Abstract
Alfalfa is very sensitive to soil acidity and its yield and stand duration are compromised due to inhibited root growth and reduced nitrogen fixation caused by Al toxicity. Soil improvement by liming is expensive and only partially effective, and conventional plant breeding for Al tolerance has had limited success. Because tobacco and papaya plants overexpressing Pseudomonas aeruginosa citrate synthase (CS) have been reported to exhibit enhanced tolerance to Al, alfalfa was engineered by introducing the CS gene controlled by the Arabidopsis Act2 constitutive promoter or the tobacco RB7 root-specific promoter. Fifteen transgenic plants were assayed for exclusion of Al from the root tip, for internal citrate content, for growth in in vitro assays, or for shoot and root growth in either hydroponics or in soil assays. Overall, only the soil assays yielded consistent results. Based on the soil assays, two transgenic events were identified that were more aluminum-tolerant than the non-transgenic control, confirming that citrate synthase overexpression can be a useful tool to help achieve aluminum tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil acidity is a global problem that limits crop productivity (von Uexkull and Mutert 1995). At low pH, aluminum (Al) becomes soluble and available to plants in the Al3+ and Al(OH)2+ forms (Kinraide 1991). Micromolar concentrations of Al3+ can inhibit root growth and, as a consequence, yield is severely reduced as a result of insufficient uptake of water and other nutrients (Kochian 1995). It has been estimated that 56% of the soils in the humid tropics and 40% of the soils worldwide are affected by Al toxicity. Acid soils also cover vast areas in Northern Europe (Buol and Eswaran 1993).
The physiological mechanisms of Al toxicity and resistance have been studied by several groups. Organic acid secretion is one of the principle mechanisms involved (Delhaize et al. 1993; Ryan et al. 1995; de la Fuente and Herrera-Estrella 1999; Lopez-Bucio et al. 2000; Matsumoto 2000; Yang et al. 2000; Ma et al. 2001; Pineros et al. 2002; Zhao et al. 2003). It is hypothesized that the Al-chelating ability of some organic acids, namely citrate, malate and oxalate, confers tolerance through the formation of stable complexes with Al3+ that are not toxic to plants or that cannot enter the roots. In some Al-tolerant plants, one or more of these organic acids are secreted by the root tip as a response to toxic Al, although organic acid exudation does not appear to be the only tolerance mechanism (Pineros et al. 2005; Hoekenga et al. 2006).
Improvement of Al tolerance in plants is a major plant-breeding objective worldwide (reviewed by Samac and Tesfaye 2003). Alfalfa (Medicago sativa L.) is very sensitive to Al3+, and its yield and stand duration in acid soils are compromised because of inhibited root growth and reduced nitrogen fixation due to the acid sensitivity of the symbiotic bacteria (Hartel and Bouton 1989). Several approaches have been suggested to increase the yield of alfalfa in acid soils, including both soil and crop improvement. Soil improvement by liming is expensive and only partially effective (Devine et al. 1990). In addition, surface-liming is not very effective at raising the soil pH below the plow layer (Dall’Agnol et al. 1996).
Conventional plant breeding has had limited success in addressing Al susceptibility in alfalfa. A breeding line improved for yields in acid soils (“GA–AT”) yields significantly more than unimproved ones in the presence of toxic Al, but the yields remain too low, only 20% of normal (Bouton and Sumner 1983; Bouton et al. 1986; Hartel and Bouton 1989; Bouton and Parrott 1997). An Al-tolerant diploid M. coerulea genotype from a Turkish germplasm (PI 464724) was selected using callus production on tissue culture medium containing toxic Al as a screening test (Parrott and Bouton 1990; Dall’Agnol et al. 1996). Tolerance was mapped in a cross with an Al-sensitive M. coerulea (PI 440501), and two RFLP markers were found to be associated with the trait in F2 and backcross populations, using both tissue culture and soil tests. This Al tolerance was then introgressed into tetraploid cultivated alfalfa using 2n gametes and marker-assisted selection (Sledge et al. 2002). Nevertheless, this breeding strategy will still require additional sources of Al tolerance to achieve agronomically useful levels of tolerance. Genetic engineering provides another opportunity to enhance Al tolerance through the expression of endogenous or foreign genes associated with the biosynthesis of organic acids involved in the Al-chelating and detoxifying process.
