Abstract
A prerequisite for biotechnological improvements of storage roots is the availability of tissue-specific promoters enabling high expression of transgenes. In this work, we cloned two genomic fragments, pMe1 and pDJ3S, controlling the expression of a gene with unknown function from cassava (Manihot esculenta) and of the storage protein dioscorin 3 small subunit gene from yam (Dioscorea japonica), respectively. Using β-glucuronidase as a reporter, the activities of pMe1 and pDJ3S were evaluated in independent transgenic carrot lines and compared to the constitutive CaMV35S and the previously described cassava p15 promoters. Activities of pMe1 and pDJ3S in storage roots were assessed using quantitative GUS assays that showed pDJ3S as the most active one. To determine organ specificities, uidA transcript levels in leaves, stems and roots were measured by real-time RT-PCR analyses showing highest storage root specificity for pDJ3S. Root cross sections revealed that pMe1 was highly active in secondary xylem. In contrast, pDJ3S was active in all root tissues except for the central xylem. The expression patterns caused by the cassava p15 promoter in carrot storage roots were consistent with its previously described activities for the original storage organ. Our data demonstrate that the pDJ3S and, to a lesser extent, the pMe1 regulatory sequences represent feasible candidates to drive high and preferential expression of genes in carrot storage roots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Storage roots and tubers are important in human nutrition. According to the Food and Agriculture Organization of the United Nations (FAO 2000), almost 99% of the world production of tuber crops is provided by five species with potato (Solanum tuberosum) being the most important (45%) followed by cassava (Manihot esculenta; 26%), sweet potato (Ipomoea batatas; 20%), yam (Dioscora spp.; 6%) and taro (Colocasia esculenta; 1%). In arid areas such as sub-Saharan Africa, cassava ranks first place thanks to its capability to grow on poor soils and to withstand drought. Worldwide, cassava provides food for more than 600 million people (Nweke et al. 2002).
Storage roots and tubers are rich in starch; however, they contain low levels of other nutrients. For instance, on a dry weight basis, the total protein content of cassava storage roots is only about 1–2%, while it ranges in seeds from about 10% in cereals to about 40% in soybean (Shewry 2003). Cassava roots are also very poor in micronutrients. For instance, most cultivars possess extremely low levels of provitamin A, promoting the incidence of vitamin A deficiency diseases (VAD) in populations living predominantly on cassava-based diets. In addition, cassava roots are very poor in mineral content such as iron and zinc (Montagnac et al. 2009), adding to the burden of iron deficiency anaemia (Ariza-Nieto et al. 2006). Another problem in many cultivars of cassava is their toxicity due to the presence of cyanogenic glycosides. Furthermore, the phenomenon of post-harvest deterioration severely reduces shelf life, posing constraints to marketing (Booth 1976; Chávez et al. 2006; Montagnac et al. 2009). Thus, there are numerous good reasons to invest into the improvement of cassava; however, compared to rice, maize and wheat, investments have been very small.
Unlike many other crops, improvement through breeding is difficult with cassava. This problem relates to very long breeding cycles as well as to the large genetic load and the heterozygous nature of parents and consequently, progenies. Cassava is propagated vegetatively and has therefore never been pushed to produce inbreds from the segregating progenies of a given cross rendering varietal recovery an extremely difficult task (Ceballos et al. 2004). Due to these reasons and because of the multitude of possible options for improvement, cassava must be considered as a major target for genetic transformation. Ideally, target cultivars (landraces) would be transformed directly to produce the desired trait(s). However, genetic transformation requires a toolset of enabling technology, which is not sufficiently developed for cassava. One important component would be to have a suite of validated promoters providing storage root specificity of transgene expression to avoid unwanted side effects such as those that may arise upon enhancing contents of provitamin A carotenoids. For instance, the constitutive expression of the carotenogenic enzyme phytoene synthase often needed to achieve high β-carotene (provitamin A) levels (Fray et al. 1995; Shewmaker et al. 1999; Ye et al. 2000; Lindgren et al. 2003; Paine et al. 2005; Maass et al. 2009) resulted in dwarfism in tomato, most likely caused by redirection of the precursor geranylgeranyl diphosphate, which gibberellins and carotenoid pathways have in common (Fray et al. 1995). Similarly, constitutive expression of the bacterial carotene desaturase CrtI led to significant changes in the carotenoid pattern of rice and potato leaves, which may impact photosynthetic performance negatively (Al-Babili et al. 2006; Diretto et al. 2007).
