Abstract
The transit peptide sequence of ibAGP2 (TP2) was found to be capable of targeting protein into the chloroplast in the Arabidopsis protoplasts. TP2 was fused to a β-glucuronidase (GUS) reporter gene and expressed in Arabidopsis under the control of the ibAGP2 promoter with the aim of dissecting the effect of the transit peptide in elevating foreign protein accumulation in the transgenic plant. β-glucuronidase protein levels were determined at three different developmental stages and in assorted tissues. TP2 dramatically elevated GUS protein accumulation regardless of developmental stage, but the level of the enhancing effect was developmental stage-dependent. This enhancing effect was strongest at the seedling stage (16-fold) and relatively moderate at the vegetative (tenfold) and reproductive (11-fold) stages. TP2 also elevated GUS protein accumulation to varying degrees (4 to 19-fold) in assorted tissues, with the effect being highest in the primary inflorescence stem and petiole (19-fold) and weakest in the root (fourfold). Although TP2 also increased GUS mRNA levels, the increased levels were not large enough to account for the elevated GUS protein levels, suggesting that the enhancing effect of TP2 does not solely result from increased levels of transcripts. Taken together, our results reveal that the TP2 significantly increased the levels of protein accumulation and that its effectiveness was developmental stage- and tissue-type-dependent in transgenic Arabidopsis. Possible differential targeting efficiencies of different transit peptides are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to introduce foreign genes into plant species has provided researchers with the tool to explore the possibility of developing commercially valuable transgenic plant production systems. The use of transgenic plants as host organisms for the production of useful recombinant proteins has a wide range of advantages. These include the cost-effectiveness of transgenic plant production systems and the processing of plant material, the ability of the plant to modify proteins, which enables post-translational modifications, and the absence of a requirement for biosafe and sterile fermentation systems (Hellwig et al. 2004; Daniell et al. 2005; Dus Santos and Wigdorovitz 2005).
The key factor in controlling the yield of recombinant protein in transgenic plants is the optimum design of an expression vector. A strong promoter is a very important factor affecting the productivity of the recombinant protein. One of the most frequently used promoters in recombinant protein production systems in plants is the cauliflower mosaic virus (CaMV) 35S promoter, or its enhanced version. (Sijmons et al. 1990; Fischer et al. 1999; James et al. 2000). Another important factor is the use of an appropriate signal peptide, such as a transit peptide and/or apoplast targeting sequence, which facilitates the transfer of recombinant proteins from the cytoplasm where they are synthesized to subcellular organelles and are accumulated (Hellwig et al. 2004; Jayaraj et al. 2007). A number of introns have also been shown to increase foreign protein yield in transgenic Arabidopsis (Rose and Last 1997; Kim et al. 2006).
The yield of recombinant proteins can be increased by targeting foreign proteins into plastids through the mediation of a transit peptide. Herminghaus et al. (1991, 1996) were able to obtain the expression of a bacterial lysine decarboxylase (LDC) gene fused to the transit peptide of Rubisco small subunit (rbcS) in tobacco hairy root culture and transgenic tobacco plants. These researchers noted that the activity of LDC increased markedly in both transgenic tobacco plants and hairy root culture when the LDC protein was transported into chloroplasts. Bae et al. (2006) reported that the xylanase expressed in the cytosol was targeted either to the chloroplasts or the peroxisomes, or to both organelles simultaneously. When xylanase was targeted only to chloroplasts, the amount of xylanase accumulation in that organelle was increased by 250% than that found in the cytosol, and when xylanase was targeted to both the chloroplasts and peroxisomes simultaneously, the accumulation of xylanase in these organelles was increased by 400% than that in the cytosol.
The most commonly used transit peptide for transporting recombinant protein into the plastids of transgenic plants is rbcS (Comai et al. 1988; Herminghaus et al. 1991; Wong et al. 1992; Corbin et al. 2001). Transit peptides of granule-bound starch synthase (gbss) (Di Fiore et al. 2002; Hoppmann et al. 2002), waxy (Klösgen and Weil 1991), and chlorophyll a/b binding protein (Cab) (Kavanagh et al. 1988) have also been used for targeting foreign proteins into chloroplasts.
