Abstract
Signaling pathways, specifically calcium and calcium-dependent protein kinase (CDPK), have been implicated in the regulation of stress and developmental signals in plants. Here, we reported the isolation and characterization of an orchid, Phalaenopsis amabilis, CDPK gene, PaCDPK1, by using the rapid amplification of cDNA ends (RACE)-PCR technique. The full length cDNA of 2,310 bp contained an open reading frame for PaCDPK1 consisting of 593 amino acid residues. Sequence alignment indicated that PaCDPK1 shared similarities with other plant CDPKs. PaCDPK1 transcripts were expressed strongly in labellum but not in leaves and roots. In addition, the PaCDPK1 gene was transcriptionally activated in response to low temperature, wounding, and pathogen infection. To identify the regulatory role of the PaCDPK1 promoter, a construct containing the PaCDPK1 promoter fused to a β-glucuronidase (GUS) gene was transferred into Arabidopsis by Agrobacterium-mediated transformation. GUS staining revealed that PaCDPK1/GUS expression was induced by cold, wounding, and pathogen challenge in leaves and stems of transgenic Arabidopsis. These results suggested that this PaCDPK1 gene promoter could be used as an endogenous promoter for biotechnological purposes in orchids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orchidaceae is one of the largest families of flowering plants. In several tropical countries, orchids have become a major ornamental export crop, and the demand for their cut flowers has increased rapidly over the years. Despite numerous studies involving the physiological aspects of this large group, a molecular understanding of the stress signaling mechanisms in orchids remains unknown.
Plants are exposed to biotic or abiotic stress because of unfavorable changes in their environment and respond with a rapid activation of signal transduction pathways. Among the earliest cellular responses to stress stimuli, such as cold, wounding, or pathogen infection are changes in the cytosolic calcium concentration. The second messenger Ca2+ is a ubiquitous intracellular signaling molecule that regulates many growth and developmental processes (Braun and Schulman 1995; Helper and Wayne 1995; Snedden and Fromm 2001).
Protein kinases belong to the large family of serine/threonine kinases, which are divided into several groups on the basis of structural, biolochemical, and physiological properties. In plants, two classes of protein kinases, mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) have been reported to respond to many environmental stresses and undergo rapid biochemical activation upon exposure to biotic and abiotic stresses (Stone and Walker 1995; Jonak et al. 2002; Ren et al. 2002; Kim et al. 2003). The CDPKs are widespread in plants, but have not been identified in yeast and animal systems (Robert and Harmon 1992). At present, CDPKs comprise a large gene family, 34 members in Arabidopsis (Harmon et al. 2001), and 29 members in rice (Asano et al. 2005). It suggests that CDPKs have different functions and participate in multiple diverse signaling pathways.
The evidence suggests that CDPKs show specific calcium activations and substrate specificities as well as a typical inducement of plant hormones (Abo-El-Saad and Wu 1995; Martin and Busconi 2000). CDPKs also play essential roles in both biotic and abiotic stress signal transduction pathways. Exposure to non-specific elicitors and wounding were reported to cause an increase of NtCDPK1 in tobacco (Yoon et al. 1999). In Arabidopsis, cold, salinity, and drought stresses caused an increase in the transcript level of CDPKs (Urao et al. 1994; Tähtiharju et al. 1997). Recently, genetic engineering of signal transduction mechanisms is starting to emerge (Vinocur and Altman 2005). Saijo et al. (2000) found that over-expression of rice OsCDPK7 increased the tolerance to cold, salinity and draught. Currently, Zhang et al. (2005) suggest that NtCDPK4 might play a vital role in plant development and responses to environmental stress.
In this report, we applied the rapid amplification of cDNA ends (RACE)-PCR technique for full-length cloning of the cDNA, encoding a novel CDPK named PaCDPK1 by using degenerated primers. The expression of PaCDPK1 was tissue-specific and was induced by cold, wounding, and pathogen infection. Therefore, the promoter region of PaCDPK1 has a clear biotechnological interest. A 1,301 bp 5′-flanking DNA fragment of the CDPK gene was isolated with a genome walking method. Then, the promoter sequence was fused with a glucuronidase (GUS) reporter gene, and the construct was delivered into Arabidopsis by Agrobacterium-mediated transformation. We showed that several stress challenges were able to induce in vivo expression of PaCDPK1 promoter.