Yam is the name applied to about 500 species of the genus Dioscorea, including widely grown species like D. cayenensis, D. rotundata and D. alata. In general, yam tubers contain dioscorins as the major storage protein accumulated in the vacuoles and accounting for about 85% of total protein (for review see Shewry 2003). Dioscorins are present as two classes, with about 70% similarity to each other and with significant homology to α-carbonic anhydrase from various sources. This enzymatic activity was demonstrated for dioscorins purified from D. batatas, D. alata and D. pseudojaponica (for review see Shewry 2003). However, a comprehensive, more recent study on D. batatas tuber storage proteins and their enzymatic functions led to the isolation of four different dioscorins designated as DB1, DB2, DB3 and DB4, and revealed DB1 and DB3 as lectins binding mannose and maltose, respectively. In addition, it was shown that DB3 is composed of a large 66-kDa protein consisting of two 31-kDa subunits (DB3L¸ 242 amino acids) cross linked by disulfide bonds, and two small 31-kDa subunit (DB3S; 241 amino acids). The two subunits show a sequence identity of 72% to each other and exhibit high homology to dioscorin A and B from D. alata (Gaidamashvili et al. 2004).
Underground storage organs can originate from stems (potato, taro), hypocotyls (yam and sweet potato) and roots (cassava, carrot). The development of the storage root in cassava is initiated by rapid cambial activity along the root axis and by differentiation of secondary xylem towards the inside, with starch accumulation restricted to ray parenchyma cells, thereby replacing secondary xylem fibres (Lowe et al. 1982). The carrot storage root is formed mainly from secondary phloem containing storage parenchyma cells (Surles et al. 2004). In contrast to carrot and cassava, the storage organ of sugar beet develops from both hypocotyl and primary root by secondary growth initiated by the generation of consecutive rings of secondary cambium producing xylem and phloem (Elliott and Weston 1993).
To date, the genetic transformation of cassava relies on heterologous promoters, such as the cauliflower mosaic virus CaMV35S promoter (González et al. 1998; Zhang et al. 2000; Jørgensen et al. 2005) and the cassava vein mosaic virus promoter (Verdaguer et al. 1996). Apart from these constitutive promoters, the tuber-specific potato class I patatin promoter was used to engineer starch synthesis (Ihemere et al. 2006). However, little is known about its tissue specificity in the cassava background. By differential screening of a storage root cDNA library of cassava Mcol1505, Zhang et al. (2003) identified the genes, c15 and c54, encoding a cytochrome P450 and a glutamic acid-rich protein, respectively. Using uidA as reporter gene, evaluation of the p15 and p54 promoters demonstrated that both of them were highly active in the starch-rich parenchyma cells of cassava transgenic storage roots. However, both promoters were also active in phloem, cambium and xylem vessels of vascular tissues of leaves, stems and roots (Zhang et al. 2003).
To identify novel storage root specific promoters, we relied on GenBank data representing genes preferentially expressed in cassava roots. In addition, we considered candidate genes expressed in underground storage tissues homologous with the cassava root, i.e. yam tuber. As a first candidate, we cloned a promoter fragment (pMe1) from cassava controlling the expression of a gene with unknown function, showing preferential storage root expression and encoding a member of the α-amylase inhibitors (AAI), lipid transfer (LT) and seed storage (SS) protein family (AAI_LTSS). As a second candidate, we isolated a promoter fragment (pDJ3S) of the D. japonica DJ3S gene encoding the small subunit of the DJ3 dioscorin. In this work, the activities of these two promoters were assessed in carrots as a model system producing storage roots. This was deemed necessary because cassava transformation is very time consuming. As comparators, we used the cassava p15 (Acc. no. AY217352) and the constitutive CaMV35S promoters. Our data demonstrate that the yam pDJ3S and, to lesser extent, the pMe1 promoters represent suitable candidates to drive high and preferential expression of genes in carrot storage roots.