In an earlier study (Noh et al. 2004), we reported the isolation of two small subunit ADP-glucose pyrophosphorylase (AGPase) genes (ibAGP1 and ibAGP2) from sweetpotato. Based on a comparison of the open reading frames (ORFs) of sweetpotato and spinach obtained by Edman degradation, we proposed that the N-terminal 73 and 74 amino acid residues of ibAGP1 and ibAGP2 are putative transit peptides (Bae and Liu 1997). The use of ibAGP1 transit peptide (TP1) was subsequently found to be highly effective in increasing the yield of foreign protein, and its enhancing effect was organ-specific (Kwak et al. 2007).
Schaaf et al. (2005) reported that endogenous moss signal peptides demonstrated a secretion efficiency that was up to fivefold higher than that of signal peptides of human origin in transiently transformed moss protoplasts. They also demonstrated that five different moss signal peptides retained differential secretion efficiency. This suggests the possibility that various transit peptides originated from distinct genes and/or organisms might also possess a differential targeting efficiency into plastids of specific plants. However, no study has been conducted to compare transit peptides in terms of their targeting efficiencies and, consequently, for their effects on increasing the accumulation of foreign protein according to various developmental stages and tissue types.
The aim of the study reported here was to compare the targeting efficiency of TP1, reported in our earlier study (Kwak et al. 2007), with that of the ibAGP2 transit peptide (TP2). We therefore have analyzed the effect of TP2 on the accumulation of β-glucuronidase (GUS) protein according to developmental stages and tissue types in transgenic Arabidopsis. Our results reveal that the targeting of GUS protein into plastids with the aid of TP2 dramatically elevated the amount of GUS accumulation. This elevating effect of TP2 was developmental stage- and organ-specific, and the specific effects did not resemble those of TP1.
Materials and methods
In vivo targeting experiment
The 5′-end of ibAGP2 (284 bp), including 31 bp of the 5′ untranslated region, 222 bp of the putative transit peptide, and 31 bp of the mature peptide region, was PCR-amplified using primers 5′-acaggatccATAGCGATCGAAGAG-3′ and 5′-gacggatccGCGAATTCTGGGAGTCGG-3′, which contained BamHI restriction sites to facilitate cloning. The resulting PCR product was fused in frame to the coding region of soluble-modified green fluorescent protein (smGFP). The resulting construct was designated as 35S:TP2:GFP. The fusion construct was introduced into Arabidopsis protoplasts using the polyethylene glycol-mediated transformation method (Jin et al. 2001). Leaf tissues were harvested from 2-week-old Arabidopsis plants and incubated with 50 ml cellulose enzyme solution [1% cellulose R-10, 0.25% macerozyme R-10, 200 mM mannitol, 0.1% bovine serum albumin (BSA), 8 mM CaCl2, 5 mM (N-morpholino)ethanesulfonic acid (MES)-KOH, pH 5.6] at 22°C for 12 h with gentle agitation. The protoplasts were resuspended in 5 ml W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 1.5 mM MES-KOH, pH 5.6), overlaid on top of 20 ml of 21% sucrose, and centrifuged for 5 min at 500 rpm. The intact protoplasts at the interface were transferred to a new conical tube containing 20 ml of W5 solution and incubated on ice for 1 h. Plasmid DNA (10 μg) was added to 300 μl protoplast suspension, followed by the addition of 325 μl polyethylene glycol (PEG) solution [1 M mannitol, 1 M Ca(NO3)2, 40% PEG 8000], and the suspension was then incubated at 25°C for 24 h in the dark. The transformed protoplasts were monitored with a fluorescent microscope (Olympus BX51 TRF) at 475 nm excitation and 510 nm emission.