In alfalfa, overexpression of a nodule-enhanced form of malate dehydrogenase induced a significant increase of the concentration of malate and other organic acids in tissues of some transgenic lines, resulting in Al tolerance (Tesfaye et al. 2001).
Tobacco and papaya plants overexpressing a bacterial citrate synthase (CS) gene exhibited citrate overproduction and enhanced tolerance to Al (de la Fuente et al. 1997). A subsequent study by Delhaize et al. (2001) on the same and other tobacco lines expressing CS at higher levels was unable to confirm the findings. These authors also reported that two CS-expressing alfalfa events did not show improved Al tolerance.
Given the contradictory nature of the information, additional information was necessary to further assess the usefulness of citrate synthase overexpression. Accordingly, a similar genetic engineering approach to Al tolerance was carried out in alfalfa by introducing the CS gene from Pseudomonas aeruginosa controlled by the Arabidopsis actin 2 (Act2) constitutive promoter or the tobacco RB7 root-specific promoter. The results presented show that some alfalfa plants expressing CS can have significantly better root and plant growth in Al toxic soil in a greenhouse test, possibly due to exclusion of Al3+ from the root tip.
Materials and methods
Construction of transformation vectors
The coding region of the P. aeruginosa CS was amplified from the plasmid pPKB (provided by H. W. Duckworth, University of Manitoba, Canada) with the primers forward 5′-ATAGGATCCCATCATGGCTGAC-3′ and reverse 5′-GAGAGCTCAGCCGCGATCCTTG-3′. Inclusion of BamHI and SacI restriction sites (in italic) in the primers allowed the PCR products to be ligated (Fast-Link DNA ligation kit, http://www.epicentre.com) into the BamHI and SacI site of the vector pAPCK between the Act2 constitutive promoter (An et al. 1996) and the NOS terminator. Vector pAPCK contains the NptII gene as selectable marker for kanamycin resistance under the control of the potato ubiquitin3 promoter and terminator (Garbarino and Belknap 1994).
A second CS expression cassette was obtained by substituting the Act2 promoter with the tobacco RB7 root-specific promoter (Yamamoto et al. 1991). This was excised from the pBluescript-TobRB7 plasmid (provided by M. A. Conkling, North Carolina State University) with BamHI and XbaI and ligated into pAPCK–Act2–CS from which the Act2 promoter had been removed with BamHI and SpeI. Each expression cassette was separately subcloned into the AscI and PacI sites of the binary vector pPZP201BK (Covert et al. 2001), thus obtaining the transformation vectors pPZP201BK–Act2–CS and pPZP201BK–RB7–CS (Fig. 1). The integrity of the cassettes was assessed by sequencing, after which they were separately introduced into Agrobacterium tumefaciens strain LBA4404 via electroporation (Gene pulser, http://www.biorad.com) according to the manufacturer’s instructions.
T-DNAs used for alfalfa transformation. UbiT, potato ubiquitin3 terminator; NptII, neomycin phosphotransferase coding sequence; UbiP, potato ubiquitin3 promoter; Act2P, Arabidopsis Actin 2 promoter; CS, citrate synthase coding sequence; NosT, Nopaline synthase terminator; RB7P, tobacco RB7 gene promoter
Alfalfa transformation
One hundred pieces approximately 0.25 cm2 in size, from young, fully expanded leaves of the highly regenerable RSY1 genotype of Regen-SY germplasm (Bingham 1991) were transformed for each construct, using the media, procedures and conditions as described by Austin et al. (1995). About 3 g l−1 of GelRite (http://www.caissonlabs.com) were used as the gelling agent in all tissue culture media; 250 mg l−1 cefotaxime were used for Agrobacterium control. Mature embryos were obtained on selective (25 mg l−1 kanamycin) growth medium, isolated, and subjected to partial desiccation by placing them in groups of 5–15 into a Petri dish containing a piece of solidified B5HO medium in the middle (approx. 0.5 cm3, not touching the embryos) and sealed with Nescofilm (http://www.karlan.com). After 2 days, the embryos were transferred to germination medium (1/2 MS basal medium with B5 vitamins) in Petri dishes for 2 weeks; single plants were then transferred to Magenta GA-7 boxes (http://www.magentacorp.com).