Materials and methods
Plant material and growth
Tubers of Dioscorea japonica were obtained from the local market and roots from Manihot esculenta Crantz were obtained form CIAT (landrace MPer 183). Wild white carrot seeds (Daucus carota subsp. carota, Queen Anne’s Lace) were obtained from Richters Herbs (Goodwood, Canada). Carrot plants were grown in soil under long day conditions. Roots of 9-week-old carrot plants were used for histological dissection and RNA and protein extraction.
Southern blot analysis
Manihot esculenta Crantz genomic DNA was isolated according to Sambrook and Russell (2001). 40 μg of genomic DNA were digested with EcoRI, HindIII, NcoI and XbaI and fractioned electrophoretically through a 0.8% (w/v) agarose gel before capillary transfer and immobilization on nylon membrane (Hybond N+, Amersham Biosciences Europe, Freiburg, Germany). A probe covering the cloned genomic fragment was obtained from pMe1-Gen1 using the enzymes BamHI and HindIII. Radiolabeling, blotting and hybridization were performed according to Sambrook and Russell (2001).
Construction of binary vectors
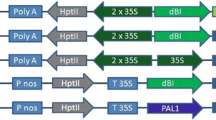
To generate pCA-pMe1-uidA the pMe1 promoter was amplified from pST-ext-gene using the primers Extgen1 and Extgen2 carrying a SphI and XbaI site, respectively. PCR was performed with the proofreading PWO DNA Polymerase (Peqlab, Erlangen, Germany), and the obtained promoter was digested with SphI, filled-in with T4 DNA polymerase and treated with XbaI. To replace the CaMV35S promoter driving the uidA expression in the binary pCAMBIA1305.2 (CAMBIA, Australia), the plasmid was digested with NcoI, filled-in with T4 DNA polymerase and treated with XbaI. After dephosphorylation, the plasmid was ligated with the pMe1 fragment to yield pCA-pMe1-uidA.
To generate pCA-pDJ3S-uidA, the yam promoter was amplified from pYam-Gen using the Pfx DNA polymerase (Invitrogen, Paisley, UK) with the primers Yam G(i)1 and MutGUS that carries a NcoI site. The obtained product was then digested with NcoI and EcoRV and ligated into NcoI and Ecl136II treated pCA-CP1, a pCAMBIA1305.2 derivative, to yield pCA-pDJ3S-uidA. A 1 kb portion was then excised from the 5′-end region of the pDJ3S promoter of the pCA-pDJ3S-uidA plasmid using EcoRI and Ecl136II restriction sites. The plasmid was then religated to form pCA-pDJ3S-s-uidA.
pCA-p15-uidA binary vector was kindly provided by ETH (Zhang et al. 2003).
Inverse PCR
To produce circular DNA fragments, 10 μg of genomic DNA from cassava and yam were digested in a total volume of 100 μl with ClaI and EcoRI, respectively. Digested DNA was purified using GFXTM PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ) and eluted with 100 μl of 60°C pre-warmed water. 40 μl of purified genomic fragments were then ligated for 2 h at room temperature followed by 20 h at 16°C in a total volume of 300 μl, using 100 U of T4 DNA ligase. The enzyme was then deactivated by heating for 10 min at 60°C, and circularised genomic DNA was precipitated with EtOH and resuspended in 100 μl water. Inverse PCR was then performed in a Mastercycler gradient (Eppendorf, Hamburg, Germany) using 1 μl of the circularised DNA, 100 μM dNTPs, 50 ng of each primer and 0.5 μl Advantage® cDNA Polymerase Mix (DB Bioscience, CA, USA) in the buffer provided, as follows: 2 min initial denaturation at 94°C followed by 35 cycles (30 s 94°C, 30 s annealing, 5 min 68°C) and 10 min final polymerization at 68°C. For annealing, a temperature gradient of 1°C, ranging from 58 to 68°C, was applied.
Promoter sequence analysis
To detect putative cis-acting regulatory elements in the promoters sequences cloned, the database PLACE was used (Higo et al. 1999). The database is accessible at http://www.dna.affrc.go.jp/PLACE.