Generation of transgenic Arabidopsis plants
The ibAGP2 genomic clone had been previously isolated from the genomic library of sweetpotato (Noh et al. 2004). A 2,257-bp fragment of the ibAGP2 5′ flanking region [1,962 bp of promoter region + 49 bp of 5′ untranslated region (UTR) + 246 bp of transit peptide region] was PCR-amplified with gene-specific primers [ibAGP2(-2308)SalF (5′-cgagtcgacCCCTGTGGCGATATCAAC) and ibAGP2(+284)BamR(5′tgcggatccCTGCGAATTCTGGGAGTC)] under the following conditions: 30 cycles of 92°C for 1 min, 58°C for 1 min, and 72°C for 1 min 40 s. The PCR-product was then inserted into the SalI and BamHI sites located at the 5′ of the GUS reporter gene in pBI101 (Clontech, Palto Alto, CA). The resulting construct was designated as ibAGP2:TP2:GUS and transformed into Arabidopsis by the floral dip method (Clough and Bent 1998). Transgenic plants were selected on MS solid medium (Murashige and Skoog 1962) supplemented with kanamycin (50 μg/μl). Transgenic Arabidopsis plants with ibAGP2:GUS were already available (Kwak et al. 2006). More than ten independent T3 homozygous transgenic plants from three different lines transformed with each construct were employed for quantification of GUS activity.
Histochemical and fluorometric analysis of GUS activity
The histochemical and fluorometric analyses of GUS activity were performed as described in Jefferson et al. (1987) with minor modifications. The samples were incubated in 100 mM sodium phosphate, pH 7.0, containing 20 mM 5-bromo-4 chloro-3 indolyl-β-d-glucuronide (X-gluc), 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 10 mM EDTA, and 0.1% Triton X-100 (v/v) at 37°C for 12 h. The stained tissues were washed in 70% ethanol to clear chlorophyll from the tissues. The image was photographed using a SZ61 microscope (Olympus, Japan). For quantification of GUS activity, each tissue was harvested and homogenized in 150 μl of extraction buffer (50 mM phosphate buffer, pH 7.2, 10 mM EDTA, 0.1% Triton X-100, 0.1% sarcosyl, 10 mM β-mercaptoethanol). After centrifugation of the homogenate at 10,000g for 10 min, an aliquot (25 μg proteins) of each supernatant was incubated in 1 mM 4-methylumbelliferyl β-d-galactoside (MUG) at 37°C for 1 h. The reaction was stopped by adding 0.2 M Na2CO3, and fluorescence was determined at 455 nm emission and 365 nm excitation using a fluorescence spectrophotometer (model Mithras LB 940; Berthold Technologies, Bad Wildbad, Germany).
Microscopic examination
Samples were soaked in a fixative containing 2% (w/v) paraformaldehyde and 2.5% (v/v) glutaldehyde in 25 mM phosphate buffer (pH 7.0) at 4°C for 12 h and then dehydrated in a graded ethanol series. The dehydrated samples were embedded in acrylic resin (LR White Resin; London Resin Company, Thales, Berkshire, UK) and sliced into 2-μm-thick sections with an ultra-microtome (model Bromma 2088; LKB, Sweden). Following staining with safranin-O, the tissue sections were observed under a light microscope (Olympus model BX51 TRF).
Northern blot analysis
Total RNAs were isolated from the rosette leaves (100 mg) of transgenic Arabidopsis plants with Nucleospin® (Machery-Nagel, Duren, Germany) according to the manufacturer’s instructions. RNA gel blot analysis was carried out according to the methods described by Sambrook et al. (1989) with following modifications. A 20-μg aliquot of total RNA was run on 1.2% formaldehyde agarose gel and the products were transferred onto a Tropilon-Plus™ membrane (Tropix, Bedford, MA). A biotin-labeled probe (dCTP-biotin; Invitrogen, Carlsbad, CA) was prepared by PCR using GUS-specific primers (forward: 5′-GGGCAGGCCAGCGTATCG-3′; reverse: 5′-CCTTCACCCGGTTGCCAG-3′). Prehybridization and hybridization were conducted in 0.25 M sodium phosphate (pH 7.2), 7% sodium dodecyl sulfate (SDS), and 1 mM EDTA at 68°C. The membrane was then washed in 0.1× SSC and 0.1% sodium dodecyl sulfate (SDS) at 65°C, and the signal was detected with Southern-Star™ (Tropix; Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions.