One leaflet was aseptically excised from each in vitro plantlet, and divided into two parts that were used for in vitro regeneration as described, but omitting Agrobacterium treatment; one part was placed on selective medium (50 mg l−1 kanamycin), the other on kanamycin-free medium. Multiple plants from each event were obtained through this second cycle of regeneration under selective conditions (T0-II). One or two plants per transformation event were transferred to soil and then to a greenhouse.
Preliminary screening of transformants by PCR
DNA was isolated with a CTAB protocol (Murray and Thompson 1980) from leaf tissue of the putative transformants. Equal amounts of DNA (200 ng) of the putative transformed and control plants and 1 ng of the binary vector pPZP201BK–Actin–CS were used for PCR with the primers NptIIA (5′-AGAGGCTATTTCGGCTATGAC-3′) and NptIIB (5′-CGAATATCATGGTGGAAAATGG-3′), giving an expected 553-bp amplicon, and with the primers CS-1F (5′-TGA TCA TCG AGG GCT CAG CCC CCG-3′) and CS-1R (5′-CTT GAG GGC GGT GAA GTC GCG CTG-3′), giving an expected 1,252-bp amplicon. The reactions were performed as follows: 5 min at 94°C, followed by thirty-five 45 s cycles at 94°C, 45 s at 67°C (CS) or 55°C (NptII), 90 s at 72°C, and a final extension of 5 min at 72°C.
Southern analysis
Total plant genomic DNA (10 μg per sample) and plasmid pPZP201BK–Actin–CS (10 pg) digested with BamHI (that cuts the T-DNAs between the Act2 or the Rb7 promoter and the CS coding sequence, Fig. 1), were separated by electrophoresis on a 0.8% TAE agarose gel and transferred to a positively charged nylon membrane (Hybond+, http://www.amersham.com) according to Sambrook et al. (1989). A 576-bp PCR fragment of the CS gene was amplified with the primers CS-2F (5′-GGACATCAATAACCCGAAG-3′) and CS-2R (5′-TTCATCGCCAGTTCCAGT-3′) and used as a probe. The probe was labeled with α-[32P] dCTP (http://las.perkinelmer.com) using the Rediprime labeling system (http://www.gelifescience.com). Prehybridization and hybridization were performed at 65°C The membranes were washed twice in 2× SSC 0.1% SDS at 65°C for 30 min each, once in 0.5× SSC, 0.1% SDS at 65°C for 15 min, and once in 0.1× SSC, 0.1% SDS at room temperature for 30 min. The membrane was subjected to autoradiography using Kodak BioMax Film at −80°C for 3 days.
