Abstract
Two pathways are used by higher plants for the biosynthesis of isoprenoid precursors: the mevalonate pathway in the cytosol and a 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway in the plastids, with 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase (HDR) catalyzing the last step in the MEP pathway. In order to understand the contribution of MEP pathway in isoprenoid biosynthesis of Oncidium orchid, a full-length cDNA corresponding to HDR from the flower tissues of Oncidium Gower Ramsey was cloned. The deduced OncHDR amino acid sequence contains a plastid signal peptide at the N-terminus and four conserved cysteine residues. RT-PCR analysis of HDR in Oncidium flowering plants revealed ubiquitous expression in organs and tissues, with preferential expression in the floral organs. Phylogenetic analysis revealed evolutionary conservation of the encoding HDR protein sequence. The genomic sequence of the HDR in Oncidium is similar to that in Arabidopsis, grape, and rice in structure. Successful complementation by OncHDR of an E. coli hdr − mutant confirmed its function. Transgenic tobacco carrying the OncHDR promoter-GUS gene fusion showed expression in most tissues, as well as in reproductive organs, as revealed by histochemical staining. Light induced strong GUS expression driven by the OncHDR promoter in transgenic tobacco seedlings. Taken together, our data suggest a role for OncHDR as a light-activated gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orchids (Orchidaceae) represent the second largest family of plants, and the number of species may exceed 20,000–25,000 (Pridgeon and Chase 1995). Hybrids of the genus Oncidium and its hybridization compatible alliance genera Odontoglossum and Miltonia are grown widely for production of commercial cut flowers and potted plants because of their showy and colorful flowers. The pigments in the Oncidium flower consists of anthocyanins and carotenoids, the latter being the main component in the labellum or lips in a flower (Hieber et al. 2006). Currently, only limited information on the biosynthesis of carotenoids in Oncidium flowers is available. Hieber et al. (2006) reported on the mixtures of carotenoids and anthocyanin pigments present in Oncidium flowers; with the isomers of violaxanthin representing the predominant yellow pigment in the lip tissue.
Carotenoids are derived from isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP), the universal precursors of all isoprenoid compounds (Cunningham and Gantt 1998; Lange et al. 2000). There are two pathways leading to biosynthesis of both IPP and DMAPP in higher plants, namely, the mevalonic acid (MVA) pathway, which produces cytosolic IPP; and the plastid 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway (for review see Eisenreich et al. 2001; Lichtenthaler 1999; Rodríguez-Concepción and Boronat 2002).
The MVA pathway provides precursors for many isoprenoid compounds (Edwards and Ericsson 1999), and some have been shown to be essential for growth and development. As well, MVA biosynthesis appears to be essential for cell cycle progression (Hemmerlin and Bach 1998) and in response to wounding (Maldonado-Mendoza et al. 1997).
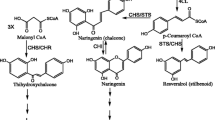
The plastidial MEP pathway has been studied by a combination of biochemical and genomic approaches in some plants (Rodríguez-Concepción 2006; Rodríguez-Concepción and Boronat 2002). The precursors for monoterpenes, diterpenes, carotenoids, tocopherols, and the prenyl moiety of chlorophyll are all derived from the MEP pathway (Eisenreich et al. 2001) (Fig. 1).