Results
Amino acid sequence analysis of TP2
An amino acid sequence comparison of TP2 with transit peptides of four other small subunit AGPase sequences, ibAGP1 from Ipomoea batatas, AGPB1 from Solanum tuberosum, VfAGPC from Vicia faba, and PfagpS1 from Perilla frutescens, revealed a 60, 47, 41, and 37% sequence identity, respectively (Fig. 1). The amino acid sequence of TP2 showed relatively high levels of identity with that of TP1, but a relatively low sequence identity was detected in the amino acid sequences of the transit peptides from other species. The amino acid sequences were highly conserved in the N-terminal domain (amino acid positions 1–13), central region (amino acid positions 32–50), and C-terminal end (amino acid positions 66–74) of five transit peptide sequences. Hydrophobic residues (A, I, G, L and P) were abundantly distributed in the N-terminal domain. The 5′ end of the central region (amino acid positions 32–40) possesses sequential hydrophobic (I, F, and A) and hydrophilic (S) amino acid residues, and there is an abundance of hydrophilic amino acid residues (N, S, D, and K) in the 3′ end of the central region (amino acid positions 41–50). Hydrophobic (A, P, I, and V) residues were abundantly distributed in the C-terminal end. Hydrophobic alanine and lysine residues accounted for 23% of the total amino acid frequency of TP2 and hydroxylated residues, such as serine and threonine, for 20% (Fig. 1).
Amino acid sequence alignment of AGPase transit peptides from sweetpotato and other plant species. Amino acid sequence of TP2 was aligned with TP1 from sweetpotato (Ipomoea batatas; GenBank accession no. AAS66988) and three other small subunit AGPase sequences from Solanum tuberosum (GenBank accession no. P23509), Vicia faba (GenBank accession no. P52417), and Perilla frutescens (GenBank accession no. AAF66434). The black, gray, and light-gray backgrounds indicate 100, 80, and 60% identity, respectively. Alignments were performed using GeneDoc software. Arrowheads indicate the serine and threonine residues. Asterisks indicate hydrophobic alanine and leucine residues
Protein targeting properties of TP2
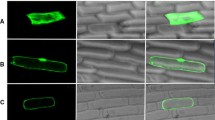
An in vivo targeting experiment aimed at examining the targeting properties of TP2 was carried out using PEG-mediated transformation of heterologous Arabidopsis protoplasts (Fig. 2) and smGFP as a fluorescent marker. The 284-bp fragment of the TP2 and processing site was fused in frame to the coding region of smGFP. The fusion constructs were introduced into the protoplasts of Arabidopsis and their transient expression was observed on epifluorescence images. The fused protein (35S:TP2:GFP) was found to be strongly expressed in chloroplasts, and little or no fluorescence was detected in the cytoplasm. Green fluorescent protein without the transit peptide (35S:GFP) was distributed throughout cytoplasm. This result indicates that TP2 is capable of targeting protein into the chloroplast.
Spatial activity of the ibAGP2 promoter
An initial investigation of the spatial activity of the ibAGP2 promoter in ibAGP2:TP2:GUS transgenic Arabidopsis tissue of three independent transgenic lines revealed that the GUS staining pattern was similar for all three lines (Fig. 3). In young plants, the GUS staining was strong in almost all plant tissues, including the lamina, cotyledons, hypocotyls, petioles, and primary and secondary roots. Microscopic observations revealed that there was a high level of GUS expression in the epidermis, cortex, endodermis, and vascular tissue of the root. In mature plants, GUS expression was strong in the rosette leaves, inflorescence stems, and lateral inflorescence stems; it was also high in anther, stigma, sepal, and filament but relatively low in the petal and style. Closer microscopic examination of the major veins in the lamina revealed strong GUS staining in the epidermis, collenchyma, vascular bundle sheath, phloem, and xylem cells. Strong GUS staining was also detected in the pollen grains of the anther tissue, but GUS expression was relatively low in the epidermis and endothecium cells. These results indicate that the ibAGP2 promoter activity was high in almost all the tissues of Arabidopsis.