Reverse transcriptase-PCR
Total RNA was extracted from tissue of young leaves of transgenic and control plants with TRIZOL Reagent (http://www.gibcobrl.com) according to the manufacturer’s instructions and treated with RNase-free DNase to remove contaminating DNA. Then, cDNAs were synthesized from 1 μg of total RNA by reverse transcription in a 50-μl reaction containing 12 units of AMV reverse transcriptase (http://www.usbweb.com), 0.3 μg oligo dT primer, 10 mM of each dNTP and 5 μl of 10× reverse transcriptase buffer. Residual RNA was removed by incubating the cDNA in 2 units of E. coli RNase H (http://www.invitrogen.com) for 20 min at 37°C. One twentieth of the reaction was amplified with the citrate synthase-specific primers CS-1F and CS-1R (see above), with 3 min at 94°C, 35 cycles at 94°C, 45 s; 67°C 45 s; 72°C 90 s. The reaction outcome was visualized by ethidium bromide staining after 1% agarose gel electrophoresis. A PCR reaction on total RNA treated with RNase-Free DNase was performed with the same primers without the reverse transcriptase step to confirm the absence of any genomic DNA contamination in the samples. The 18S RNA gene was used as an internal control to normalize for sample to sample variations in total RNA amounts and for reaction efficiency. A 210-bp fragment of the 18S gene was amplified using primers 18S-F 5′-AAGGAATTGACGGAAGGGCACCA-3′ and 18S-R 5′-TAAGAACGGCCATGCACCACC-3′.
Aluminum tolerance assessment
In vitro root growth
To obtain a preliminary estimate of Al tolerance, ten somatic embryos from each transgenic plant and from control plants obtained in a second regeneration cycle were converted into plants on Al-toxic and Al-free modified Blaydes medium (Parrott and Bouton 1990) in 10-cm Petri plates. When the first true leaf was completely expanded, the length of the longest root was measured on each plantlet.
Exclusion of Al from the root tip
Roots from three in vitro T0-II plants per each of seven transgenic events (all the events for which we had three vigorous in vitro plants) and three control plants were excised and treated as described by Polle et al. (1978). The roots were rinsed in distilled water and immersed into 5 ml of one of the three liquid growth media: 1/2 MSO, 1/2 MSO + Al (400 μM AlCl3) or modified Blaydes + Al (40 μM AlCl3) in 50-ml plastic tubes. After 2 h, the roots were rinsed five times in distilled water and treated in the same tubes with 5 ml staining solution (2 g l−1 hematoxylin and 0.2 g l−1 NaIO3) for 15 min. After five rinses in distilled water, the roots were left in water overnight with gentle agitation. The root tips were then excised, placed on blotting paper and photographed immediately.
Evaluation in soil
Rooted cuttings of the 15 transgenic plants confirmed by molecular analysis were used in a greenhouse trial to test for Al tolerance; five of the plants were lost due to sensitivity to bacterial wilt. The plants were grown in a soil mixture containing toxic levels of Al due to its acidity, with and without liming, as described by Dall’Agnol et al. (1996) in 0.72-l polystyrene cups using a randomized complete block design with six replicates. After 6 weeks, the plants were washed free of soil, the root biomass was scored in a 1 (minimum) to 5 (maximum) scale, and the plants were dried and weighed. Data were subjected to the analysis of variance using the SAS GLM procedure (SAS Institute, Cary, NC, USA, 1995).
Citrate concentrations in roots of transgenic plants
Eight transgenic lines and one control plant (RSY1) were grown in aerated hydroponic culture using Hoagland’s solution, with three clonal replications and the roots excised. One gram of roots was ground to powder in liquid nitrogen, extracted in two volumes of 0.6 M perchloric acid, and neutralized with 2 M KOH before assaying for citrate using the enzymatic procedure described in Bergmeyer et al. (1974).
Results
Alfalfa transformation and molecular analyses of transgenic plants
The CS gene sequence was identical to that from the complete genome of P. aeruginosa (Stover et al. 2000, GenBank AE004091.2), revealing a few errors in the originally published sequence (GenBank M29728.1, Donald et al. 1989).
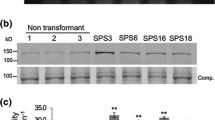
Forty-one putative transgenic alfalfa plants were regenerated, of which 20 were retained following PCR and a functional test of the NptII gene expression through regeneration of leaf explants on kanamycin-containing medium as described. Of these plants, 15 were confirmed to be transgenic for the gene of interest by Southern analysis. A high percentage of escapes was reported previously in RSY1 alfalfa transformation with this protocol (Rosellini et al. 2007). Five of these contained the Actin-CS (labeled with an “A” prefix) construct, and the other ten contained the RB7-CS T-DNA (labeled with a “T” prefix). The CS copy number per event varied between 1 and 6 (Fig. 2). No correlation was observed between the CS gene copy number and the results of Al tolerance tests (not shown).
