-overexpressing hairy root lines. This is in consistence with a recent study in which the authors explained that ORCA3 is one of the dominant regulators of STR and SLS genes in the JA signaling cascade, and pointed the possibility of different regulatory mechanisms of TIA biosynthesis between cell suspension cultures and differentiated tissues such as hairy roots (Peebles et al. 2009).
The production of catharanthine paralleled the STR expression level of OG transgenic lines under normal conditions, but showed no correlation following ABA or cold treatment, suggesting that the STR gene plays an important role but not as a bottleneck in the TIA biosynthesis pathway. This was also confirmed by the GH2 line in which a highly increased catharanthine amount corresponds to a low expression of the STR gene. Strictosidine β-d-glucosidase (SGD) catalyzes the transformation from strictosidine to cathenamine, and the latter is the immediate precursor substance of the TIAs. As the SGD gene showed a large variable expression in the non-transgenic cell suspension lines (Van der Fits and Memelink 2000), it was not selected for analysis in this study. However, it was presumed recently that a decrease in SGD transcripts could limit the flux through the TIA biosynthesis pathway, causing the feeding of upstream metabolites to be ineffective in the ORCA3-overexpressed hairy root line (Peebles et al. 2009). Thus, further study on the SGD expression would be helpful for understanding the difference of catharanthine amount in the transgenic hairy root lines of this study.
In this study, the amount of catharanthine is increased through the overexpression of G10H or both G10H and ORCA3 genes in C. roseus hairy roots. An increase of 6.5-fold was obtained when compared with the control lines. However, the increased catharanthine accumulation in transgenic hairy roots is still less than that in C. roseus plants (data not shown). The lack of vinblastine and vincristine in C. roseus hairy roots has been ascribed to an absence of vindoline (Bhadra et al. 1998). This may be due to the undetectable expression of the D4H and DAT genes in the transgenic hairy roots. In C. roseus plants, D4H and DAT mRNAs are preferentially expressed in idioblast and laticifer cells of leaves, stems, and flower buds, but not detectable in roots (St-Pierre et al. 1999). Thus, a more detailed study of the genes involved in the TIA biosynthesis pathway as well as a deep understanding of the differentiation mechanism between C. roseus plants and hairy roots would be helpful for improving bisindole alkaloid production in hairy root cultures by biotechnological methods.
References
Behnam B, Kikuchi A, Celebi-Toprak F, Kasuga M, Yamaguchi-Shinozaki K, Watanabe KN (2007) Arabidopsis rd29A:DREB1A enhances freezing tolerance in transgenic potato. Plant Cell Rep 26:1275–1282
Bhadra R, Morgan JA, Shanks JV (1998) Transient studies of light-adapted cultures of hairy roots of Catharanthus roseus: growth and indole alkaloid accumulation. Biotechnol Bioeng 60:670–678
Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K, Sharma KK (2007) Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep 26:2071–2082
Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508:215–220
Da Costa e Silva O, Klein L, Schmelzer E, Trezzini GF, Hahlbrock K (1993) BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J 4:125–135
Favretto D, Piovan A, Filippini R, Caniato R (2001) Monitoring the production yields of vincristine and vinblastine in Catharanthus roseus from somatic embryogenesis. Semiquantitative determination by flow-injection electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 15:364–369
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hong SB, Peebles CA, Shanks JV, San KY, Gibson SI (2006) Terpenoid indole alkaloid production by Catharanthus roseus hairy roots induced by Agrobacterium tumefaciens harboring rol ABC genes. Biotechnol Bioeng 93:386–390
Hughes EH, Hong SB, Shanks JV, San KY, Gibson SI (2002) Characterization of an inducible promoter system in Catharanthus roseus hairy roots. Biotechnol Prog 18:1183–1186
Hughes EH, Hong SB, Gibson SI, Shanks JV, San KY (2004) Metabolic engineering of the indole pathway in Catharanthus roseus hairy roots and increased accumulation of tryptamine and serpentine. Metab Eng 6:268–276
Magnotta M, Murata J, Chen J, De Luca V (2007) Expression of deacetylvindoline-4-O-acetyltransferase in Catharanthus roseus hairy roots. Phytochemistry 68:1922–1931
Pasquali G, Porto DD, Fett-Neto AG (2006) Metabolic engineering of cell cultures versus whole plant complexity in production of bioactive monoterpene indole alkaloids: recent progress related to old dilemma. J Biosci Bioeng 101:287–296
Pauw B, Hilliou FA, Martin VS, Chatel G, de Wolf CJ, Champion A, Pre M, van Duijn B, Kijne JW, van der Fits L, Memelink J (2004) Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem 279:52940–52948
Peebles CA, Hughes EH, Shanks JV, San KY (2009) Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab Eng 11:76–86
Pino MT, Skinner JS, Park EJ, Jeknic Z, Hayes PM, Thomashow MF, Chen TH (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5:591–604
Rahman L, Kouno H, Hashiguchi Y, Yamamoto H, Narbad A, Parr A, Walton N, Ikenaga T, Kitamura Y (2009) HCHL expression in hairy roots of beta vulgaris yields a high accumulation of p-hydroxybenzoic acid (pHBA) glucose ester, and linkage of pHBA into cell walls. Bioresource Technol 100:4836–4842
Runguphan W, O’Connor SE (2009) Metabolic reprogramming of periwinkle plant culture. Nat Chem Biol 5:151–153
Shadwick FS, Doran PM (2007) Propagation of plant viruses in hairy root cultures: a potential method for in vitro production of epitope vaccines and foreign proteins. Biotechnol Bioeng 96:570–583
St-Pierre B, Vazquez-Flota FA, De Luca V (1999) Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11:887–900
Suttipanta N, Pattanaik S, Gunjan S, Xie CH, Littleton J, Yuan L (2007) Promoter analysis of the Catharanthus roseus geraniol 10-hydroxylase gene involved in terpenoid indole alkaloid biosynthesis. Biochim Biophys Acta 1769:139–148
Tikhomiroff C, Jolicoeur M (2002) Screening of Catharanthus roseus secondary metabolites by high-performance liquid chromatography. J Chromatogr A 955:87–93
Van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
Wise AA, Liu Z, Binns AN (2006) Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol Biol 343:43–53
Woods RR, Geyer BC, Mor TS (2008) Hairy-root organ cultures for the production of human acetylcholinesterase. BMC Biotechnol 8:95
Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Zhang L, Ding R, Chai Y, Bonfill M, Moyano E, Oksman-Caldentey KM, Xu T, Pi Y, Wang Z, Zhang H, Kai G, Liao Z, Sun X, Tang K (2004) Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc Natl Acad Sci USA 101:6786–6791
Acknowledgment
The authors are grateful to Professors J. Memelink (Leiden University, Netherlands) for providing the vectors containing G10H or ORCA3 genes, and Dr. Wang Yan-zhang (Shanghai Institutes for Biological Sciences, China) for providing A. rhizogenes MSU440, and Chen Jun (Shanghai Institutes for Biological Sciences, China) for HPLC analysis. This work was supported by the following grants: the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Grant No. 2007KIP304); the National Basic Research Program of China (Grant No. 2006CB100106; 2010CB126600); the National Natural Science Foundation of China (Grant No. 30571196; 0933ZF11C1; 0933Z411C1); The Ministry of Science and Technology of China (Grant No. 2007AA10Z187); Shanghai Science and Technology Commission (Grant No. 08DZ2270800).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
Rights and permissions
About this article
Cite this article
Wang, CT., Liu, H., Gao, XS. et al. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep 29, 887–894 (2010). https://doi.org/10.1007/s00299-010-0874-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0874-0