The pathway for carotenoid biosynthesis in plants. GAP glyceraldehyde 3-phosphate, DXP deoxyxylulose 5-phosphate, MEP methylerythritol 4-phosphate, CDP-ME 4-diphosphocytidyl-methylerythritol, CDP-MEP 4-diphosphocytidyl-methylerythritol 2-phosphate, ME-cPP methylerythritol 2,4-cyclodiphosphate, HMBPP hydroxymethylbutenyl 4-diphosphate, IPP isopentenyl diphosphate, DMAPP dimethylallyl diphosphate, GPP geranyl diphosphate, GGPP geranylgeranyl diphosphate, ABA abscisic acid. Enzymes are indicated in bold: DXS DXP synthase, DXR DXP reductoisomerase, CMS CDP-ME synthase, CMK CDP-ME kinase, MCS ME-cPP synthase, HDS HMBPP synthase, HDR HMBPP reductase, GGDS GGPP synthase, PSY phytoene synthase, PDS phytoene desaturase, ZDS ζ-carotene desaturase, CRTISO carotenoid isomerase, LCYB lycopene β-cyclase, LCYE lycopene ε-cyclase, CHYB carotenoid β-ring hydroxylase, CHYE carotenoidε-ring hydroxylase, ZEP zeaxanthin epoxidase, VDE violaxanthin de-epoxidase, NSY neoxanthin synthase
Currently, there is no information available for MEP pathway to operate in orchids. We are working on isolating and characterizing the genes from this pathway in Oncidium. In this work, we aimed to clone the Oncidium HDR gene of the MEP pathway and characterize its expression and regulation, due in part to more complete data available for this gene. A second reason for choosing the HDR gene is its importance being considered as the controlling point for MEP pathway and limiting step for isoprenoid biosynthesis in E. coli (Cunningham et al. 2000; Rodríguez-Concepción 2006). We analyzed HDR expression pattern by RT-PCR and promoter reporter fusion, and subsequent transformation into the tobacco genome. HDR was expressed in most tissues of Oncidium plants and OncHDRp::GUS in transformed tobacco seedlings. We further confirmed the function of the Oncidium HDR by complementation study in a bacteria mutant defective in the HDR gene.
Materials and methods
Plant materials and RNA isolation
Plants of Oncidium Gower Ramsey orchid were grown in the fan-and-pad greenhouse of National Pingtung University of Science and Technology (Pingtung, Taiwan) under shade light and controlled temperature ranging from 23 to 27°C. For all experiments, young flower buds and other tissues were harvested and frozen in liquid nitrogen, then stored at −80°C until use. Total RNAs were isolated and purified by use of TRIZol reagent according to the manufacturer’s instructions (Invitrogen). RNA precipitates were resuspended in DEPC-treated sterile water and precipitated with LiCl (at a final concentration of 2 M). The supernatant was treated with RNase-free DNase (Promega) at 37°C for 30 min to remove residual genomic DNA. Purified RNAs were quantified by spectrophotometry (U2000, Hitachi) and quality was checked by agarose gel electrophoresis (Chen et al. 2005).
Cloning of Oncidium Gower Ramsey HDR cDNA
Total RNA was isolated from unopened flower buds of Onicidium. An amount of 1 μg RNA was used to synthesize double-stranded (ds) cDNA by use of the SMART cDNA PCR Synthesis Kit (Clontech). Synthesized dscDNA fragments were used as templates for PCR. HDR degenerate primers (Supplementary Table 1), designed from conserved amino acid sequences found in GenBank, were used to amplify regions of interest. The amplified fragment was cloned into pGEM-T Easy vector (Promega) and sequenced. Gene-specific primers (Supplementary Table 1) were designed from the known sequence of the OncHDR fragment for 5′ and 3′ rapid amplification of cDNA ends (RACE) by use of the SMART RACE cDNA kit (Clontech) according to the manufacturer’s instructions. The 5′-RACE and 3′-RACE products were cloned into the pGEM-T Easy vector and sequenced. On the basis of the RACE sequence data, the Onc.HDR-F and Onc.HDR-R primers (Supplementary Table 1) were designed for amplification of full-length cDNAs by use of Ex Taq (Takara, Tokyo).
DNA blot analysis
Genomic DNA (30 μg/sample) was extracted from young Oncidium leaves by the CTAB DNA extraction method (Doyle and Doyle 1990) and digested overnight with EcoRI and HindIII (BioLabs). The product was fractionated by 0.8% agarose gel electrophoresis, transferred to Hybond N+ membranes (Amersham International, Buckinghamshire, UK), and hybridized with a 32P-labeled 500-bp fragment containing the partial OncHDR coding region excised from plasmid DNA, purified by gel electrophoresis and labeled by random priming of hexamers. Pre-hybridization and hybridization followed standard protocols (Sambrook and Russell 2001).
Transcript analysis by semi-quantitative RT-PCR
Total RNA was isolated from various tissues of Oncidium as described above. For cDNA synthesis, total RNA (1 μg) was reverse-transcribed in a 20 μL reaction mixture by use of the ImProm-II™ Reverse Transcription System (Promega). The primers used were vHDR-F and vHDR-R (Supplementary Table 1). As an internal control, actin 1 gene was amplified with the primers Actin-2 and Actin-3 (Supplementary Table 1). Five microliters of cDNA sample from the RT reaction was used for 26 cycles of PCR: denaturation at 94°C (30 s), annealing at 60°C (30 s), and extension at 72°C (40 s). The PCR product (25 μL) from each reaction was analyzed by electrophoresis in 1.2% agarose gel.