Materials and methods
Plant materials and treatment with pathogen infection
Phalaenopsis orchid “TS97K” [Phalaenopsis Amabilis W1-10X P. Amabilis W1-22] were purchased by the Taiwan Sugar Company, Tainan, Taiwan. Orchids were grown in a greenhouse under natural light and a controlled temperature ranging from 23 to 27°C.
The Erwinia chrysanthemi (a gift from Mr. Kuo-Ching Wei, Taiwan Sugar Company) was cultured overnight at 28°C in LB medium. To evaluate symptoms in pathogen infected orchid leaves, the leaves were infiltrated with 30 μl of the bacterial culture containing 8–9 × 107 CFU as an original concentration. The inoculated plants were incubated in the growth chamber at 28°C. As a control, mock inoculation was carried out similarly except that a sterile inoculation buffer rather than a bacterial suspension was used.
CDPK fragment production by PCR
Total RNA was isolated from orchid flower buds. The first-stranded cDNA for reverse transcriptase (RT)-PCR was synthesized with Acess RT-PCR (Promega, Madison, WI, USA) following the manufacturer’s instructions. PCR was performed with 50 ng of cDNA and 100 ng of degenerate primers: 5′-AGT GG(C/T) GT(A/C/G) CC(C/G/T) CC(A/C/G/T) TT(C/T) TGG GC-3′ and 5′-C(G/T)(C/G/T) CCA TC(A/C/G) TT(A/G) TC(C/T) TAA TC-3′ corresponding to the conserved amino acid sequences SGVPPFWA and DQDNDGR, respectively. PCR parameters consisted of 2 min at 94°C for denaturing, 1 min at 50°C for annealing, and 1 min at 72°C for extension of 30 cycles, and a final extension step of 10 min at 72°C. PCR products were separated on a 1.8% agarose gel, eluted and sequenced.
3′-RACE (rapid amplification of cDNA ends) of PaCDPK1
3′-RACE-ready cDNA synthesis was performed with the 3′-RACE System (Clontech, Palo Alto, CA, USA). Essentially, RNA was reverse-transcribed with the 3′-RACE CDS Primer A (5′-AAG CAG TGG TAT CAA CGC AGA GTA C(T)30N−1N-3′, N = A,C,G, or T, N−1 = A, G or C). 3′-RACE was performed with the Advantage 2 Polymerase Mix (Clontech). PCR reaction was performed by using a GSP (gene-specific primer) 1 (5′-GAT GTT GAC AAC AGC GGC ACC ATC GAC T-3′) and Universal Primer A Mix (UPM; Long: 5′-CTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA TCA ACG CAG AGT-3′; Short: CTA ATA CGA CTC ACT ATA GGG C-3′) under the following conditions: cDNA was denatured at 94°C for 5 min followed by five cycles (94°C for 5 s, 50°C for 10 s, and 72°C for 2 min), five cycles (94°C for 5 s, 48°C for 10 s, and 72°C for 2 min), and 25 cycles (94°C for 5 s, 46°C for 10 s, and 72°C for 2 min), and by 7 min at 72°C. The PCR product was purified and cloned into pGEM-T vector (Promega) for sequencing.
5′-RACE of PaCDPK1
5′-RACE-ready cDNA synthesis was performed with the 5′-RACE System (Clontech). RNA was reverse-transcribed with the 5′-RACE CDS Primer A [5′-(T)25N−1N-3′, N = A, C, G or T, N-1 = A, G or C] and SMART II A Oligonucleotide (5′-AAG CAG TGG TAT CAA CGC AGA GTA CGC GGG-3′). 5′-RACE was performed with the Advantage 2 Polymerase Mix (Clontech). PCR reaction was performed by using a GSP2 (5′-GCT CGG TTT CCG CCC AAA ATG GAG GCA C-3′) and UPM under the following conditions: cDNA was denatured at 94°C for 5 min followed by five cycles (94°C for 5 s, 62°C for 10 s, and 72°C for 2 min), five cycles (94°C for 5 s, 60°C for 10 s, and 72°C for 2 min) and 25 cycles (94°C for 5 s, 58°C for 10 s, and 72°C for 2 min), and by 7 min at 72°C. The PCR product was purified and cloned into pGEM-T vector (Promega) for sequencing.