Histochemical analysis of ibAGP2 promoter in transgenic Arabidopsis. a Seedling. b Leaf of seedling. c Petiole of seedling. d Hypocotyl of seedling. e Root of seedling. f Cross section of root of seedling. Ep epidermis, Ct cortex, Ed endodermis, Vt vascular tissue. g Inflorescence. h Mature rosette leaf. i Cross section of mature leaf. Ep epidermis, Vb vascular bundle, Co collenchyma, Xy xylem, Vbs vascular bundle sheath, Ph phloem, Enlarged vascular bundle shown in the box. j Stamen and carpel. k Petal. l Sepal. m Anther. n Stigma. o Cross section of anther. Ep epidermis, St Septum, Et Endothecium cell, Pg Pollen grain, Enlarged pollen grain is shown in the box
Effect of TP2 on GUS accumulation
To investigate the effect of TP2 on the accumulation of GUS protein, we quantified the GUS protein in the rosette leaves at various growth stages of transgenic Arabidopsis transformed with ibAGP2:GUS and ibAGP2:TP2:GUS, respectively (Table 1). β-Glucuronidase protein levels increased progressively as Arabidopsis plants matured. At the seedling, vegetative, and reproductive stages, GUS accumulation in ibAGP2:TP2:GUS Arabidopsis was much higher than that in ibAGP2:GUS Arabidopsis. The levels of the enhancing effect of TP2 were distinct among three developmental stages, with TP2 increasing GUS accumulation by 16-, 11-, and 10-fold at the seedling, reproductive, and vegetative stage, respectively. These results indicate that use of TP2 was highly effective in increasing foreign protein accumulation at each developmental stage, with this effect being most prominent at the seedling stage.
The effect of TP2 on GUS accumulation was also determined in various tissues at the reproductive stage (Table 2). At this stage, GUS levels in ibAGP2:GUS Arabidopsis were relatively high in the lamina, moderate in the inflorescence, cauline leaf, and lateral inflorescence stem, and low in the petiole, primary inflorescence stem, and root. β-Glucuronidase accumulation in ibAGP2:TP2:GUS Arabidopsis was higher than that in ibAGP2:GUS Arabidopsis in all the tissues. However, the effectiveness of TP2 was variable in different tissues: GUS protein levels increased dramatically in the petiole (19-fold), primary inflorescence stem (19-fold), and lateral inflorescence stem (16-fold), whereas the root showed only moderately elevated GUS levels (fourfold). These results indicate that the transit peptide of ibAGP2 increased foreign protein accumulation in almost all the tissues of Arabidopsis, but its effectiveness was tissue type-dependent.
Effect of TP2 on GUS transcript levels
To investigate whether the effect of TP2 was attributable to the elevated levels of GUS transcripts, GUS activity and mRNA levels were compared in transgenic Arabidopsis plants transformed with ibAGP2:GUS, ibAGP2:TP2:GUS, and 35S:GUS, respectively. β-Glucuronidase activity was determined in the rosette leaves of transgenic Arabidopsis at the vegetative stage (Fig. 4a). Similar levels of GUS activity were observed in ibAGP2:TP2:GUS and 35S:GUS Arabidopsis plants, while GUS activity in ibAGP2:TP2:GUS Arabidopsis was 11-fold higher than that of ibAGP2:GUS Arabidopsis. An RNA gel blot revealed that the GUS mRNA level in ibAGP2:TP2:GUS Arabidopsis was higher than that in ibAGP2:GUS Arabidopsis (Fig. 4b). The difference in mRNA level, however, was less than 11-fold and, consequently not large enough to account for the difference in GUS activities between these two transgenic lines. The GUS mRNA level in ibAGP2:TP2:GUS Arabidopsis was also significantly lower than that in 35S:GUS Arabidopsis despite both these two transgenic lines showing a similar level of GUS activity. These results imply that the enhancing effect of TP2 in GUS protein accumulation only partially results from elevated levels of mRNA.