Quantification of transcript levels by real-time PCR assay
Quantitative real-time PCR and data analysis were performed in the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocol. Total RNA was isolated from Oncidium flowers. Total RNA (5 μg) was treated with DNase I and used for first-strand cDNA synthesis by priming with oligo d (T15) and catalyzed with Superscript III Reverse Transcriptase (Invitrogen) at 42°C for 1.5 h. The gene-specific forward and reverse primers (HDRr-F and HDRr-R) (Supplementary Table 1) were predicted using the Primer Express 2.0 (Applied Biosystems). The real-time PCR conditions were: 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Before running real-time PCR, primer efficiency was evaluated by using both HDR and 26S rRNA at 50, 150 and 300 nM combinations. The 150 nM for both gene was chosen as most suitable combination. Each sample was amplified in triplicate and all PCR reactions were performed on ABI PRISM 7000 Sequence Detection System. With a housekeeping gene 26S rRNA, the relative amount of the OncHDR transcript is presented as 2−ΔCT according to the ΔCT method described in the Real-Time PCR Applications Guide (Applied Biosystems).
Pigment isolation and quantification
Fifteen milliliters of hexane/acetone/ethanol (2:1:1) was added to 0.3–0.6 g of fresh Oncidium tissues (flower buds, leaves and roots) and mixed until the tissue was bleached. The organic solvent phase was separated by the addition of 2 mL water and hexane fraction dried in a stream of nitrogen gas at room temperature and resuspended in acetone. Total carotenoids and chlorophylls were determined spectrophotometrically according to Arnon et al. (1954).
Complementation of the E. coli HDR mutant with OncHDR
The E. coli HDR mutant strain MG1655 ara< >ispH was maintained on Luria–Bertani (LB) medium containing 50 mg L−1 kanamycin and 0.2% (w/v) arabinose (Ara) (McAteer et al. 2001). The pQE-30-OncHDR plasmid was transformed into the E. coli HDR mutant and selected on LB plates containing 50 mg L−1 kanamycin, 50 mg L−1 ampicillin, 0.2% glucose (Glc), and 0.5 mM IPTG. The surviving colonies containing the pQE-30-OncHDR plasmid were identified. The empty pQE-30 vector was transformed into the E. coli HDR mutant as a control and selected on LB plates containing 50 mg L−1 kanamycin, 50 mg L−1 ampicillin, and 0.2% Ara. The transformants containing the control vector could not grow on 0.2% Glc-containing medium.
Sequence analysis
Protein sequences of HDR homologues from other species were retrieved from GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) by the TBLASTN algorithm. Multiple alignments of amino acid sequences involved use of the ClustalW program (http://www2.ebi.ac.uk/clustalw). Plastid-targeting peptides were predicted by the ChloroP program (http://www.cbs.dtu.dk/services/ChloroP).
Isolation of OncHDR promoter by genomic walking
The OncHDR promoter was isolated according to the instructions of the GenomeWalker Universal Kit (Clontech). Genomic DNA was extracted from young Oncidium leaves by the CTAB DNA extraction method (Doyle and Doyle 1990). Orchid genomic DNA was digested with four restriction enzymes (DraI, EcoRV, PvuII, and StuI), and the products were ligated to a Genome Walker adaptor before being used as templates for PCR reactions. The first PCR involved use of an adaptor primer AP1 and one gene-specific primer, HDRpGSP1 (Supplementary Table 1), designed on the basis of the OncHDR full-length cDNA sequence. In addition, the other adaptor primer AP2 and the second gene-specific primer, HDRpGSP2 (Supplementary Table 1), were used in secondary PCR. The amplification conditions for the first PCR were seven cycles at 94°C (for 10 s) and 72°C (for 3 min), 32 cycles at 94°C (for 10 s) and 67°C (for 3 min), then a final extension at 67°C (for 7 min). The conditions for the second PCR were the same as for the first, except that the number of cycles was reduced to 25. The secondary PCR products were analyzed by electrophoresis on 1% agarose gels. The major product band (s) from independent second PCR reactions was gel-purified with use of QIAEXII Gel Extraction Kit (Qiagen), cloned into pGEM-T Easy vector and sequenced.