DNA blot analysis
Frozen leaf tissue of P. amabilis was ground to a fine powder under liquid nitrogen. Total DNA was isolated as previously described protocol (Sambrook et al. 1989) with minor modifications. In brief, to 2 g of frozen tissue 15 ml of extraction buffer (100 mM Tris–HCl, 1.4 M NaCl, 20 mM Tris–HCl, 2% CTAB, and 1% PVP) and 200 ml β-mercaptoethnol was added followed by 65°C vortexing for 2 h. The sample was re-extracted with 10–15 ml of chloroform followed by centrifugation at 5,000×g for 15 min. DNA was recovered by the addition of an equal volume of isopropanol and incubated at room temperature for 10 min followed by centrifugation at 5,000×g for 15 min. The pellet was resuspended with 1.3 ml NaCl for 10–30 min at 65°C. After the addition of RNase A (10 mg/ml), the samples were incubated at 37°C for 1 h.
For Southern blot analysis, 15 μg of genomic DNA isolated from leaves of P. amabilis was digested with various restriction enzymes EcoRI and EcoRV. The digested DNA was then separated on 2% agarose gel and then transferred to nylon membrane (Hybond XL, Amersham, Buckinghamshire, UK). PCR amplified PaCDPK1 3′ UTR fragment radiolabeled with P32-dCTP according to manufactures instructions was used as the probe for hybridization (Rediprime Labeling System, Amersham). After hybridization the DNA blot was washed twice in 2× SSC, 0.1% SDS for 10 min and 0.1× SSC and 0.1% SDS for 10 min at 65°C. Subsequently the blot was exposed to X-ray film (Kodak, Rochester, NY, USA) with intensifying screens for signal detection at −80°C.
RNA gel blot hybridization and RT-PCR
Total RNA was isolated from orchids by using the RNeasy kit (QIAGEN, Hilden, Germany). Total RNA was fractionated on a 1.0% gel and transferred to MAGNA nylon transfer membranes (MSI, Cambridge, MA, USA) with the TransVac vacuum blotting unit (Hoefer, San Francisco, CA, USA) and UV cross-linked with the Stratalinker UV box (Stratagene, La Jolla, CA, USA). The blots were prehybridized for 2 h in the hybridization buffer (1.3 M NaCl, 1× TPSE, 1× Dehardt’s solution, 10% dextran sulfate, and 50% formamide), and hybridization was carried out at 42°C for 16 h. The membrane was rinsed twice in 2× SSC and 0.1% SDS for 10 min at room temperature, and washed once in 0.1× SSC and 0.1% SDS for 10 min at 42°C followed by 0.1× SSC and 0.1% SDS for 10 min at 60°C. The membrane was exposure to X-ray film (Kodak) with intensifying screens for signal detection at −80°C. As the PaCDPK1-specific probe, a 224 bp fragment was amplified by PCR using the following primers: 5′-GAT GCA GAA GGG TAA TGC AGG-3′ and 5′-CAT TAT GTA TAT GTG GTG AAC-3′. As a standard, a partial fragment of Phalaenopsis rRNA gene was hybridized to all RNA blots (a gift from Dr. Y. Y. Kao, Institute of Molecular and Cellular Biology, National Taiwan University). Probes were labeled with -P32 dCTP using Rediprime Kit (Amersham).