Southern blot analysis of some of the transgenic plants. The probe was part of the CS coding sequence, as shown in Fig. 1. RSY1: non-transgenic control. The rightmost lane was loaded with a one-copy equivalent amount of the pPZP201BK–Act2–CS vector
When the transgenic plants were tested for CS gene expression by RT-PCR, a 1,252-bp fragment was amplified from all transgenic plants, while no fragment was detected in the non-transgenic control (Fig. 3). Because Northern blot assays failed to detect the transcript (not shown), it is probable that the transcription level or the transcript stability was low.
RT-PCR analyses. a The expected 1.252-kb amplicon from the CS gene was obtained from eight of the transgenic plants, whereas no amplification was detected in the controls. b A 210-bp fragment from the 18S gene was amplified in the same conditions. M (a): 100-bp DNA ladder; A4-T14#2: transgenic plants; 622 and RSY1: untransformed controls; P plasmid pPZP201BK–Act2-CS. M (b): 25-bp DNA ladder; B water
Al tolerance assays
The internal citrate content of roots as estimated in hydroponic culture was generally lower in transgenic than in the non-transgenic control plants (Fig. 4), though the differences were not statistically significant. Different cuttings of the same transgenic events (A4 and A4#2; T14 and T14#2) differed markedly, suggesting that either gene expression is highly variable, or the citrate levels within the cells are highly variable.
All plant cells produce citric acid as part of the citric acid or tricarboxylic acid (TCA) cycle, the regulation of which is still poorly understood (Siedow and Day 2000). Many of the TCA enzymes are subject to negative feedback by high levels of ATP or a high ratio of NADH/NAD+. Such feedback would be triggered by increased levels of citric acid in transgenic cells, and lead to dynamic levels within the cell. Such cell-level regulation of internal citrate concentration could easily explain the variability observed in the levels of citric acid, also reported in the literature.
Likewise, in vitro root length in the presence of toxic Al levels was not significantly higher than that of the non transgenic control for any of the transgenic plants, even though the difference between transgenic plants and controls in the Al-toxic growth medium was much higher than in the control medium (Table 1).
Exclusion of Al from the root tip was suggested by the consistent lack of hematoxylin staining of the root tips shown by some of the transgenic plants (T5 and T8 in Fig. 5). However, root-tip staining was variable (A3, T25) in some transgenic events, and was comparable to that of the non-transgenic control in some others (A4, T7).
The soil test showed that root and shoot biomass did not significantly differ between any of the transgenic plants and the control at a neutral pH (limed soil), which is as expected. However, in the absence of lime (i.e., in acid, aluminum toxic soil), four transgenic plants had significantly higher root mass (Table 1; Fig. 6), and three had significantly higher shoot biomass with respect to the non-transgenic control (Table 1), suggesting Al tolerance. Two plants, A3 and A4, were significantly better than the control for both traits: showing 62 and 37% greater root biomass, and 56 and 121% greater shoot biomass, respectively. Lines A5 and A7 appeared to be Al-tolerant at the root level, but this did not result in increased shoot biomass. In contrast, the increased shoot biomass of T8 was not accompanied by higher root biomass.
Because aluminum toxicity affects all aspects of plant growth, and because the different transgenic events can behave differently, absolute shoot or root mass comparisons to a non-transgenic control are not always appropriate. Instead, each transgenic event can serve as its own best control if its performance is measured as a ratio of the amount of growth a given event has in aluminum-toxic soil as compared to the growth of that same event in soil with a neutral pH in replicated trials.
These ratios are provided in Table 1. A ratio of 100 would indicate the transgenic event performed as well under limed as under unlimed conditions. Most events were comparable to the non-transgenic control, and only produced about half the root mass in unlimed soil as they did in limed soil. Notable exceptions were events A4 and T4, which essentially did as well in limed soil as in unlimed soil.