Construction of the OncHDR promoter reporter gene fusion
The 1.7 kb promoter fragment of OncHDR spanning the coding region was amplified by PCR with the primers HDRp-F and HDRp-R (Supplementary Table 1), which contained HindIII and BamHI restriction enzyme sites at their 5′ and 3′ ends, respectively. The PCR products were first digested by HindIII and BamHI and then cloned into the HindIII and BamHI sites in a promoter-less binary vector pCAMBIA1391Z (Cambia GPO, Canberra, Australia) that contained the intron-GUS gene. The construct was designated pCAMBIA1391Z-OncHDRp.
Tobacco transformation
The constructed plasmid pCAMBIA1391Z-OncHDRp was transformed into A. tumefaciens LBA4404 by electroporation (Nagel et al. 1990). Leaf discs of tobacco (Nicotiana tabacum cv. Petit-Havana SR1) were transformed as described previously (Horsch et al. 1985). Transformed plants were selected and regenerated on MS medium containing cefotaxime (250 mg L−1) and hygromycin (40 mg L−1).
Analysis of GUS gene expression
Histochemical localization of GUS gene expression was analyzed according to Jefferson et al. (1987). Briefly, tissues of in vitro-grown seedlings of transgenic tobacco were immersed in GUS assay buffer containing X-gluc. The GUS-stained tissues and seedlings were photographed by use of a digital camera.
Results
Characterization of OncHDR cDNA and deduced OncHDR protein
We cloned the full-length OncHDR cDNA from Oncidium Gower Ramsey flower tissues using degenerate primers designed on the basis of seven plant HDR proteins conserved in a core region. A cDNA fragment of about 500 bp was amplified by RT-PCR, with total RNA isolated from young floral buds being used as templates. On the basis of this sequence, we designed two gene-specific primers for RACE, which produced two PCR products of about 956 bp and 588 bp. A full-length cDNA (Accession no. EU908200) consisting of a 1,392 bp open reading frame (ORF) was obtained by combining the sequence information from 3′ and 5′ RACE. The deduced protein sequence of OncHDR consisted of 463 amino acid residues, of 52.1 kDa. The amino acid sequence of the HDR of Oncidium Gower Ramsey showed high homology throughout the entire coding region to that of Hevea brasiliensis (77.5%), Oryza sativa (77.3%), Vitis vinifera (77.3%) Lycopersicon esculentum (76.6%), Aquilegia (76.5%), Arabidopsis thaliana (73.8%) and Ginkgo biloba (71.1%) (Fig. 2). The deduced OncHDR sequence contains an N-terminal chloroplast signal peptide consisting of 33 residues predicted by the ChloroP program (Emanuelsson et al. 1999), as do those of putative HDR homologues of other plants (Fig. 2). All plant HDRs, including that of Oncidium orchid, showed four conserved cysteine residues, which supposedly participate in the coordination of the iron–sulfur bridge proposed to be involved in enzymatic catalysis (Grawert et al. 2004; Rohdich et al. 2002; Wolff et al. 2003). The position of one of these cysteine residues is not conserved in bacterial proteins and the protein of one plant species, Picrorhiza kurrooa (Fig. 2).
Multi-alignment of amino acid sequences of Onc.HDR and other plant HDRs. Synechococcus (YP_172141), Synechocystis (Q55643), Anabaena (YP_323455), Adonis (AAG21984), Aquilegia (TC9243), Lactuca (TC12240), Stevia (ABB88836), Lycopersicon (TC124188), Solanum (ABB55395), Arabidopsis (AAN87171), Gossypium (TC27556), Oryza (ABF98702), Triticum (TC232642), Oncidium (EU908200), Ginkgo (ABC84344), Chlamydomonas (TC48149), Picrorhiza (EF199770), Vitis (GSVIVT00036436001) and E. coli (P22565) sequences deposited in the TIGR, NCBI, Grape Genome Browser and Swiss-Prot databases. Conserved residues are highlighted (with black when present in all sequences). The plastid-targeting peptide cleavage site predicted by the ChloroP algorithm is marked with a black arrow. Black circles mark the position of cysteine residues
Phylogenetic relationship of OncHDR
We constructed a phylogenetic tree using ClustalW (http://www.ebi.ac.uk/clustalw/). The tree (Fig. 3) shows the eukaryotic HDR homologues clustered into two clades, with dicots and monocots; cyanobacteria into two subgroups; and green algae into another group. The result suggests that the OncHDR is conserved evolutionarily with other HDRs.