cDNA was synthesized from the total RNA (1 g) by ImProm-II™ Reverse Transcription System for RT-PCR (Promega) with the mix of oligo (dT)18 and (dT)20 as a primer. PCR reactions were set up as follows: 0.5 μl template cDNA; 10×buffer; 10 mM each dNTP; 1 μM PaCDPK primer; 1 U Taq polymerase (Promega); water to 20 μl. The sequence 5′-AAA CCT CAA GGA GCA GAG-3′ was used as the forward primer, while 5′-AGC TAC CGC GTC TTC ATA-3′ was used as the reverse primer. The basic thermocycling parameters were as follows: an initial incubation at 94°C for 2 min; followed by 94°C for 1 min, 48°C (annealing temperature for PaCDPK1) for 1 min, and 72°C for 1.5 min for 35 cycles. The final extension was performed at 72°C for 10 min. The PCR product was sub-cloned into pGEMT-vector (Promega), and sequenced. Experiments were repeated at least twice and reproducibility was confirmed. The orchid actin gene (Actin 9) was used as a positive internal control with primers 5′-GGC TAA CAG AGA GAA GAT GAC C-3′ and 5′-AAT AGA CCC TCC AAT CCA GAC -3′ (Li et al. 2005). The expected sizes of the amplified DNA products were 351 bp—PaCDPK1, 696 bp—actin.
Vector construction and plant transformation
We amplified the PaCDPK1 promoter region using the forward primer: (5′-CGG GAT CCA AAA GCA GGG GTA ATA TT-3′) and the reverse primer (5′-CAT GCC ATG GCC CCA TCA GCT TCA A-3′). The resultant construct was digested with BamHI and NcoI. The 1,301 bp PaCDPK1 promoter was sub-cloned upstream of the coding region of the GUS gene in pCAMBIA1303 between the BamHI and NcoI sites to generate the PaCDPK1 promoter/GUS fusion construct. The PaCDPK1 promoter/GUS fusion was sequenced to verify the reading frame of the gene constructs. The construct was then mobilized into Agrobacterium tumefaciens strain LBA 4404 by electropotation (Equibio, CA, USA) and was selected for resistance to kanamycin (50 mg/l) and streptomycin (50 mg/l). Arabidopsis plants (Col-0 ecotype) were transformed by the floral-dip method (Clough and Bent 1998) and transgenic seedlings were selected on hygromycin medium (50 mg/l).
Histochemical GUS staining
For histochemical GUS analysis, samples were immersed in a staining solution (0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronicacid; 0.1 M NaPO4, pH 7.5; 0.5 mM K3[Fe(CN)6]; 3 mM K4[Fe(CN)6]; 10 mM EDTA, pH 7.5; 0.1% Triton X-100) at 37°C overnight. The samples were then cleared of chlorophyll using 70% ethanol and viewed under a microscope.
Statistical analyses
For the RT-PCR expression analysis, all data were analyzed by software Microsoft Excel (Microsoft Corp., Seattle, WA, USA). Data were expressed as mean ± SD. Differences between treatments were assessed by analysis of variance. P-values <0.1 were considered to be statistically significant.
Results
Cloning and sequencing of PaCDPK1 from Phalaenopsis amabilis
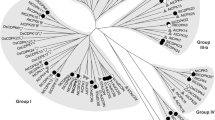
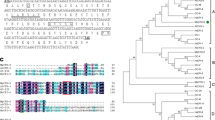
In order to clone the orchid CDPK gene, total RNA was isolated from orchid flower buds and reverse-transcribed into cDNA. A full length cDNA (designed as PaCDPK1) from an orchid was isolated using the RACE approach. The PaCDPK1 (accession number EF555574) cDNA was 2,285 bp long including 3′ UTR, an ORF and the part of 5′ UTR. The PaCDPK1 cDNA contained a 1,779 bp ORF which encoded a 593 amino acid polypeptide with a predicted molecular mass of 65.6 kDa. The deduced protein sequence of PaCDPK1 was highly homologous to other plant CDPKs, including Arabidopsis AtCPK1 (accession number At5g04870, 71% identity), Arabidopsis AtCPK2 (accession number At3g10660, 67% identity), maize ZmCDPK9 (accession number D85039, 61% identity), and tomato LeCDPK1 (accession number AF363784, 59% identity). The sequence alignment indicated that the PaCDPK1 protein possessed all of the structural characteristics of CDPKs (Fig. 1a). The CDPKs have a variable N-terminal domain, a central kinase domain, an auto-inhibitory domain, and a C-terminal CaM-like domain. The kinase domain was a typical serine/threonine kinase containing the eleven sub-domains. The calmodulin-like region contained four EF-hand Ca2+-binding motifs at its C-terminus. In addition, the normally well conserved auto-inhibitory domain connecting the kinase domain to the CaM-like domain in CDPKs was degenerated in PaCDPK1. A phylogenetic tree was constructed with deduced amino acid sequences from cloned CDPKs. The phylogenetic tree based on the amino acid sequence of the entire polypeptide also suggested that PaCDPK1 was related both to CDPKs and to CDPK-related kinases (CRKs) (Fig. 1b). Although CRKs belong to the CPK superfamily, they have the degenerated EF-hand motifs, and their activities do not require calcium. The C-terminal of PaCDPK1 contains the four EF-hands characteristic of all CPKs.
