Abstract
A highly efficient and convenient method for the Agrobacterium rhizogenes-dependent production of transformed roots of Saponaria vaccaria L. (Caryophyllaceae) is described. The parameters tested and optimized include S. vaccaria cultivar, explant type, Agrobacterium rhizogenes strain and culture conditions. For cotransformation using additional recombinant T-DNA-containing A. rhizogenes strains, use of neomycin phosphotransferase and enhanced green fluorescent protein genes as selectable markers were tested alone and in combination. Optimal results, yielding a minimum of one transformed root per explant, were obtained using the cultivar Pink Beauty, the A. rhizogenes strain LBA9402 and internode explants precultured on a phytohormone mixture. Selection of cotransformed roots by observation of enhanced green fluorescent protein fluorescence alone was highly effective and convenient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saponaria vaccaria L. (syn. Vaccaria segetalis and Vaccaria hispanica), also known as cow cockle, is a widely distributed member of the Caryophyllaceae family (Sang et al. 2003). Seeds of this annual herb have been used in traditional Chinese medicine for the treatment of amenorrhea, breast infections and the stimulation of lactation (Sang et al. 2003). Included in the bioactive compounds found in S. vaccaria are triterpene saponins, similar to those found in the bark of Quillaja saponaria (Jia et al. 2002; Sang et al. 2003). Since such saponins have a variety of industrial and health-related applications, there is value in understanding the biochemistry and genetics of their biosynthesis in the plant. As a prelude to the relevant gene identification studies, it was of interest to us to establish a platform to carry out gene identification through a combination of RNA interference methods and a convenient genetic transformation system. For plants in which the traits of interest are expressed in roots, the use of Agrobacterium rhizogenes-transformed roots provides an efficient system for such gene identification (Kumagai and Kouchi 2003).
The laboratory use of A. rhizogenes to generate transformed root cultures as model systems and for natural product production is well established (Tepfer 1984; Christey 1997). The A. rhizogenes bacterium contains an Ri plasmid which includes T-DNA regions. Infection of plant tissue with A. rhizogenes results in the integration of T-DNA into the plant genome and the production of adventitious roots. These roots, which can be cultured indefinitely without exogenous hormones, typically show a distinct morphology which includes plagiogeotropism and enhanced lateral branching and root hair formation–—the “hairy root” phenotype (Tepfer 1984).
It is possible to produce hairy root cultures with additional genetic modifications by cotransformation with an additional Agrobacterium strain which includes recombinant T-DNA in an appropriate plasmid. For example, expression of a cytochrome P450 in plants was tested by transferring the gene into Atropa belladonna roots (Banerjee et al. 2002). Similarly, the role of endogenous genes can be tested through the use of RNA interference combined with the use of transformed roots (Kumagai and Kouchi 2003). In both instances, the uses of transformed roots obviated the need for the generation of whole plants, a time-consuming and potentially very inefficient process. On the other hand, for some systems, the possibility exists to regenerate whole transformed plants from transformed roots, after a relative rapid test for the desired phenotype (Tepfer 1984).
In this paper, with a view to future gene identification studies, we describe the development of a highly efficient system for A. rhizogenes-mediated transformation of S. vaccaria. Parameters including S. vaccaria cultivar, A. rhizogenes strain, culture conditions and antibiotic and fluorescence selection regimes were tested and optimized.
Materials and methods
Plant material
Seed of S. vaccaria ‘Scott WT’ was obtained from Eric Johnson (Agri-Food and Agriculture Canada, Scott Experimental Farm, Scott, Saskatchewan). Seed of S. vaccaria cv ‘Pink Beauty’ and ‘White Beauty’ were obtained from CN Seeds, Denmark House, Pymoor, Ely, Cambridgeshire CB6 2EG United Kingdom U.K. and accession. PI597629 originating from Mongolia was obtained from the North Central Regional Plant Introduction Station, USDA-ARS.