Carrot transformation
Transformation was carried out using pieces of cotyledons and hypocotyls of 1-week-old light-grown QAL seedlings and the A. tumefaciens GV3101 strain, according to Hardegger and Sturm (1998), with some modifications. Instead of claforan and geneticin, vancomycin (200 mg/l) and hygromycin (10 mg/l) were used to arrest bacterial growth and to select transgenic calli, respectively.
GUS assays
β-Glucuronidase (GUS) activity in carrot tap roots of primary transformants was determined as described by Jefferson (1987) and Jefferson et al. (1987). For the fluorometric GUS assay 100 mg tissue was homogenized in 1 ml GUS reaction buffer consisting of 50 mM NaPO4, pH 7.1 mM EDTA; 0.1% (v/v) Triton X-100; 0.1% (v/v) sodium sarcosine and 10 mM β-mercaptoethanol. After centrifugation, 10 μl of the supernatant was added to 190 μl GUS reaction buffer containing 0.56 mM 4-methylumbelliferyl-β-d-glucuronide (4-MUG) and incubated at 37°C. The amount of 4-methylumbelliferone (4-MU) produced was determined after 5 min, and the measurement was repeated five times in 15 min intervals. For this purpose, 20-μl aliquots were mixed with 180 μl water and measured in a luminescence spectrometer (LS 50, Perkin Elmer) at excitation 365 nm, emission 455 nm. 0.0267 fluorescent units corresponded to 1 pmol 4-MU. Protein content was determined with Bio-Rad protein assay (Bio-Rad, Munich, Germany), and enzyme activity was calculated as pmol 4-MU min−1 (µg protein)−1.
For histochemical staining, carrot tap root dissections were incubated for 5 min at 37°C in X-Gluc buffer [1 mM 5-bromo-4-chloro-3-indolyl-b-d-glucuronide (X-gluc), 50 mM Na2HPO4, pH 7.0, 0.01% (v/v) Triton X-100, 10 mM EDTA, 0.5 mM K4Fe(CN)6 and 0.5 mM K3Fe(CN)6]. After staining, dissections were treated for 10 s with 70% (v/v) ethanol.
TaqMan real-time RT-PCR assay
RNA was isolated using the plant RNA isolation reagent (Invitrogen, Heidelberg, Germany). RNA purification and on-column DNaseI digestion were performed using the Qiagen RNeasy mini kit (Qiagen, Hilden, Germany). After first-strand cDNA synthesis, Taq-Man real-time RT-PCR assays were carried out with an ABI Prism 7000 (Applied Biosystems) using 18S rRNA levels for normalization. The relative quantity of the transcripts was calculated using the comparative threshold cycle method (Livak 1997). Data were first normalized to the corresponding 18S rRNA levels quantified using the eukaryotic 18S rRNA endogenous control kit (Applied Biosystems) and then calculated relative to a specific sample. Data represent averages of six transgenic lines per construct. Primers and 6FAM-labelled probes for uidA and Me1 genes were designed using Primer Express software (Applied Biosystems). The measurements were performed with primer/probe combinations GUS-Fw, GUS-Rv/GUS–probe for uidA and Ext-Fw, Ext-Rv/Ext-probe for Me1 (see Table S1 for primers/probes sequences).
Results and discussion
Cassava Me1 gene is encoded by a single copy gene preferentially expressed in storage roots
Nucleotide sequences with preferential root expression annotation reported in the GenBank of the NCBI (http://www.ncbi.nlm.nih.gov/Genbank) were selected and subsequently blasted against a collection of cassava ESTs described by Lopez et al. (2004). This process led to the identification of an EST (Acc. no. CK651140), here called Me1, preferentially expressed in storage roots and encoding a member of the AAI_LTSS protein superfamily (see “Introduction”). According to the NCBI conserved domains database (Marchler-Bauer et al. 2009; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), the AAI_LTSS protein family is characterized by a common pattern of eight cysteines forming four disulfide bridges.