Phylogenic relationship between the HDR of Oncidium Gower Ramsey and those of other organisms. The numbers on the branches represent bootstrap support for 1,000 replicates. The species and corresponding accession number are as follows: Synechococcus (YP_172141), Synechocystis (Q55643), Anabaena (YP_323455), Adonis (AAG21984), Aquilegia (TC9243), Lactuca (TC12240), Stevia (ABB88836), Lycopersicon (TC124188), Solanum (ABB55395), Arabidopsis (AAN87171), Gossypium (TC27556), Oryza (ABF98702), Triticum (TC232642), Oncidium (EU908200), Ginkgo (ABC84344), Chlamydomonas (TC48149), Picrorhiza (EF199770), Vitis (GSVIVT00036436001) and E. coli (P22565)
Expression of OncHDR correlates with carotenoid accumulation in Oncidium flowers
To investigate the expression profile of OncHDR in different tissues of Oncidium Gower Ramsey plants, total RNA isolated from roots, leaves, peduncle, labellum (lip), whole flowers was analyzed by two-step RT-PCR, and flower buds at different stages (Fig. 4a) by real-time RT-PCR. OncHDR expression could be detected in all tissues examined, with weaker expression in roots (Fig. 4b). The transcript of OncHDR reached their highest level at the half-open state, and then decreased in fully open flowers (Fig. 5a). The trend of transcript expression level is well correlated with the accumulation of total carotenoid at different developmental stages of flower buds (Fig. 5a, b).
a Transcript levels of OncHDR in Oncidium floral stages analysed by real-time PCR. Flower bud stages: Fb1: <0.2 cm buds, Fb2: >0.2 cm bubs; Fb3: half-open flower, Fb4: fully open flower. b Chlorophyll a, b and carotenoid contents in roots, leaves and flower buds of Oncidium Gower Ramsey. Fb1 <0.2 cm Buds, Fb2 >0.2 cm bubs, Fb3 half-open flower, Fb4 fully open flower
Genomic structure of OncHDR in Oncidium similar to other plants
A genomic sequence of the Oncidium HDR gene was isolated after PCR amplification of genomic DNA with specific primers (Onc.HDR-F and Onc.HDR-R, Supplementary Table 1) based on the OncHDR cDNA sequence. The OncHDR genomic sequence spans approximately 5.2 kb (Accession no. EU908201), whereas the length of Arabidopsis (Guevara-García et al. 2005; Hsieh and Goodman 2005) and rice homologues is 2.4 and 2.8 kb, respectively. These gene homologues all possess nine introns, and their relative distribution is similar among the four species compared. The only difference in these homologues is the variation in their intron size (Fig. 6a). Introns 2, 7, and 9 of OncHDR are longer than those of Arabidopsis and rice homologues (Fig. 6a, b). All introns in OncHDR followed the GT/AG splicing rule (Brendel et al. 1998).
To examine the copy number of OncHDR in Oncidium, we used DNA blot analysis of the Oncidium genomic DNA digested with two different restriction enzymes. Use of a partial cDNA fragment, spanning from 400 to 920 bp, of OncHDR as a probe revealed more than three hybridized bands in the blot (Supplementary Fig. 1), which suggests that more than one copy of OncHDR exists in the Oncidium Gower Ramsey genome.
Oncidium HDR complements the E. coli hdr (IspH) mutant
To further characterize the function of OncHDR, we use a complementation assay with an E. coli hdr − mutant lacking HDR activity (Hsieh and Goodman 2005) to determine whether the deduced OncHDR is functional in the bacteria. In E. coli hdr mutant strain MG1655 ara < > ispH, a kanamycinR cassette was used to replace the endogenous HDR gene, and a single copy of ispH was present on the bacterial chromosome under control of the araBAD promoter (McAteer et al. 2001). Since the HDR gene is essential for bacterial survival, the E. coli hdr mutant strain could grow only in medium containing arabinose but not in glucose-containing medium (Fig. 7). Transforming the bacterial mutant with the pQE30-OncHDR plasmid, but not the empty pQE-30 vector, enabled the transformants to grow on LB medium containing glucose, which demonstrates the catalytic competence of the expressed OncHDR protein in the bacterial cells (Fig. 7).