Seeds were surface sterilized in sodium hypochlorite (5.6% w/v) and approximately 0.003% (v/v) Tween 20 for 30 min, followed by 15 min in mercuric chloride (0.025% w/v) and washed four times in sterile water. The seeds were germinated on half strength MS medium (Murashige and Skoog 1962) pH 5.8, with 1% sucrose, 0.05 mg/l BA, 0.05 mg/l NAA and 0.8% Phyto Agar in the dark at 4°C for approximately 10 days. After this cold synchronization, the plants were transferred into sterile magenta jars and maintained at 25°C with 16 h light photoperiod for 9 days.
To generate non-sterile plants for stem inoculations, surface sterilized germinated seedlings of PI597629, Pink Beauty, Scott WT and White Beauty varieties were moved to redi-earth containing cells and grown under a 16°C, 16 h day and 6°C, 8 h night for 5 weeks. Healthy seedlings were then transferred to pots and grown at 22°C, 16 h day and 17°C, 8 h night for 7 weeks. Leaves from node 3 to node 8 were trimmed back almost to the base of the stem before inoculation.
To generate axenic plants, as an internode source and occasional leaf source, the seedlings were moved individually to magenta jars containing 50 ml half strength MS medium, pH 5.8 with 1% sucrose, and 0.8% Phyto Agar and maintained at 25°C, 16 h photoperiod, from 3 weeks to 2 months.
Transformation and tissue culture
Agrobacterium rhizogenes wild type strains A4, 15834, and LBA9402 were grown on solid YMB medium (Hooykaas et al. 1977). The binary vector pK7GWIWG2D(II) (Karimi et al. 2005) was introduced into LBA9402 by electroporation and selected on YMB medium with 100 mg/l spectinomycin. The vector contains T-DNA that includes the selectable marker genes neomycin phosphotransferase (nptll) and enhanced green fluorescent protein (egfp) (Cormack et al. 1996).
Stem inoculations were performed in a fashion similar to Komarnytsky and coworkers (Komarnytsky et al. 2004). Suspensions totaling approximately 500 μl A. rhizogenes grown for 48 h in YMB (or 500 μl of YMB for negative controls) were injected into each single stemmed plant with a 25 gauge needle using 50 injections per stem. Plastic overhead sheets, Parafilm and tape were used to make an approximately 8 cm long humidity chamber to cover the inoculation area and maintain high humidity. Roots became visible starting at 2 weeks and were scored 3 weeks after inoculation.
To obtain seedling explants for tissue culture, 9-day-old seedlings were cut at the point where they emerged from the MS medium and the aerial parts were transferred to water-soaked filter paper. A cut was made to free the cotyledons and leaves. If hypocotyls were used, a second cut was made in the middle of the hypocotlyl. Leaves from axenic plants were excised at the petiole.
Internode explants were prepared from axenic plants by transferring the aerial parts to water-soaked filter paper. The internodes were dissected away from the nodes. Each internode was cut transversely into approximately 2 mm disks.
For tissue culture protocol A (Table 1), the excised explants were immersed in A. rhizogenes inoculum (strain 15834 or LBA9402) containing 100 μM acetosyringone and in the case of cotyledons and leaves, wounded by scoring five times across the rib on filter paper. Uninfected control explants were scored in sterile ddH2O. The explants were blotted to remove excess moisture and placed on co-cultivation medium (1× MS, 3% sucrose, 50 μM acetosyringone, 0.8% Phyto Agar, pH 5.8) for 2 days at 25°C, 16 h photoperiod. After co-cultivation the explants were placed on culture medium (1× MS medium with 3% sucrose, 0.8% Phyto Agar, pH 5.8 and 300 mg/l Timentin) and left at 25°C (16 h photoperiod) until adventitious roots developed (2–3 weeks).
For Protocol B, Protocol A was modified as follows. Explants were precultured on pre-culture medium (containing 1 × MS, 3% sucrose, 0.2 mg/l BA, 1.0 mg/l NAA, and 0.8% Phyto Agar, pH 5.8) for 4 days, prior to wounding and inoculation. For internodes, 2 mm disks were scored three times on face. After inoculation, explants were transferred to filter paper soaked with 50 μM acetosyringone solution and placed at 25°C, 16 h photoperiod for 2 days, followed by transfer to culture medium, which in some cases included 100 mg/l paromomycin.