a Comparison of the deduced amino acid sequence of PaCDPK1 with other CDPKs. The deduced amino acid sequence of PaCDPK1 was aligned with the amino acid sequences of CDPKs from AtCPK1 (At5g04870), AtCPK2 (At3g10660), LeCDPK1 (AF363784), and ZmCDPK9 (D85039). Black and gray shaded backgrounds indicated amino acids that were identical or similar, respectively, to the PaCDPK1 sequence. Roman numerals designed the position of kinase sub-domains. The auto-inhibitory domain and EF-hand motifs were also indicated. b Phylogenetic tree made with MegaII software showing the relationship between PaCDPK1 and other kinases. Accession numbers: AtCDPK1 (D21805), AtCDPK2 (D21806), OsCDPK1 (P53682), NtCDPK1 (AF072809), LeCDPK1 (AF363784), ZmCDPK9 (D85039), StCPK1 (AF030879), LeCPK1 (AJ308296), ZmCRK (D84507), and DcCRK (X83869)
Southern blot hybridization was carried out to assess the copy number of the PaCDPK1 gene in the P. amabilis genome, using PaCDPK1 3′ UTR as the probe. Orchid genomic DNA was digested with EcoRI and EcoRV, which cut within the PaCDPK1 cDNA, and blotted for Southern blot. EcoRV-digested products generated only two hybridizing bands (Fig. 2), suggesting that only one copy of PaCDPK1 gene is present in the orchid genome.
Genomic Southern blot analysis of PaCDPK1. Orchid genomic DNA was digested completely EcoRI (1), and EcoRV (2), respectively, which cut only once in the PaCDPK1 cDNA coding region, separated by 2.0% agarose gel electrophoresis, and the blotted onto Hybond XL nylon membrane. The blot was hybridized to a coding region PaCDPK1 cDNA probe. Molecular size markers are indicated on the right
Expression of PaCDPK1 in different parts of orchid
In order to analyze PaCDPK1 expression by Northern blot hybridization, total RNAs were isolated from different organs of orchid and hybridized with the 3′-untranslational region of the PaCDPK1 cDNA. The results indicated that PaCDPK1 was expressed at high levels in labellum and peloric flowers (Fig. 3a, b) but not in other organs tested, including roots, leaves, sepals, petals, columns, or flower buds.
a Northern blot analyses for PaCDPK1 expression in different organs. Total RNA was extracted from orchid plants, including (1) sepal, (2) petal, (3) labellum, (4) column, (5) flower bud, (6) root, and (7) leaf. For each Northern blot analysis, equal loading of the RNA samples on the gel was checked by ethidium bromide-staining of the ribosomal RNA (rRNA). The arrow showed the wounded or inoculated sites. b RT-PCR analyses for PaCDPK1 expression in different parts of flowers. Total RNA was extracted from wild type and peloric flowers, including petal, and labellum. The actin9 transcript was amplified as RT-PCR control. The histograms showed relative intensity mRNA levels in percentage considering the expressed PaCDPK1 mRNA level at wild-type petal as 1. Asterisk, significantly differs from the expression of wild-type petal PaCDPK1 mRNA level at P ≤ 0.1, according to paired t-test. One representative experiment was shown of two separate experiments
PaCDPK1 expression in response to low temperature, wounding, and pathogen infection
To elucidate the role of PaCDPK1 in stress responses, we investigated whether PaCDPK1 could be induced by various stresses (Fig. 4a, b). The level of PaCDPK1 transcript in leaves was significantly higher than the controls after Erwinia inoculation. A time-course of PaCDPK1 induction by pathogen stress was also analyzed (Fig. 4c). The PaCDPK1 transcripts started to accumulate 6 h after Erwinia inoculation, and then increased until 24 h and declined after 48 h.