Shoot mass ratios for most events were also similar to those of the non-transgenic control, and under unlimed conditions only produced about 20% of the shoot mass they produced under limed conditions. Again, events A4 and T4 were the notable exceptions, as their shoot masses under unlimed conditions were about 67% of what they were under the limed conditions.
Given all these criteria, event A4 showed resistance when compared to the non-transgenic control under soil conditions and when compared to itself under limed and unlimed conditions. Event T4 only showed resistance when compared to itself under limed and unlimed conditions.
Discussion
Two conclusions are evident from the results presented here. First, it is clear that enhanced plant growth under acidic, aluminum-toxic conditions can be obtained in transgenic alfalfa plants expressing citrate synthase. Secondly, the type of assay used is critical to properly evaluate the transgenic phenotype.
The widely differing results between de la Fuente et al. (1997) and a subsequent study by Delhaize et al. (2001) have called the strategy of citrate overproduction into question. Overexpression of mitochondrial CS has also been carried out, with encouraging results. Increased CS activity and/or exudation with a concomitant increase in Al tolerance was observed in Arabidopis thaliana (Koyama et al. 2000), cultured carrot cells (Koyama et al. 1999) and in canola plants (Anoop et al. 2003), but not in Aspergillus niger (Ruijter et al. 2000). It is not clear to what extent, if any, the Aspergillus results apply to plants.
The results presented here agree with those of Delhaize et al. (2001), but differ markedly from those of de la Fuente et al. (1997), in that there was no consistent detection of increased citrate content within the tissues, leading to inconsistent results between the in vitro assays and the performance of plants in soil assays. For example, the events that performed best in acid soil, A3 and A4, did not clearly differ from the non-transgenic control for root-tip staining or for in vitro root growth in the presence of toxic Al (Fig. 5; Table 1). Plant T8 showed both decreased root-tip staining and improved shoot biomass, but its roots were not larger than those of the control. The in vitro staining test samples roots during a short, 2-h period, while the soil assay tests the root performance over several weeks. Likewise, the use of root growth in a medium designed for plant cell culture may not accurately represent root growing conditions in soil, illustrating how the type of assay used can have a great influence on the results obtained.
In addition, the negative results obtained by Delhaize et al. (2001) may have been due to the fact that they only presented the analysis for two transgenic alfalfa events. The work presented here found great variability between the 15 different events tested. The two events tested by Delhaize et al. (2001) may simply be too few to capture the full extent of phenotypes possible. Furthermore, their characterization was based on Western blot ands root elongation in hydroponic culture in the presence and absence of Al. In contrast, a soil assay was more reliable than a hydroponic one to measure aluminum tolerance in the work reported here.
Root citrate secretion was not tested in this study, so we cannot exclude that Al tolerance is due to increased citrate exudation by the root tip. Increased citrate secretion in plants overexpressing P. aeruginosa CS was demonstrated in tobacco (de la Fuente et al. 1997). Overexpression of mitochondrial CS genes also resulted in increased citrate efflux in cultured carrot cells (Koyama et al. 1999), Arabidopsis (Koyama et al. 2000), and canola (Anoop et al. 2003) plants. The enhanced aluminum tolerance found in a few of the transgenic events here suggests that this CS overexpression technique is promising, and germplasm with agronomically useful levels of tolerance might be obtained with further refinements. Additional results from the published literature suggest that citric acid may be stabilized if there were a mechanism whereby the cells could excrete the elevated levels of citric acid. Recently, it was demonstrated that overexpression of the ALMT1 gene of wheat, encoding a malate transporter, induced Al tolerance in barley through Al-activated efflux of malate (Delhaize et al. 2004). A strategy that combines citrate overproduction and a way to facilitate its excretion might provide a strategy that can stabilize citrate levels within the cell and lead to increased aluminum tolerance levels.