Effect of TP2 on β-glucuronidase (GUS) protein and mRNA levels in Arabidopsis plants. a GUS activity in Arabidopsis plants transformed with ibAGP2:TP2:GUS, ibAGP2:GUS, and 35S:GUS, respectively. Nine independent transgenic plants from three different lines transformed with each construct were employed for GUS quantification. Assays were repeated three times. Data are mean ± SD of 27 rosette leaves. b RNA gel blot analysis of GUS transcripts. Twenty micrograms of total RNA from ibAGP2:GUS, ibAGP2:TP2:GUS, and 35S:GUS transgenic lines was loaded per lane and hybridized with biotin-labeled GUS probe
Discussion
To dissect the effect of TP2 in increasing the foreign protein accumulation, we characterized assorted tissue types of ibAGP2:TP2:GUS transgenic Arabidopsis at various developmental stages.
A comparison of the amino acid sequence of the TP2 with that of four other small subunit AGPase transit peptides showed relatively low levels of sequence identity (Fig. 1). This is in agreement with the finding that transit peptide sequences share very little homology in terms of primary amino acid sequences (von Heijne et al. 1989; Bruce 2001; Zhang and Glaser 2002). Our comparison of the amino acid sequence, however, identified three highly conserved domains in the N-terminal, central, and C-terminal regions, respectively, of the AGPase transit peptide sequences, suggesting the possibility that these three domains may be involved in the targeting properties of those transit peptides. It has been reported that transit peptides contain multiple motifs that provide either distinct or possibly overlapping functions (Bruce 2001; Lee et al. 2006). Transit peptides are known to be rich in hydrophobic residues and hydroxylated amino acids, such as serine and threonine (von Heijne et al. 1989; Zhang and Glaser 2002). We found that hydrophobic alanine and lysine residues accounted for 23% of the total amino acid frequency of TP2 and serine and threonine residues for 20%. This feature of TP2 leads us to conclude that TP2 is most likely a transit peptide. This conclusion is supported by the fact that TP2 directly targeted GFP to plastids of Arabidopsis protoplasts (Fig. 2).
TP2 was highly effective in increasing the foreign protein accumulation regardless of developmental stage and tissue type (Tables 1, 2), a property it shares with TP1, which has also been shown to significantly increase GUS accumulation in transgenic Arabidopsis (Kwak et al. 2007). However, TP1 and TP2 were found to have different enhancing levels, with TP1 increasing GUS accumulation by seven- to eightfold in rosette leaves of the seedling, vegetative, and reproductive stages (Kwak et al. 2007), and TP2 elevating GUS protein levels by 10 to 16-fold. TP1 and TP2 have also been found to have different levels of effectiveness in assorted tissues, with TP1 raising GUS levels by twofold to ninefold in various tissues, including lamina, primary inflorescence stem, petiole, root, inflorescence, lateral inflorescence stem, and cauline leaf (Kwak et al. 2007), and TP2 increasing GUS levels by 4 to 19-fold. Although the effect of TP1 and TP2 was determined with two different promoters (ibAGP1 promoter for TP1 and ibAGP2 promoter for TP2), these results suggest that TP2 is more effective than TP1 in elevating foreign protein accumulation in plastids. The effect of TP1 and TP2 could be more precisely compared when they are controlled by an identical promoter.
TP2 increased GUS accumulation in various tissues, but the levels of the enhancing effect were tissue-specific. TP1 also showed tissue-specific strength in its enhancing effect (Kwak et al. 2007). These results suggest that the possible causes of this tissue-specific effect may result from different numbers of plastids in the cells of the different tissues and/or differential sink strengths in different tissues. The increasing effects of TP1 and TP2 were equally weak in the root, which is known to contain relatively low numbers of plastids. This observation provides further evidence that the tissue-specific enhancing effects of TPs may possibly reflect an uneven distribution of plastids in the cells of different tissue types and distinct sink strengths in specific tissues. TP1 and TP2, however, exhibited different tissue-specific effects, in that the effect of TP2 was greatest (19-fold) in the primary inflorescence stem and petiole, while the effect of TP1 was strongest (ninefold) in the inflorescence. These results strongly suggest another possibility that various transit peptide sequences may retain differential targeting efficiencies according to specific tissue types. In the case of signal peptides, five different signal peptides from different species have been shown to have different secretion efficiencies in transiently expressed moss protoplasts (Schaaf et al. 2005).