Complementation assay of Oncidium HDR with the E. coli HDR mutant. The E. coli HDR mutant strain MG1655 ara< >IspH transformed with Oncidium HDR but not the control mutant carrying pQE30 was able to grow in LB medium containing 0.2% glucose (right). The E. coli mutant was able to grow on medium with 0.2% arabinose but not glucose (left)
Sequence characterization of the OncHDR promoter
The promoter region, about 1.7 kb long, of the Oncidium HDR gene was isolated from Oncidium genomic DNA by genome walking. We searched for a promoter motif to define putative cis-elements in the OncHDR promoter sequence using the software programs PLACE (Higo et al. 1999) and PlantCARE (Lescot et al. 2002; Rombauts et al. 1999) and found a number of potential regulatory motifs corresponding to known cis-elements of eukaryotic genes (Fig. 8). A potential TATA-box and CAAT-box were detected at positions −111 to −106 and −254 to −250 upstream, respectively, of the translation start codon ATG (Fig. 8, Table 1); these boxes may function as basal promoter elements for transcription. Furthermore, several other potential regulatory elements with roles in regulating gene expression are present in the OncHDR promoter (Table 1). These include a circadian element (CAACATTATC) at positions −26 to −17 (Piechulla et al. 1998), a WRKY-type transcription factor recognition sequence (TGAC) at positions −128 to −125 (Xie et al. 2005), a pyrimidine box (TTTTTTCC) at positions −569 to −562 (Cercós et al. 1999), a GARE-motif (AAACAGA) at positions −732 to −726 (Ogawa et al. 2003), and a MADS factor recognition site (CAATTTAAAG) at positions −949 to −940 (Tang and Perry 2003). In addition, the OncHDR promoter contains two MYB binding sites (Abe et al. 2003), three light-responsive elements and three GT-1-like transcription factor recognition sites (Terzaghi and Cashmore 1995; Park et al. 2004).
OncHDR promoter sequence and its putative cis-elements. Numbers indicate the positions relative to the translation start site. The translation initiation codon is shaded and indicated by an arrow. The putative TATA box, CAAT box, and other important putative cis-elements are shown in bold and labeled
Light up-regulates OncHDRp::GUS expression in transgenic tobacco seedlings
To investigate the temporal and spatial regulation of the OncHDR promoter, transgenic tobacco plants carrying the GUS reporter gene fused to the OncHDR promoter fragment were grown in the greenhouse, and flowering plants and in vitro T1 seedlings were histochemically assayed for GUS activity. Positive GUS staining of whole seedlings revealed ubiquitous expression of the OncHDR promoter driving GUS in most tissues, including cotyledons, true leaves, petioles, hypocotyls, and roots (Fig. 9). Root tips did not show any GUS activity, but weak GUS expression was detected in the root hairs (Fig. 9d). In 6- and 15-day-old seedlings, strong GUS staining appeared in the vascular tissues of hypocotyls, cotyledons, true leaves and petioles (Fig. 9a–c, f). In the reproductive organs, strong GUS activity was observed in stigma (Fig. 9g). GUS staining was also visible in petals and sepals (Fig. 9g, h).