To verify the Me1 expression pattern, real-time RT-PCR analyses were performed. These demonstrated that the transcript levels of Me1 are markedly higher in 3-month-old roots than in leaves or stems and steadily increased during storage root development (Fig. 1a). The copy number of the Me1 gene was determined by Southern blot hybridization and revealed one band with the XbaI- and HindIII-digested DNA (Fig. 1b), indicating the presence of a single copy gene. This was confirmed by the presence of two fragments upon NcoI- and EcoRI-digestion (Fig. 1b), arising from corresponding restriction sites in the gene (Fig. S1a; Acc. no. GU324673). Taken together, the cassava Me1 is represented by a single copy, which is preferentially expressed in storage roots. These features predict the Me1 promoter as candidate to allow storage root specific expression of transgenes.
Characterization of Me1 gene from cassava. a mRNA levels of the Me1 gene were determined by real-time RT-PCR in leaves (L), stems (S) and storage roots from 3- (R3M), 6- (R6M) and 9- (R9M) month-old cassava plants. mRNA values are given relative to the levels determined in leaves (L). Bars represent the mean of 18S rRNA-normalized mRNA levels + SD of duplicate samples from three biological replicates, each. b Southern Blot analysis of pMe1 gene using a Me1 PCR fragment as a probe
Cloning of the cassava pMe1 promoter
To clone the Me1 promoter, inverse PCR on ClaI-digested and circularised genomic DNA was performed using the primer pair Ex-In1/Ex-In2 (for sequence, see Table S1) deduced from the corresponding EST sequence (Acc. no. CK651140). A 1,748-bp gene fragment (Me1) was then obtained by PCR on genomic DNA using the primer pair Ex-Prom1/Ext2 (for sequence, see Table S1), which were designed according to the sequence of the inverse PCR product. As shown in Fig. S1a, the obtained gene fragment Me1 consists of 1,378 bp upstream of the start-ATG (pMe1), containing promoter sequence and unidentified 5′ untranslated region (5′ UTR), followed by 370 bp of the coding region (see Fig. S1a; Acc. no. GU324673). The TATA box of the pMe1 is 120 bp upstream from the start-ATG. According to the PLACE software program (http://www.dna.affrc.go.jp/PLACE), the pMe1 promoter contains transcription factor binding sites reported for storage proteins, in addition to several putative motifs mediating hormone, biotic and abiotic stress responses (Table 1).
Cloning of a dioscorin DJ3S promoter from D. japonica
As first approach, we used genomic PCR to clone fragments corresponding to dioscorin mRNA sequences available in the NCBI database. Several sets of primers were designed based on the mRNAs of Dioscorea batatas DB3S storage protein (Gaidamashvili et al. 2004; Acc. no. AB178473); Dioscorea alata dioscorin B and A (Acc. no. AF243526 and AF245019) and of dioscorin from D. cayenensis (Acc. no. X76187). The usage of the primer pair Yam1/Yam3 (for sequence, see Table S1) led to the cloning of a ca. 1.7 kb genomic fragment encoding a D. japonica discorin 3 small subunit (DJ3S), as suggested by sequence comparisons with the D. batata DB3S and to the D. japonica 3 discorin sequences (Acc. no. AB178473 and AM849818, respectively). Inverse PCR was then performed on EcoRI-digested and circularised genomic DNA using the primer pair Y-Inv1/Y-Inv3 (for sequence, see Table S1) deduced from the genomic fragment. The sequences of the 5′ and 3′ ends of the ca. 3 kb inverse PCR product were used to design the primers Yam-G(I)1 and Yam-G(I)2 (for sequence, see Table S1), which allowed the cloning of a 2,309 bp genomic fragment consisting of 379 bp coding region and 1,930 bp upstream of the start-ATG (pDJ3S; see Fig. S1b; Acc. no. GU324672), containing promoter sequence and unidentified 5′ UTR. A putative TATA box of the pDJ3S was identified 75 bp upstream from the start-ATG. According to the PLACE software, the pDJ3S promoter contains transcription factor binding sites reported for storage proteins and root-specific genes, in addition to several putative motifs mediating expression in root nodules and response to hormones, biotic and abiotic stress (Table 1). A short version pDJ3S-s (891 bp) was produced by cutting down 1,039 bp from the 5′-end of the 1930 promoter region (using Ecl136II restriction enzyme. See Fig. S1b).