Selection and analysis of transformed roots: initial transformation and derived root cultures
In order to test antibiotic and fluorescence selection regimes, selection was performed during the initial transformation phase. Subsequently, selected adventitious roots derived from infected explants were cultured and the resulting cultures were tested for genotype by PCR and for antibiotic resistance and fluorescence phenotype as follows. Three weeks after infection, 1–2 cm long adventitious roots were chosen from separate explants either at random (for no selection and for 100 mg/l paromomycin selection alone), or according to positive GFP fluorescence (for fluorescence selection and for combined fluorescence and antibiotic selection). The roots were excised and transferred to individual plates containing MS medium with 3% sucrose, 0.8% Phyto Agar and 300 mg/l Timentin either with or without 50 mg/l paromomycin. The root cultures were grown for 2 weeks in the dark at 25°C after which they were scored for GFP fluorescence and growth on paromomycin and 50–100 mg samples were taken for PCR analysis. As a negative control, clones were also obtained from roots resulting from transformation with the A. rhizogenes strain LBA9402 (without the pK7GWIWG2D(II) vector).
Fluorescence microscopy and spectroscopy
GFP fluorescence in plant tissues was observed using a Leica MZFLIII stereomicroscope equipped for epifluorescence using the GFP1 [425(60) nm excitation, >480 nm emission] and GFP3 [470(40) nm excitation, 525(50) nm emission] filter sets. Photomicrographs were taken using an Optronics DEI-750 CCD camera.
Fluorescence spectroscopy was performed using a Perkin Elmer model LS50 Luminescence Spectrometer and FL Data Manager software. Four-week-old roots were pulled away from solid growth media with forceps and immediately placed in a quartz cuvette. Fluorescence emission spectra were digitally recorded from 460–700 nm, with a slit width corresponding to ±5 nm, and an excitation wavelength of 425 ± 10 nm.
PCR analysis
DNA was extracted (Bekesiova et al. 1999) from a 50 to 100 mg sample of each root culture, dissolved in 50 μl ddH2O and subjected to multiplex PCR analysis to simultaneously score for the presence or absence of the rolC, virD1, egfp and nptII genes. The multiplex PCR was designed to include a primer set that generates a 490 bp fragment of the rolC gene (Bonhomme et al. 2000) plus three primer sets (virD1, egfp and nptII) all selected for minimal cross-interaction and for production of distinct fragments after separation of PCR reaction products on a 1.2% agarose gel. This includes a 441 bp fragment of the virD1 gene (5′-ATGTCGCAAGGCAGTAAG-3′ and 5′-CAAGGAGTCTTTCAGCATG-3′), a 311 bp fragment of the egfp gene (5′-CCTCGTGACCACCCTGACC-3′) and 5′-CCTTGATGCCGTTCTTCTGC-3′) and a 698 bp fragment of the nptII gene (5′-GGATTGCACGCAGGTTCTC-3′ and 5′-CTCTTCAGCAATATCACGGGTAG-3′).
PCR reaction mixtures (25 μl) contained 1 μl of DNA extract and 1× Invitrogen “PCR Buffer minus Mg2+”, 2 mM MgCl2, 0.2 mM of each dNTP, 0.4 μM of each of the six rolC, egfp and nptII primers, 200 ng of each of the two vird1 primers and 2.5 U Invitrogen Taq DNA polymerase. Amplification was performed in a Stratagene Robocycler Gradient 96 using the following program: 95°C for 4 min, 29 cycles of 95°C for 45 s, 50°C for 45 s, and 72°C for 1 min, followed by 72°C for 10 min.