Northern blot analyses for PaCDPK1 expression in a wounding or b pathogen infection treatment. a Total RNA was extracted from wounded leaves and W1 to W4 revealed different distance to wounding site. b Orchid leaves were inoculated with Erwinia and total RNA was extracted from P1 to P4 and revealed differents distance to the pathogen infection site. For each Northern blot analysis, equal loading of the RNA samples on the gel was checked by ethidium bromide-staining of the ribosomal RNA (rRNA). The arrow showed the wounded or inoculated sites. c The time course of PaCDPK1 expression upon infection. Total RNA was extracted from the leaves of orchid harvested at 0, 6, 12, 24, and 48 h post-inoculation. The actin9 transcript was amplified as RT-PCR control. The histograms showed relative intensity mRNA levels in percentage considering the expressed PaCDPK1 mRNA level at 0 h as 1. Asterisk, significantly differs from the expressed PaCDPK1 mRNA under treatments for 0 h at P ≤ 0.1, according to paired t-test. One representative experiment was shown of two separate experiments
To test possible involvement of PaCDPK1 in response to low temperature, orchids were grown at 28°C for 2 weeks and then either kept at 28°C (control plants) or moved to 4°C (cold-treated plants) or 37°C (heat-treated plants). PaCDPK1 transcript levels increased 3 h after low temperature treatment (4°C; Fig. 5a) but not after 24 h at 37°C (Fig. 5b). Symptom spread following Erwinia inoculcation was also faster at 28°C than at 4°C (Fig. 5c).
a Northern blot analyses for PaCDPK1 expression in response to low temperature. Total RNA was extracted from orchid plants and the number showed different lengths of time after 4°C treatment. For each Northern blot analysis, equal loading of the RNA samples on the gel was checked by ethidium bromide-staining of the ribosomal RNA (rRNA). The arrow showed the wounded or inoculated sites. b RT-PCR analyses for PaCDPK1 expression with different temperature. Total RNA was extracted from orchid leaves kept at 4, 28, and 37°C for 24 h. The acint9 transcript was amplified as RT-PCR control. The histograms showed relative intensity mRNA level in percentage considering the expressed PaCDPK1 mRNA level at 28°C as 1. Asterisk, significantly differs from the expressed PaCDPK1 mRNA level under 28°C at P ≤ 0.1, according to paired t-test. One representative experiment was shown of two separate experiments. c The effect of different concentrations of pathogen infection (1.original 2. 1/10 original 3. 1/100 original 4. 1/1,000 original 5. 1/10,000 original 6. 1/100,000 original, respectively), on orchid leaves pretreated 4°C or 28°C for 24 h
Cloning and structural features of the PaCDPK1 promoter
The promoter region of the orchid PaCDPK1 gene were isolated from orchid genomic DNA by the Genome Walking strategy. The 5′ 1,301 bp upstream of the translation start codon were analyzed for putative cis-elements in the promoter region by using PLACE (a database for plant cis-acting regulatory DNA elements located at http://www.dna.affrc.go.jp). We found one ethylene-responsiveness element (ERE), one WB-box, two low-temperature-responsive elements (LTREs), and three W-box element (accession number EF587760, Table 1).
Activity analysis of the PaCDPK1 promoter during environmental stresses
For functional analysis of this promoter region of the orchid PaCDPK1 gene, the 1,301 bp promoter fragment was amplified by PCR and cloned into pCAMBIA 1303 resulting in transcriptional fusion with the gus gene. We produced transgenic Arabidopsis plants and GUS activity of Arabidopsis seedlings containing PaCDPK1/GUS construction was analyzed under three environmental stresses. PaCDPK1/GUS transgenic Arabidopsis seedlings subjected to low temperature, wounding, and pathogen infection exhibited deep blue GUS staining (Fig. 6). When seedlings were subjected to various temperatures between 4 and 28°C for 24 h, the induction of PaCDPK1/GUS was detected at 4–16°C (Fig. 7).