Evaluation of the pMe1 and the pDJ3S fragments in carrot
Since the transformation of carrot is more feasible and less time consuming than of cassava, we decided to test the activities of the fragments pMe1, the pDJ3S and its short version pDJ3S-s in the former species. To estimate the eligibility of carrot as a model system for testing storage root specificity, we also assessed the activity of the cassava p15 promoter, which was previously evaluated in its respective storage tissue (Zhang et al. 2003). The constitutive CaMV35S promoter was employed as a constitutive comparator.
For this purpose, binary plasmids harbouring pMe1::uidA and pDJ3S::uidA fusions were constructed (junction sequences are shown in Fig. S2). At least six independent transgenic lines for each promoter were then generated and grown for 9 weeks to allow the formation of storage roots. The activities of pMe1 and pDJ3S in transgenic carrot roots were measured by quantitative GUS assays. This revealed pronounced differences (Fig. 2) with the yam pDJ3S as the most active, followed by, the cassava pMe1, the constitutive CaMV35S, the cassava p15 and finally by the short pDJ3S-s fragment.
Quantitative GUS assays with transgenic carrot roots. GUS assays were performed using 4-methylumbelliferyl-β-d-glucuronide (4-MUG) as substrate with storage roots from transgenic carrots transformed with constructs harbouring the various promoter::uidA fusions investigated pDJ3S and its short version (pDJ3S-s) from yam, pMe1 from cassava, p15 from cassava and CaMV35S. Bars represent the mean + SD of the specific activities of six different transgenic lines per construct
To evaluate organ specificity of expression, the corresponding leaves and stems were also subjected to GUS assays. However, the GUS enzymatic activities determined in leaves were negligibly low (data not shown), also in the CaMV35S::uidA transformants. Such low GUS enzymatic activities might be caused by interfering secondary metabolites or by the instability of the enzyme in these carrot tissues. Similar difficulties have been reported in carrot old leaves (Hardegger and Sturm 1998) and wheat where GUS activities in transgenic leaf and root tissues were supposed to be inhibited by an unidentified non-proteinaceous compound (Bahieldin et al. 2005). In carrot roots where GUS activity could be determined, the rank-order given in Fig. 2 showed highest activity with the yam pDJ3S.
Because of the problems given above, we resorted to transcript levels to assess the organ specificities of the promoters tested. For this purpose, real-time RT-PCR analyses were carried out using RNA isolated from roots, stems and leaves of six transgenic lines for each promoter (Fig. 3). With the exception of the previously described cassava promoter p15 that showed comparatively higher activity in leaves, all tested regulatory sequences exhibited better storage root specificity when compared to the constitutive CaMV35S promoter. The highest expression levels were again obtained with the pDJ3S from yam, as seen with the GUS enzymatic assays. In addition, the pDJ3S from yam provided the best storage root specificity. The cassava pMe1 showed intermediate root specificity expression levels in the carrot system.
Expression levels of reporter uidA gene in different organs of transgenic carrots with various promoter::uidA constructs. Total RNA from 9-week-old roots, stems and leaves from transgenic carrots were used for real-time RT-PCR. Transcript levels are expressed after normalization to 18S rRNA concentrations of the corresponding samples. Bars represent the mean + SD of six different transgenic lines
In Fig. 4, cross sections of the GUS-stained storage roots from the diverse promoter::uidA transgenic carrot are shown. The constitutive CaMV35S promoter (Fig. 4a) did not stain the residual primary xylem and was not active in large parts of the secondary xylem. Similar to previously published data (Hardegger and Sturm 1998), GUS staining was mainly observed in the younger xylem and phloem ray cells and in adjacent parenchymatic cell. In contrast to the CaMV35S promoter, the cassava p15 promoter, used as a comparator in this study, led to very intense GUS staining of the vascular cambium; staining of the xylem and phloem rays was restricted to the youngest cells (Fig. 4b). This is in agreement with the results obtained in cassava roots where the strongest GUS activity was observed in the cambium (Zhang et al. 2003). A completely different expression pattern was obtained with the cloned pMe1 from cassava (Fig. 4c), showing the highest expression in the secondary xylem. This appears favourable since the cassava storage root develops mainly from this tissue. Another difference to the p15 pattern is that both xylem and phloem rays were not stained rather appearing as “negatively stained”. Instead, the pMe1 activity was restricted to areas within the functional secondary xylem and phloem leading to a “spotted” appearance.