Results
Root production in S. vaccaria transformed with A. rhizogenes
As an initial test of the ability of A. rhizogenes to transform S. vaccaria, in planta stem inoculations were performed. These included four S. vaccaria varieties (Pink Beauty, White Beauty, PI597629 and Scott WT) and three A. rhizogenes strains (LBA9402, 15834 and A4). While not all combinations were tested, Pink Beauty showed the highest level of root formation with all three A. rhizogenes strains, with LBA9402 being the most infectious strain (data not shown). None of the strains induced root formation in inoculated stems of the Scott WT variety. Thus, the best combination of variety and strain tested was Pink Beauty and LBA9402 (data not shown). Clearly, hairy roots could be formed in S. vaccaria, albeit in a plant and bacterial genotype-specific manner. It is notable that chemical analysis of the S. vaccaria transformed root cultures indicated the presence of saponins (J. Balsevich et al. unpublished) similar to those present in the seed (Sang et al. 2003), indicating that saponin biosynthesis occurs in the roots of this species.
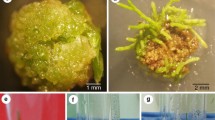
As a next step in the development of a method for A. rhizogenes-dependent genetic transformation of S. vaccaria, tests were done in axenic tissue culture using Protocol A (see Materials and methods). This involved five types of seedling explants (roots, upper hypocotyls, lower hypocotyls, cotyledons and leaves) from the Pink and White Beauty varieties and two strains of A. rhizogenes (Table 1 and data not shown). Three explant types were found to be unsuitable for hairy root formation. Root explants showed consistently high levels of fungal contamination. The top half of hypocotyls showed a very high frequency of shoot and root formation in the absence of A. rhizogenes infection. The bottom half of hypocotyls showed a very low frequency of root formation. However, as indicated in Fig. 1b and Table 1, a good frequency of A. rhizogenes-dependent root formation from cotyledon and leaf explants was found using protocol A. Figure 1 demonstrates the low frequency of root formation in uninfected Pink Beauty cotyledons protocol A (Fig. 1a) and moderate frequency of hairy root formation upon A. rhizogenes infection (Fig. 1b). Table 1 illustrates the effect of variety, explants and A. rhizogenes strain on the frequency of root formation. In agreement with stem inoculations (data not shown), Pink Beauty showed better root formation than White Beauty (Table 1). White Beauty showed some increases in root formation over uninfected explants with LBA9402. Pink Beauty leaves infected with LBA9402 gave the highest root production relative to uninfected explants. However, this effect was highly variable. Based on consistently high root formation, the Pink Beauty/cotyledon/LBA9402 combination was chosen for further study. The extent of true T-DNA-mediated transformation of roots was determined by rolC PCR (see below) for this combination and was found to be 59 ± 18% (data not shown; all roots were found to be PCR-negative for virD1, indicating that A. rhizogenes contamination did not account for the rolC PCR result).
Root production from S. vaccaria Pink Beauty seedling explants using A. rhizogenes strain LBA9402. Adventitious root formation on cotyledons using Protocol A (a, b) and Protocol B (c, d) and on internodes using protocol B (e, f). Uninfected (a, c and e) and infected (b, d and f) explants are shown. Eight-week-old non-transformed (spontaneous) (g) and transformed (h) root cultures are also shown. See Materials and methods for details
In an attempt to improve the efficiency of A. rhizogenes transformation, the tissue culture method was modified to Protocol B (see Materials and methods). This included the pre-culture of explants for 4 days on phytohormone-containing medium and co-cultivation with acetosyringone solution, similar to a protocol reported for carnation A. tumefaciens transformation by Nontaswatsri and coworkers (Nontaswatsri et al. 2004). In an additional modification of the transformation method, internodes were investigated. Given a supply of axenic plants, the use of internodes makes it possible to avoid the three weeks delay in harvesting cotyledons after seed germination.
Protocol B proved to be highly efficient at generating hairy roots from A. rhizogenes-infected cotyledons and internodes. Figure 1 illustrates the very high frequency of A. rhizogenes-dependent root formation after two weeks for cotyledons (Fig. 1d) and internodes (Fig. 1f), for which no spontaneous root formation was observed (Fig. 1e). In triplicate experiments using 24 Pink Beauty explants and A. rhizogenes LBA9402, 100% of cotyledons and 95±6% of internodes showed A. rhizogenes-dependent root formation, in comparison to spontaneous root formation of 20 ± 5% for cotyledons and 0% for internodes. Subsequent culture of excised roots was found to be straightforward and cultures, which were subsequently confirmed to be transformed (see below), were similar in appearance to non-transformed root cultures (Fig. 1g, h).