Histochemical localization of GUS gene expression driven by the OncHDR promoter in transgenic tobacco. a hypocotyl. b cotyledon. c lower hypocotyl. d root. e, root tip. f, OncHDRp::GUS expression in 15-day-old seedling. g–h, OncHDRp::GUS expression in flower tissue. i Left are 15-day-old seedlings grown in constant dark; right are 15-day-old seedlings grown in 16-h/8-h light/dark. j 1 is 15-day-old seedling and dark adaptive for three days; 2 is a 3-day dark-adaptive seedling, then exposed to 1 h of light treatment; 3 is dark-adaptive seedling, then exposed to 6 h of light treatment; 4 is a dark-adaptive seedling, then exposed to 24 h of light treatment; 5 is a 15-day-old seedling treated with 24 h of continuous light
The OncHDR promoter harbors three I-boxes (Table 1) and an ATCTA motif at positions −541 to −546 relative to the translational start site that were predicted to be light-responsive elements. To determine the effect of light on GUS activity driven by the OncHDR promoter in transgenic tobacco, we compared GUS expression in 15-day-old seedlings grown in constant dark and those grown under 16-h/8-h light/dark conditions. The activity of OncHDRp::GUS was higher under light–dark conditions than under dark conditions (Fig. 9i), which indicated a strong activation of the OncHDR promoter by light. GUS staining was associated only with stems and petioles of seedlings grown in constant dark (Fig. 9i left). To compare the light-mediated induction kinetics of the OncHDR promoter, transgenic 15-day-old tobacco seedlings were grown in the dark for 3 days, then exposed to white light for 0, 1, 6, and 24 h, and light–dark-grown seedlings were exposed to light for 24 h. GUS activity was observed in stems and petioles but not roots of seedlings grown in the dark (Fig. 9j–l). After 1, 6, and 24 h of light treatment, OncHDRp::GUS expression was evident in leaves and roots of transgenic tobacco plants (Fig. 9J-2–4). These results indicate that the inducibility of OncHDR promoter was significantly promoted in light-grown seedlings.
Discussion
HDR genes have been cloned and characterized from organisms such as bacteria, cyanobacteria, and a few plants, but no report did exist on the cloning of HDR genes from orchid species such as Oncidium. Our report probably is the first to describe the cloning and characterization of the Oncidium gene encoding HDR.
Among all the MEP pathway enzymes found in the Arabidopsis and rice genomes, only DXS might be encoded by more than one gene (Rodríguez-Concepción and Boronat 2002). To determine the genomic organization of OncHDR, our DNA blot analysis suggested at least two OncHDR genes in Oncidium (Supplementary Fig. 1), which indicates that OncHDR belongs to a small gene family.
The deduced amino acid sequence of OncHDR is similar to that in other plant species. The functional activity of OncHDR was confirmed via complementation assay with the E. coli HDR mutant, MG1655 ara< >ispH, whereby expression of the OncHDR could rescue the lethal phenotype of the E. coli ispH mutant (Fig. 7). Thus, OncHDR and E. coli IspH (HDR) proteins may have similar enzymatic activity in catalyzing the formation of IPP and DMAPP.
We were able to detect OncHDR expression in different tissue types of Oncidium Gower Ramsey, with moderate higher transcription level in most tissues and lower level in root tissues (Fig. 4b). Low HDR transcript level was also observed in roots of Arabidopsis (Hsieh and Goodman 2005). The expression of OncHDR in leaf, peduncle, and flower bud, and to some extent in roots, might explain the gene’s basic physiological functions in chloroplast biogenesis and metabolism for the building blocks of carotenoid in chromoplasts (Guevara-García et al. 2005; Hsieh and Goodman 2005; Tambasco-Studart et al. 2005). Because Oncidium roots possess some photosynthetic capacity (Hew and Yong 2004), OncHDR may help protect the plant under natural light conditions. The Arabidopsis ispH (HDR) null mutant showed albino phenotype (Guevara-García et al. 2005; Hsieh and Goodman 2005) due to defect in carotenoids biosynthesis. Complementation of the null mutant with HDR gene suggests its role in synthesizing isoprenoid units to produce carotenoid (Guevara-García et al. 2005; Hsieh and Goodman 2005). In tomato, the strong expression of LeHDR was related to carotenoid production during tomato fruit ripening and Arabidopsis seedling deetiolation process (Botella-Pavía et al. 2004). Similarly, our results suggest that OncHDR is also involved in carotenoid biosynthesis of Oncidium plants, as we detected an increase of total carotenoids during flower development (Fig. 5a), and this pigment accumulation was correlated with the OncHDR expression pattern (Fig. 5b).