Histochemical localization of GUS activity in carrot storage roots. The smaller pictures show selected details (see text). Roots of 9-week-old transgenic carrots (diameter ca. 2 cm) were used to produce 360 μm cryo-sections. a CaMV35S promoter, b p15 promoter from cassava, c pMe1 from cassava, d pDJ3S from yam, e carotenoid formation driven by CrtB (phytoene synthase) under control of pDJ3S, f untransformed white root control from e. PX primary xylem, SX secondary xylem, SX secondary phloem, VC vascular cambium, PR phloem ray, XR xylem ray
The pDJ3S fragment from yam (Fig. 4d) was active in all root tissues except in the central xylem. The secondary xylem appeared somewhat stronger coloured in most cases. Strongest expression was, however, observed for those parenchyma cells aligning the xylem and phloem rays (seen at higher magnification in Fig. 4d) leading to highly parallel appearance of the stain at the given plane of focus, feinting ray multiplication. The short version of this fragment (pDJ3S-s) abolished this specificity and caused a staining that was similar to the one observed with the CaMV35S promoter (not shown).
To further confirm its tissue specificity, the pDJ3S fragment was fused to the bacterial CrtB gene (from Pantoea ananatis coding for phytoene synthase) known to be able to drive carotenoid expression in white carrot roots (Maass et al. 2009), thereby obtaining a second reporter gene construct. This led to a similar pattern of activity visualized by carotenoid deposition with strongly labelled rays and a predominance of colour in the secondary xylem (Fig. 4e and f).
Based on these studies and assuming that the carrot model system holds for cassava, one would recommend the complete pDJ3S fragment from yam for use in biotechnological approaches aiming at improving traits of storage roots. It showed the highest levels of expression, the best storage root specificity and activity in the secondary xylem constituting by large the main tissue of cassava roots, albeit being less active and less storage root specific. Since the CrtB gene is capable of driving carotenoid formation in white cassava roots (unpublished data), fusions with this pDJ3S will be made and tested in stably transformed cassava plants.
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- DJ :
-
Dioscorea japonica
- GUS:
-
β-Glucuronidase
- Me :
-
Manihot esculenta
- uidA :
-
β-Glucuronidase gene
References
Al-Babili S, Hoa T, Schaub P (2006) Exploring the potential of the bacterial carotene desaturase CrtI to increase the beta-carotene content in Golden Rice. J Exp Bot 57:1007–1014
Ariza-Nieto M, Sanchez T, Heller L, I Hu Y, Welch M, Glahn P et al (2006) Cassava (Manihot esculenta) has high potential for iron biofortification. FASEB J 20:A624
Bahieldin A, Eissa F, Mahfouz H, Dyer E, Madkour A, Qu R et al (2005) Evidence for non-proteinaceous inhibitor(s) of b-glucuronidase in wheat (Triticum aestivum L.) leaf and root tissues. Plant Cell Tiss Org 82:11–17
Booth R (1976) Storage of fresh cassava (Manihot esculenta) I. Post-harvest deterioration and its control. Exp Agric 12:103–111
Ceballos H, Iglesias A, Pérez C, Dixon G (2004) Cassava breeding: opportunities and challenges. Plant Mol Biol 56:503–516
Chávez L, Ceballos H, Rodriguez-Amaya D, Nestel P, Ishitani M (2006) Reduction or delay of post-harvest physiological deterioration in cassava roots with higher carotenoid content. Sci Food Agric 86:634–639
Diretto G, Al-Babili S, Tavazza R, Papacchioli V, Beyer P, Giuliano G et al (2007) Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PloS ONE 2:e350
Elliott MC, Weston GD (1993) Biology and physiology of the sugar-beet plant. In: Cooke DA, Scott RK (eds) The sugar beet crop: science into practice. Chapman and Hall, London, pp 37–66
FAO (2000) Production yearbook 1999. FAO statistics series No. 53. FAO, Rome
Fray G, Wallace A, Fraser D, Valero D, Hedden P, Bramley M et al (1995) Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J 8:693–701