It is notable that the age of axenic donor plant is an important factor for transformation. The frequency of root formation from explants derived from plants more than 2 months old was low. It is also notable that when nodes were used as explants, a high frequency of shoot formation was found even in the absence of A. rhizogenes infection (data not shown).
Selection and confirmation of genetically transformed roots using antibiotic resistance, fluorescence and PCR
With a view towards the generation of transgenic plant material, it is important to test the ability to select and confirm root cultures that have been derived from A. rhizogenes-dependent DNA transfer events. With this in mind, antibiotic selection was optimized for root cultures derived from transformation with A. rhizogenes strain LBA9402 containing the binary vector pK7GWIWG2D(II). This vector is designed to confer antibiotic resistance and green fluorescence to plant material into which its T-DNA is transferred.
In order to optimize antibiotic selection of transformants, root formation from internodes and cotyledons induced by A. rhizogenes strain LBA9402 (without binary vector) was tested in the presence of varying concentrations of paromomycin. This antibiotic was sufficient to stop all root formation from infected internodes at 100 mg/l. Paromomycin at 200 mg/l prevented root formation from cotyledons, with the exception of about 5–10% of the explants producing roots of less than approximately 2 mm on their upper surfaces. Growth of clonal root cultures (see Materials and methods) was completely inhibited by 50 mg/l paromomycin (data not shown). In trials with internodes transformed with A. rhizogenes strain LBA9402/pK7GWIWG2D(II) (with binary vector), root formation was 100% at both 0 and 100 mg/l paromomycin. Root formation was also very high for transformed cotyledons, with 99% (standard deviation of 2.3%) and 96% (standard deviation of 6.9%) of explants showing hairy roots at 0 and 200 mg/l paromomycin, respectively.
The relatively recent inclusion of genes encoding fluorescent proteins in plant transformation vectors offers the possibility of rapid and convenient selection of transformed tissue (Baranski et al. 2006). The usefulness of this approach for selection of transformed S. vaccaria roots was investigated using the gene encoding enhanced green fluorescent protein in pK7GWIWG2D(II). This is illustrated in Fig. 2 in which roots transformed with LBA9402 and LBA9402/pK7GWIWG2D(II) are compared. In LBA9402-transformed roots, a moderate degree of autofluorescence was observed, particularly with the use of the GFP1 filter set (Fig. 2b). Roots transformed with LBA9402/pK7GWIWG2D(II) showed relatively intense green fluorescence which was readily distinguishable from that of control roots using either GFP1 or GFP3 filters.
Use of GFP fluorescence as a marker for genetic transformation of S. vaccaria root cultures. Micrographs of cotyledons are shown two weeks after infection with A. rhizogenes LBA9402 (a, b, c) and LBA9402/pK7GWIWG2D(II) (d, e, f) using 125 ms exposures with unfiltered light (a, d), and GFP1 (b, e) and GFP3 (c, f) filter sets
A comparison of root autofluorescence and EGFP-dependent fluorescence was made using fluorescence spectroscopy. Fluorescence emission spectra of control and EGFP-expressing roots are shown in Fig. 3. While the quantitation of the measured fluorescence is affected by sample geometry and other factors, the spectra illustrate the qualitative aspects of S. vaccaria root autofluorescence and EGFP-dependent fluorescence. As expected the roots expressing EGFP show a relatively narrow emission peak near 510 nm (Cormack et al. 1996). On the other hand, the root autofluorescence shows broad intensity in the 460–660 nm range. This suggests that the contrast between EGFP fluorescence and autofluorescence can be optimized by relatively narrow optical filtering of the emission peak near 510 nm as effected by the GFP3 filter set. This is illustrated in Fig. 2c and f.