In conclusion, we have demonstrated the potential function of the OncHDR gene by complementing to the bacterial HDR mutant, by genomic structure comparison and conserved amino acid sequence. The expression pattern of the OncHDR promoter driving the GUS reporter gene in transgenic tobacco plants indicated that the OncHDR promoter is differentially regulated in various plant organs. Strong GUS activity driven by the OncHDR promoter localized in young seedlings and petals as well as vascular bundles in transgenic tobacco (Fig. 9). Furthermore, we have demonstrated the light-induced up-regulation of the OncHDR promoter by a transgenic approach.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Arnon D, Allen M, Whatley F (1954) Photosynthesis by isolated chloroplasts. Nature 174:394
Botella-Pavía P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M (2004) Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40:188–199
Brendel V, Kleffe J, Carle-Urioste JC, Walbot V (1998) Prediction of splice sites in plant pre-mRNA from sequence properties. J Mol Biol 276:85–104
Cercós M, Gómez-Cadenas A, Ho TH (1999) Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J 19:107–118
Chen YH, Tsai YJ, Huang JZ, Chen FC (2005) Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Res 15:639–657
Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49:557–583
Cunningham FX Jr, Lafond TP, Gantt E (2000) Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J Bacteriol 182:5841–5848
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Edwards PA, Ericsson J (1999) Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem 68:157–185
Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6:78–84
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Grawert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, Schramek N, Schleicher E, Weber S, Haslbeck M, Buchner J, Rieder C, Arigoni D, Bacher A, Eisenreich W, Rohdich F (2004) IspH protein of Escherichia coli: studies on iron–sulfur cluster implementation and catalysis. J Am Chem Soc 126:12847–12855
Guevara-García A, San Román C, Arroyo A, Cortés ME, z Gutiérrez-Nava M, León P (2005) Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-d-erythritol 4-phosphate pathway. Plant Cell 17:628–643
Hemmerlin A, Bach TJ (1998) Effects of mevinolin on cell cycle progression and viability of tobacco BY-2 cells. Plant J 14:65–74
Hew CS, Yong JWH (2004) The physiology of tropical orchids in relation to the industry, 2nd edn. World Scientific, Singapore
Hieber AD, Mudalige-Jayawickrama RG, Kuehnle AR (2006) Color genes in the orchid Oncidium Gower Ramsey: identification, expression, and potential genetic instability in an interspecific cross. Planta 223:521–531
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Horsch R, Fry J, Hoffman N, Eichholtz D, Rogers S, Fraley R (1985) A simple and general method of transferring genes into plants. Science 227:1229–1231
Hsieh MH, Goodman HM (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138:641–653
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Lange BM, Rujan T, Martin W, Croteau R (2000) Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA 97:13172–13177
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lichtenthaler HK (1999) The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50:47–65
Maldonado-Mendoza IE, Vincent RM, Nessler CL (1997) Molecular characterization of three differentially expressed members of the Camptotheca acuminata 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) gene family. Plant Mol Biol 34:781–790
McAteer S, Coulson A, McLennan N, Masters M (2001) The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol 183:7403–7407
Nagel R, Elliott A, Masel A, Birch R, Manners J (1990) Electroporation of binary Ti plasmid vector into Agrobacterium tumefaciens and Agrobacterium rhizogenes. FEMS Microbiol Lett 67:325–328
Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15:1591–1604
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee SH, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135:2150–2161
Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38:655–662
Pridgeon A, Chase M (1995) Subterranean axes in three Diurideae (Orchidaceae): morphology, anatomy, and systematic significance. Amer J Bot 82:1473–1495
Rodríguez-Concepción M (2006) Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev 5:1–15
Rodríguez-Concepción M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130:1079–1089
Rohdich F, Hecht S, Gartner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W (2002) Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA 99:1158–1163
Rombauts S, Déhais P, Van Montagu M, Rouzé P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27:295–296
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Lab Press, New York
Shirsat A, Wilford N, Croy R, Boulter D (1989) Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet 215:326–331
Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102:13687–13692
Tang W, Perry SE (2003) Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem 278:28154–28159
Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46:445–474
Wolff M, Seemann M, Tse Sum Bui B, Frapart Y, Tritsch D, Garcia Estrabot A, Rodríguez-Concepción M, Boronat A, Marquet A, Rohmer M (2003) Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe–4S] protein. FEBS Lett 541:115–120
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189
Acknowledgment
This work was supported by a grant from National Science Council, Taiwan (grant number NSC 93-2313-B-020-013).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Puigdomenech.
The nucleotide sequence reported in this paper has been submitted to (NCBI) under accession numbers EU908201 and EU908200.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, JZ., Cheng, TC., Wen, PJ. et al. Molecular characterization of the Oncidium orchid HDR gene encoding 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, the last step of the methylerythritol phosphate pathway. Plant Cell Rep 28, 1475–1486 (2009). https://doi.org/10.1007/s00299-009-0747-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0747-6