Studies on transient peptides to date have obtained at least two controversial effects of transit peptides on the mRNA levels of the fused genes. Two groups of researchers have reported that the transit peptide of the Arabidopsis rbcS was equally efficient in increasing protein [CryIA(c), NPT II and PAT] levels and the corresponding mRNA levels in transgenic tobacco (De Almeida et al. 1989; Wong et al. 1992). In another study, Bae et al. (2006) reported that although the transit peptide of Arabidopsis Rubisco activase increased xylanase accumulation by 250%, the mRNA levels of xylanase were not altered in transgenic Arabidopsis. In our study, we found that TP2 significantly increased GUS protein accumulation at various developmental stages and in assorted tissues (Tables 1 and 2). TP2 also elevated GUS mRNA levels, although the difference in GUS mRNA levels was not large enough to account for the difference in GUS protein levels. This result indicates that the TP2 effect cannot solely account for the elevated levels of GUS transcripts and suggests that the TP2 effect may be the result of a synergistic interaction between the elevated steady-state mRNA levels in the cytosol and the enhanced stability of the GUS protein following its encapsulation in the plastids.
Molecular farming on an industrial scale, for example, recombinant protein production in a host plant system in a bioreactor, has received a great attention for its promising potential in terms of large-scale and economically feasible production of valuable pharmaceutical and industrial proteins. The success of any plant molecular farming system will depend on the levels of the final yield. Taking into account the fact that the effect of transit peptides is developmental stage- and tissue type-dependent and different transit peptides retain a distinct effectiveness, the selection of an optimal transit peptide will certainly contribute to a maximization of the yield of the final product.
Abbreviations
- AGPase:
-
ADP-glucose pyrophosphorylase
- TP1:
-
Transit peptide of ibAGP1
- TP2:
-
Transit peptide of ibAGP2
- 35S:
-
Cauliflower mosaic virus 35S promoter
References
Bae JM, Liu JR (1997) Molecular cloning and characterization of two novel isoforms of the small subunit of ADP-glucose pyrophospholylase from sweetpotato. Mol Gen Genet 254:179–185
Bae H, Lee DS, Hwang I (2006) Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J Exp Bot 57:161–169
Bruce BD (2001) The paradox of plastid transit peptide: conservation of function despite divergence in primary structure. Biochim Biophys Acta 1541:2–21
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Comai L, Lasrson-Kelly N, Kiser J, Mau C, Pokalsky AR, Shewmaker CK, McBirde K, Jones A, Stalker DM (1988) Chloroplast transport of ribulose bisphosphate carboxylase small subunit-5-enolpyruvyl 3-phosphoshikimate synthesis chimeric protein requires part of the mature small subunit in addition to the transit peptide. J Biol Chem 263:15104–15109
Corbin DR, Grebenok RJ, Ohnmeiss TE, Greenplate JT, Purcell JP (2001) Expression and chloroplast targeting of cholesterol oxidase in transgenic tobacco plants. Plant Physiol 126:1116–1128
Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R (2005) Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine 23:1779–1783
De Almeida ERP, Gossele V, Muller CG, Dock J, Reynaerts A, Botterman J, Krebbers E, Timko MP (1989) Transgenic expression of two marker genes under the control of an Arabidopsis rbcS promoter: Sequences encoding the rubisco transit peptide increase expression levels. Mol Gen Genet 218:78–86
Di Fiore S, Li Q, Leech MJ, Schuster F, Emans N, Fischer R, Schillberg S (2002) Targeting tryptophan decarboxylase to selected subcellular compartments of tobacco plants affects enzyme stability and in vivo function and leads to a lesion-mimic phenotype. Plant Physiol 129:1160–1169
Dus Santos MJ, Wigdorovitz A (2005) Transgenic plants for the production of veterinary vaccines. Immunol Cell Biol 83:229–238
Fischer R, Schumann D, Zimmermann S, Drossard J, Sack M, Schillberg S (1999) Expression and characterization of bispecific single-chain Fv fragments produced in transgenic plants. Eur J Biochem 262:810–816