Fluorescence emission spectra of S. vaccaria root cultures with and without GFP expression. Fluorescence spectra were recorded for a spontaneous root culture derived from a cultured, uninfected S. vaccaria axenic plant leaf explant (curve 1), a transformed root culture derived from an A. rhizogenes LBA9402-infected internode explant (curve 2), and two transformed root cultures derived from two A. rhizogenes LBA9402/pK7GWIWG2D(II)-infected internode explants (curves 3 and 4). See Materials and methods for details
In scoring for EGFP-dependent fluorescence, tissue cultures were compared to control cultures by fluorescence microscopy. Roots showing visibly higher levels of fluorescence, compared to background autofluorescence of uninfected negative control cultures within the same experiment, were scored as positive using the GFP1 filter set. In some cases, this was confirmed by observing an increase in contrast between autofluorescence and sample fluorescence using the GFP3 filter set (relative to the GFP1 filter set). In experiments in which root selection was made by fluorescence, essentially all explants formed at least one green fluorescence-positive root (data not shown).
A PCR method was established for determining the genotype of hairy root cultures. In particular, this multiplex method was designed to test for the presence of genes transferred from the Ri plasmid (rolC, 490 bp product) and the binary vector (nptII, 698 bp product and egfp, 311 bp product), as well as to rule out artifacts caused by A. rhizogenes contamination (indicated by the presence of virD1, 441 bp product). Figure 4 shows multiplex PCR results for an antibiotic-resistant green fluorescent (see below) transformed root culture and various positive and negative controls. As expected the hairy root culture transformed with LBA9402/pK7GWIWG2D(II) is positive for rolC, nptII and egfp (Fig. 4, lane 1) and the culture transformed with LBA9402 is positive only for rolC (Fig. 4, lane 2).
Agarose gel electrophoresis of PCR amplified products from genomic DNA of S. vaccaria root cultures. 1 Root culture derived from A. rhizogenes LBA9402/pK7GWIW2D transformation and antibiotic and fluorescence selection; 2 hairy root culture derived from A. rhizogenes LBA9402 transformation; 3 root culture derived from an uninfected S. vaccaria leaf explant; 4 H2O; 5 pK7GWIW2D plasmid; 6 A. rhizogenes LBA9402 colony; 7 A. rhizogenes LBA9402/pK7GWIWG2D(II) colony
With methods in place for antibiotic and fluorescence selection and genotyping by PCR, an effort was made to compare selection regimes for efficiency of transformed root production. For this work, Pink Beauty internodes were cocultivated with LBA9402/pK7GWIWG2D(II) and cultured under four selection regimes: no selection (growth without antibiotic and not scored for fluorescence), growth on 100 mg/l paromomycin, EGFP fluorescence, and both paromomycin-resistance and EGFP fluorescence. Roots from internodes infected with LBA9402 and cultured without selection were used as negative controls. After selection, 1 cm sections of individual adventitious roots were bisected transversely and subcultured on 0 and 50 mg/l paromomycin, to retest for antibiotic resistance. Subcultures grown without antibiotic were subjected to PCR tests and additional fluorescence scoring. Table 2 shows the results of these experiments. Even without selection, root formation on internodes is a fairly good indication of gene transfer from A. rhizogenes, as indicated by the presence of the rolC, egfp and nptII genes in 57–86% of the root cultures. However, the introduction of antibiotic selection raised the frequency of transferred genes detected by PCR to 93–100%. Upon subculturing of the antibiotic-selected roots, 89% were able to regrow on paromomycin. On the other hand, despite the fact that all subcultures derived from paromomycin selection were PCR-positive for egfp, only 66% of them scored positive for EGFP fluorescence. This suggests a relatively high false negative rate for EGFP scoring, or perhaps a relatively high frequency of impaired EGFP expression in PCR-positive cultures.
In contrast to antibiotic selection of adventitious root formation, all roots selected by EGFP fluorescence gave rise to subcultures which retained this phenotype. In some cases (12%), “partial” fluorescence was observed, in which some parts of the root culture showed EGFP fluorescence and others did not. When both antibiotic and fluorescence selection were used, the frequency of transferred genes, antibiotic resistance and fluorescence in the derived cultures was consistently high (96–100%), with the lowest fraction of partially fluorescent cultures (7%).