Histochemical analysis of GUS activity in transgenic Arabidopsis containing the PaCDPK1 promoter-GUS fusion gene under different environmental stimuli. GUS expression was observed mainly in the leaf region under stresses. GUS staining in low temperature, wounding, pathogen infection leaves, and stem
Discussion
Considering its status as one of the rarest and most exotic flowers in the world, a better understanding of stress signaling in orchids would undoubtedly have enormous benefits. Since the first isolation of a CDPK gene from Arabidopsis, CDPK genes have been cloned from a wide variety of plants species, including rice, maize, tobacco, and strawberries (Harper et al. 1991; Breviario et al. 1995; Berberich and Kusano 1997; Yoon et al. 1999; Llop-Tous et al. 2002). In this paper, we explored the cloning of PaCDPK1, a CDPK from orchid plants that was up-regulated in response to low temperature, wounding, and the presence of pathogen. Database searches revealed the presence of genes encoding this type of stress-inducible CDPK in both monocots and dicots. PaCDPK1 belongs to a sub-class of stress-inducible CDPKs. PaCDPK1 also showed the typical structural features of the CDPK family, including a conserved catalytic Ser/Thr kinase domain, an auto-inhibitory junction domain and a CaM-like domain with four putative Ca2+-binding EF hands.
Here, Southern analysis suggested that PaCDPK1 is a single copy gene. However, recent reports showed that there were numerous isoforms of CDPKs in the Arabidopsis genome (Cheng et al. 2002) and in other plant species (Asano et al. 2005). This suggested that individual isoforms have different functions and participate in multiple distinct signaling pathways (Romeis et al. 2001). In addition, different CDPK genes may be expressed in different tissues or at different stages during growth and development of a plant (Estruch et al. 1994; Llop-Tous et al. 2002). Here, the expression of PaCDPK1 was analyzed in the different tissues of orchid, and it was abundant in labellum but not expressed constitutively in other orchid organs. Furthermore, high-expression levels of PaCDPK1 were also detected in the petals of peloric flowers. These results suggested that PaCDPK1 may be involved in orchid flower development.
A challenge in plant signaling is to understand how cellular responses to environmental stress are activated and it is believed that CDPKs are involved in environmental and hormonal signal transduction. The expression of CDPK genes from different plant species have been implicated in general stress responses, such as low temperature, wounding, fungal elicitor, draught, hormone, and light stresses (Berberich and Kusano 1997; Frattini et al. 1999; Murillo et al. 2001). Saijo et al. (2000) have shown that over-expression of OsCDPK7 could enhance induction of some stress-responsive genes in response to salinity/drought. Here, we showed that a newly identified PaCDPK1 was up-regulated in response to low temperature, wounding, and bacterial treatments. These results suggest that this PaCDPK1 gene could be used for biotechnological purpose in orchid.
We searched the 5′ upstream region of PaCDPK1 gene for known motifs of other genes and found several potential regulatory elements (Table 1). Here, transferring the PaCDPK1 promoter/GUS gene fusion construct into Arabidopsis allowed us to further characterize the regulatory pattern of the PaCDPK1 promoter. As shown in Fig. 5, that GUS accumulated at the leaves and stem when expose to low temperature, wounding, and pathogen infection treatments. However, roots were poorly stained by GUS expression.
Our findings revealed that the PaCDPK1 gene from orchid could be regulated by environmental stresses at the transcriptional level. We also demonstrated that the cloned P. amabilis CDPK1 promoter is activated by low temperature, wounding, and pathogen challenge in transgenic Arabidopsis. The study presented here is promising, but it would be interesting to elucidate the potential tissue specific of PaCDPK1 promoter in orchid. Furthermore, this activity of the inducible promoter in orchid may be used as biotechnological tools.