References
An YQ, McDowell JM, Huang SR, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10:107–121. doi:10.1046/j.1365-313X.1996.10010107.x
Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ (2003) Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 132:2205–2217
Austin S, Bingham ET, Matthews BF, Shahan MN, Will J, Burgess RR (1995) Production and field performance of transgenic alfalfa (Medicago sativa L.) expressing alpha-amylase and manganese-dependent lignin peroxidase. Euphytica 85:381–393
Bergmeyer HU, Gawehn K Grassl M (1974) In: Bergmeyer HU (eds) Methods of enzymatic analysis. vol I, 2nd edn. Academic Press, New York, pp 442–443
Bingham ET (1991) Registration of alfalfa hybrid Regen-SY germplasm for tissue culture and transformation research. Crop Sci 31:1098
Bouton JH, Sumner ME (1983) Alfalfa, Medicago sativa L., in highly weathered, acid soils. V. Field performance of alfalfa selected for acid tolerance. Plant Soil 74:431–436
Bouton JH, Parrott WA (1997) Salinity and aluminum stress. In: McKersie rBD, Brown DCW (eds) Biotechnology and the improvement of forage legumes. CAB International, Wallingford, New York, pp 203–226
Bouton JH, Sumner ME, Hammel JE, Shahandeh H (1986) Yield of an alfalfa germplasm selected for acid soil tolerance when grown in soil with modified subsoils. Crop Sci 26:334–336
Buol SW, Eswaran H (1993) Assessment and conquest of poor soils. In: Maranville JW (ed) Adaptation of plants to soil stresses. Intsormil Publication No. 94–2, University of Nebraska, Lincoln, pp 17–27
Covert SF, Kapoor P, Lee MH, Briley A, Nairn CJ (2001) Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res 105(part 3):259–264
Dall’Agnol M, Bouton JH, Parrott WA (1996) Screening methods to develop alfalfa populations tolerant of acid, aluminum toxic soils. Crop Sci 36:64–70
de la Fuente JM, Ramirez-Rodriguez V, Cabrera-Ponce JL, Herrera-Estrella L (1997) Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 276:1566–1568
de la Fuente JM, Herrera-Estrella L (1999) Advances in the understanding of aluminum toxicity and the development of aluminum-tolerant transgenic plants. Adv Agron 66:103–120
Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L).2. Aluminum-stimulated excretion of malic-acid from root apices. Plant Physiol 103:695–702
Delhaize E, Hebb DM, Ryan PR (2001) Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiol 125:2059–2067
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101:15249–15254. doi:10.1073/pnas/0406258101
Devine TE, Bouton JH, Mabrahtu T (1990) Legume genetics and breeding for stress tolerance and nutrient efficiency. In: Baligar VC, Duncan RR (eds) Crops as enhancers of nutrient use. Academic Press, San Diego, pp 211–252
Donald LJ, Molgat GF, Duckworth HW (1989) Cloning, sequencing and expression of the gene for NADH-sensitive citrate synthase of Pseudomona aeruginosa. J Bacteriol 171:5542–5550
Garbarino JE, Belknap WR (1994) Isolation of a ubiquitin-ribosomal protein gene (ubi3) from potato and expression of its promoter in transgenic plants. Plant Mol Biol 24:119–127
Hartel PG, Bouton JH (1989) Rhizobium meliloti inoculation of alfalfa selected for tolerance to acid, aluminum-rich soils. Plant Soil 116:283–285
Hoekenga OA, Maron LG, Pineros MA, Cancado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Yamamoto Y, Koyama H, Kochian LV (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103:9738–9743
Kinraide TB (1991) Identity of the rhizotoxic aluminum species. Plant Soil 134:167–178
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260
Koyama H, Takita E, Kawamura A, Hara T, Shibata D (1999) Overexpression of mitochondrial citrate synthase gene improves the growth of carrot cells in Al-phosphate medium. Plant Cell Physiol 40:482–488
Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus limited soil. Plant Cell Physiol 41:1030–1037