Gaidamashvili M, Ohizumi Y, Iijima S, Takayama T, Ogawa T, Muramoto K (2004) Characterization of the yam tuber storage proteins from Dioscorea batatas exhibiting unique lectin activities. J Biol Chem 279:26028–26035
González E, Schöpke C, Taylor J, Beachy N, Fauquet C (1998) Regeneration of transgenic cassava plants (Manihot esculenta Crantz) through Agrobacterium-mediated transformation of embryogenic suspension cultures. Plant Cell Rep 17:827–831
Hardegger M, Sturm A (1998) Transformation and regeneration of carrot (Daucus carota L.). Mol Breed 4:119–127
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Ihemere U, Arias-Garzon D, Lawrence S, Sayre R (2006) Genetic modification of cassava for enhanced starch production. Plant Biotechnol 4:453–465
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jefferson RA, Kavanagh RA, Bevan MW (1987) GUS-fusions: β-glucuronidase as a sensitive and versatile fusion marker in higher plants. EMBO J 6:3901–3907
Jørgensen K, Bak S, Busk K, Sørensen C, Olsen E, Puonti-Kaerlas J et al (2005) Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol 139:363–374
Lindgren O, Stålberg G, Höglund A (2003) Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132:779–785
Livak KJ (1997) User bulletin No. 2: ABI PRISM 7700 sequence detection system. PE Applied Biosystems Foster City, CA, pp 11–15
Lopez C, Jorge V, Piégu B, Mba C, Cortes D, Restrepo S et al (2004) A unigene catalogue of 5700 expressed genes in cassava. Plant Mol Biol 56:41–54
Lowe S, Mahon J, Hunt L (1982) Early development of cassava (Manihot esculenta). Can J Bot 60:3040–3048
Maass D, Arango J, Wüst F, Beyer P, Welsch R (2009) Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PloS ONE 4:e6373
Marchler-Bauer A, Anderson B, Chitsaz F, Derbyshire K, DeWeese-Scott C, Fong H et al (2009) CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res 37:D205–D210
Montagnac A, Davis R, Tanumihardjo A (2009) Nutritional value of cassava for use as a staple food and recent advances for improvement. Comp Rev Food Sci Saf 8:181–194
Nweke F, Spencer D, Lynam J (2002) The cassava transformation: Africa’s best-kept secret. Michigan State University Press, East Lansing, USA
Paine A, Shipton A, Chaggar S, Howells M, Kennedy J, Vernon G et al (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487
Sambrook J, Russell W (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Shewmaker C, Sheehy J, Daley M, Colburn S, Ke D (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Shewry R (2003) Tuber storage proteins. Ann Bot 91:755–769
Surles L, Weng N, Simon W, Tanumihardjo A (2004) Carotenoid profiles and consumer sensory evaluation of specialty carrots (Daucus carotaL.) of various colors. J Agric Food Chem 52:3417–3421
Verdaguer B, Kochko A, Beachy N, Fauquet C (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31:1129–1139
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P et al (2000) Engineering the provitamin A (β-Carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Zhang P, Potrykus I, Puonti-Kaerlas J (2000) Efficient production of transgenic cassava using negative and positive selection. Transgenic Res 9:405–415
Zhang P, Bohl-Zenger S, Puonti-Kaerlas J, Potrykus I, Gruissem W (2003) Two cassava promoters related to vascular expression and storage root formation. Planta 218:192–203
Acknowledgments
This work was funded by The HarvestPlus research consortium (http://www.harvestplus.org) and by the Deutsche Forschungsgemeinschaft (DFG), Graduiertenkolleg 1305 “Signalsysteme in pflanzlichen Modellorganismen”. We are indebted to Paul Chavarriaga for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Reski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arango, J., Salazar, B., Welsch, R. et al. Putative storage root specific promoters from cassava and yam: cloning and evaluation in transgenic carrots as a model system. Plant Cell Rep 29, 651–659 (2010). https://doi.org/10.1007/s00299-010-0851-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0851-7