References
Abo-El-Saad M, Wu R (1995) A rice membrane calcium-dependent protein kinase is induced by gibberallin. Plant Physiol 108:787–793
Asano T, Tanaka N, Yang G, Hayshi N, Komatsu S (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46:356–366
Berberich T, Kusano T (1997) Cycloheximide induces a subset of low temperature inducible genes in maize. Mol Gen Genet 254:275–283
Braun AP, Schulman H (1995) The multifunctional calcium/calmodulin-dependent protein kinase: from form to function. Annu Rev Physiol 57:417–445
Breviario D, Morello L, Giani S (1995) Molecular cloning of two novel rice cDNA sequences encoding putative calcium-dependent protein kinases. Plant Mol Biol 27:953–967
Cheng SH, Willmann MR, Chen H, Sheen J (2002) Calcium signaling through protein kinase. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129:469–485
Clough S, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Estruch JJ, Kadwell S, Merlin E, Crossland L (1994) Cloning and characterization of a maize pollen-specific calcium-dependent protein kinase. Proc Natl Acad Sci USA 91:8837–8841
Frattini M, Morello L, Breviario D (1999) Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development. Plant Mol Biol 41:753–764
Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Physiol 151:175–183
Harper JF, Sussman MR, Schaller CE, Putnam-Evans C, Charbonneau H, Harmon AC (1991) A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252:951–954
Helper PK, Wayne RO (1995) Calcium and development. Annu Rev Plant Physiol 36:397–439
Jonak C, Okresz L, Bogre L, Hurt H (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5:415–424
Kim CY, Liu Y, Thorne EJ, Yang H, Fukushige H, Grassmann W, Hildebrand D, Sharp RE, Zhang S (2003) Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15:2707–2718
Li SH, Kuoh CS, Chen YH, Chen HH, Chen WH (2005) Osmotic sucrose enhancement of single-cell embryogenesis and transformation efficiency in Oncidium. Plant Cell Tissue Organ Cult 81:183–192
Llop-Tous I, Dominguez-Puigianer E, Vendrell M (2002) Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J Exp Bot 53:2283–2285
Martin ML, Busconi L (2000) Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitolation. Plant J 24:429–435
Murillo I, Jaeck E, Cordero MJ, San Segundo B (2001) Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitor and infection. Plant Mol Biol 45:145–158
Ren DT, Yang HP, Zhang SQ (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277:559–565
Roberts DM, Harmon AC (1992) Calcium-dependent proteins-targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol 43:375–414
Romeis T, Ludwig AA, Martin R, Jones JDJ (2001) Calcium-dependent protein kinase plays an essential role in a plant defense response. EMBO J 20:5556–5567
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izuki K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sambrook J, Fritch EF, Maniatis T (1989) Molecular cloning: a laboratory manuel, 2nd edn. Cold Spring Harbor Press, New York
Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal tranducer in plants. New Phytol 151:35–66
Stone JM, Walker JC (1995) Plant protein kinase families and signal transduction. Plant Physiol 108:451–457
Tähtiharju S, Sangwan V, Monroy AF, Dhindsa RS, Borg M (1997) The induction of Kim gene in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta 203:442–447
Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K (1994) Two genes that encode Ca2+-dependent protein kinase are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet 244:331–340
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Yoon GM, Cho HS, Ha HJ, Liu JR, Pai Lee HS (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39:991–1001
Zhang M, Liang S, Lu YT (2005) Cloning and functional characterization of NtCPK4, a new tobacco calcium-dependent protein kinase. Biochim Biophys Acta 1729:174–185
Acknowledgments
This study was supported by Grants NSC 93-2317-B-006-004, NSC 94-2317-B-006-003, and NSC 95-2317-B-006-002 from the National Science Council and 91-3.1.3-Z3 from the Council of Agriculture, Taiwan. We thank Ms Lorraine Rooney for assistance with English correction of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Register.
Tsung-Mu Tsai and Ying-Ru Chen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tsai, TM., Chen, YR., Kao, TW. et al. PaCDPK1, a gene encoding calcium-dependent protein kinase from orchid, Phalaenopsis amabilis, is induced by cold, wounding, and pathogen challenge. Plant Cell Rep 26, 1899–1908 (2007). https://doi.org/10.1007/s00299-007-0389-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0389-5