Lopez-Bucio J, Nieto-Jacobo MF, Ramirez-Rodriguez V, Herrera-Estrella L (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160:1–13
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278
Matsumoto H (2000) Cell biology of Al tolerance and toxicity in higher plants. Int Rev Cytol 200:1–46
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Parrott WA, Bouton JH (1990) Aluminum tolerance in alfalfa as expressed in tissue culture. Crop Sci 30:387–389
Pineros MA, Magalhaes JV, Carvalho Alves VM, Kochian LV (2002) The physiology and biophysics of an aluminum tolerance mechanism based on root citrate exudation in maize. Plant Physiol 129:1194–1206
Pineros MA, Shaff JE, Manslank HS, Alves VMC, Kochian LV (2005) Aluminum resistance in maize cannot be solely explained by root organic acid exudation. A comparative physiological study. Plant Physiol 137:231–241
Polle E, Konzak CF, Kittrick JA (1978) Visual detection of aluminum tolerance levels in wheat by hematoxylin staining of seedling roots. Crop Sci 18:823–827
Rosellini D, Capomaccio S, Ferradini N, Savo Sardaro ML, Nicolia A, Veronesi F (2007) Non-antibiotic, efficient selection for alfalfa genetic engineering. Plant Cell Rep 26(7):1035-1044
Ruijter G, Panneman H, Xu D-B, Visser J (2000) Properties of Aspergillus niger citrate synthase and effects of citA overexpression on citric acid production. FEMS Lett 184:35–40
Ryan PR, Delhaize E, Randall PJ (1995) Characterization of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta 196:103–110
Samac DA, Tesfaye M (2003) Plant improvement for tolerance to aluminum in acid soils––a review. Plant Cell Tissue Organ Cult 75:189–207
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Siedow JN, Day DA (2000) Respiration and photorespiration. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 676–728
Sledge MK, Bouton JH, Dall’Agnol M, Parrott WA, Kochert G (2002) Identification and confirmation of aluminum tolerance QTL in diploid Medicago sativa subsp coerulea. Crop Sci 42:1121–1128
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959–964. doi:10.1038/35023079
Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127:1836–1844. doi:10.1104/pp010376
von Uexkull HR, Mutert E (1995) Global extent, development and economic impact of acid soils. In: Date RA, Grundon NJ, Raymet GE, Probert ME (eds) Plant–soil interaction at low pH: principles and management. Kluwer Academic Publisher, Dordrecht, pp 5–19
Yamamoto YT, Taylor GT, Acedo GN, Cheng C-L, Conkling MA (1991) Characterization of cis-acting sequences regulating root-specific gene expression in tobacco. Plant Cell 3:371–382
Yang ZM, Sivaguru M, Horst WJ, Matsumoto H (2000) Aluminum resistance is achieved by exudation of citric acid from roots of soybean (Glycine max). Physiol Plant 110:72–77
Zhao Z, Ma JF, Sato K, Takeda K (2003) Differential Al resistance and citrate secretion in barley (Hordeum vulgare L.). Planta 17:794–800. doi:10.1007/s00425-003-1043-2
Acknowledgments
We gratefully acknowledge Monica Schmidt for generous advice, Greg Martin for valuable technical help, and Ann Bunce and Donald Wood for their help with the greenhouse plant management and soil assays. This work was funded by NATO Advanced Fellowship Programme 1999, grant #. 215.32 that supported D.R.’s stay at The University of Georgia, and by State and Hatch monies allocated to the Georgia Agricultural Experiment Stations
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Atanassov.
Pierluigi Barone and Daniele Rosellini contributed equally to this work.
Rights and permissions
About this article
Cite this article
Barone, P., Rosellini, D., LaFayette, P. et al. Bacterial citrate synthase expression and soil aluminum tolerance in transgenic alfalfa. Plant Cell Rep 27, 893–901 (2008). https://doi.org/10.1007/s00299-008-0517-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0517-x