Hellwig S, Drossard J, Twyman RM, Fischer R (2004) Plant cell cultures for the production of recombinant proteins. Nat Biotechnol 22:1415–1422
Herminghaus S, Schreier PH, McCarthy JE, Landsmann J, Botterman J, Berlin J (1991) Expression of a bacterial lysine decarboxylase gene and transport of the protein into chloroplasts of transgenic tobacco. Plant Mol Biol 17:475–486
Herminghaus S, Tholl D, Rugenhagen C, Fecker LR, Leuschner C, Berlin J (1996) Improved metabolic action of a bacterial lysine decarboxylase gene in tobacco hairy root cultures by its fusion to a rbcS transit peptide coding sequence. Transgenic Res 5:193–201
Hoppmann V, Di Fiore S, Zimmermann S, Emans N, Rademacher T, Fischer R, Schillberg S (2002) The potato granule bound starch synthase chloroplast transit peptide directs recombinant proteins to plastids. J Plant Physiol 159:1061–1067
James EA, Wang C, Wang Z, Reeves R, Shin JH, Magnuson NS, Lee JM (2000) Production and characterization of biologically active human GM-CSF secreted by genetically modified plant cells. Protein Expr Purif 19:131–138
Jayaraj J, Devlin R, Punja Z (2008) Metabolic engineering of novel ketocarotenoid production in carrot plants. Transgenic Res (in press). doi:10.1007/s11248-007-9120-0
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13:1511–1526
Kavanagh TA, Jefferson RA, Bevan MW (1988) Targeting a foreign protein to chloroplasts using fusions to the transit peptide of a chlorophyll a/b protein. Mol Gen Genet 215:38–45
Kim MJ, Kim H, Shin JS, Chung CH, Ohlrogge JB, Suh MC (2006) Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol Genet Genomics 276:351–368
Klösgen RB, Weil JH (1991) Subcellular location and expression level of a chimeric protein consisting of the maize waxy transit peptide and the beta-glucuronidase of Escherichia coli in transgenic potato plants. Mol Gen Genet 225:297–304
Kwak MS, Noh SA, Oh MJ, Huh GH, Kim KN, Lee SW, Shin JS, Bae JM (2006) Two sweetpotato ADP-glucose pyrophosphorylase isoforms are regulated antagonistically in response to sucrose content in storage roots. Gene 366:87–96
Kwak MS, Oh MJ, Lee SW, Shin JS, Paek KH, Bae JM (2007) A strong constitutive gene expression system derived from ibAGP1 promoter and its transit peptide. Plant Cell Rep 26:1253–1262
Lee DW, Lee S, Lee GJ, Lee KH, Kim S, Cheong GW, Hwang I (2006) Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol 140:466–483
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Noh SA, Kwak MS, Lee HS, Huh GH, Liu JR, Shin JS, Bae JM (2004) Genomic organization of two small subunit ADP-glucose pyrophosphorylase genes from sweetpotato. Gene 339:173–180
Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11:455–464
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schaaf A, Tintelnot S, Baur A, Reski R, Gorr G, Decker EL (2005) Use of endogenous signal sequences for transient production and efficient secretion by moss (Physcomitrella patens) cells. BMC Biotechnol 5:30. doi:10.1186/1472-6750-5-30
Sijmons PC, Dekker BM, Schrammeijer B, Verwoerd TC, van den Elzen PJ, Hoekema A (1990) Production of correctly processed human serum albumin in transgenic plants. Biotechnology (NY) 8:217–221
von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180:535–545
Wong EY, Hironaka CM, Fischhoff DA (1992) Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol 20:81–93
Zhang XP, Glaser E (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7:14–21
Acknowledgments
This work was supported by the grants (No. 20070301034017 and No. 20080401034022) from the BioGreen 21 Program funded by the Rural Development Administration, Republic of Korea, and a grant from the Plant Signaling Network Research Center, the Korea Science and Engineering Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.R. Liu.
Rights and permissions
About this article
Cite this article
Kwak, M.S., Oh, MJ., Paek, KH. et al. Dissected effect of a transit peptide of the ADP-glucose pyrophosphorylase gene from sweetpotato (ibAGP2) in increasing foreign protein accumulation. Plant Cell Rep 27, 1359–1367 (2008). https://doi.org/10.1007/s00299-008-0563-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0563-4