Discussion
Previously, A. rhizogenes-mediated transformation within members of the Caryophyllaceae has been reported for carnation (Dianthus caryophyllus) and baby’s breath (Gypsophila muralis) (Tepfer 1990). This report represents an additional species within the family to be transformed by this bacterium and for which transformation was confirmed genetically.
Genetic transformation of plant material is typically highly dependent on cultivar, explant, Agrobacterium strain and culture conditions (Christey 1997; Baranski et al. 2006). In this case, S. vaccaria is no exception, showing a wide range of transformation efficiencies and, with Pink Beauty internodes and A. rhizogenes strain LBA9402, giving very efficient transformation and a very low background of non-transformed root formation. Optimum results were obtained with protocol B which included a preculture of explants on an auxin/cytokinin-containing medium and co-cultivation on filter paper soaked with acetosyringone solution. Similar co-cultivation was thought to be responsible for achieving effective A. tumefaciens-mediated transformation efficiencies in carnation in the Caryophyllaceae (Nontaswatsri et al. 2004).
The use of the egfp gene as a marker for selection of transformed roots proved to be quite reliable, despite some autofluorescence in roots not expressing egfp. This provides for a rapid and convenient screening protocol that eliminates the need for antibiotic selection. This mode of selection has the added advantage that chimeric cultures can be identified and, where necessary, selectively subcultured to obtain genetically uniform clones.
In conclusion, a highly efficient protocol for A. rhizogenes-mediated transformation of S. vaccaria has been developed. It will be suitable for identification of genes which function in S. vaccaria roots including those involved the biosynthesis of saponins and other phytochemicals.
References
Banerjee S, Shang TQ, Wilson AM, Moore AL, Strand SE, Gordon MP, Lafferty DS (2002) Expression of functional mammalian P450 2E1 in hairy root cultures. Biotechnol Bioeng 77:462–466
Baranski R, Klocke E, Schumann G (2006) Green fluorescent protein as an efficient selection marker for Agrobacterium rhizogenes mediated carrot transformation. Plant Cell Rep 25:190–197
Bekesiova I, Nap JP, Mlynarova L (1999) Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol Biol Rep 17:269–277
Bonhomme V, Laurain-Mattar D, Lacoux J, Fliniaux M, Jacquin-Dubreuil A (2000) Tropane alkaloid production by hairy roots of Atropa belladonna obtained after transformation with Agrobacterium rhizogenes 15834 and Agrobacterium tumefaciens containing rol A, B, C genes only. J Biotechnol 81:151–158
Christey MC (1997) Transgenic crop plants using Agrobacterium rhizogenes-mediated transformation. In: Doran PM (ed) Hairy roots: culture and applications. Harwood Academic, Amsterdam, pp 99–111
Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38
Hooykaas PJ, Klapwijk PM, Nuti MP, Schilperoort RA, Rorsch A (1977) Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent agrobacteria and to explanta. J Gen Microbiol 98:477–484
Jia Z, Koike K, Sahu NP, Nikaido T (2002) Triterpenoid saponins from Caryophyllaceae family. In: Atta-ur-Rahman (ed) Studies in natural products chemistry. Elsevier, Amsterdam, pp 3–61
Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10:103–105
Komarnytsky S, Gaume A, Garvey A, Borisjuk N, Raskin I (2004) A quick and efficient system for antibiotic-free expression of heterologous genes in tobacco roots. Plant Cell Rep 22:765–773
Kumagai H, Kouchi H (2003) Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant Microbe Interact 16:663–668
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nontaswatsri C, Fukai S, Goi M (2004) Revised cocultivation conditions produce effective Agrobacterium-mediated genetic transformation of carnation (Dianthus caryophyllus L.). Plant Sci 166:59–68
Sang S, Xia Z, Lao A, Cao L, Chen Z, Uzawa J, Fujimoto Y (2003) Studies on the constituents of the seeds of Vaccaria segetalis. Heterocycles 59:811–821
Tepfer D (1984) Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell 37:959–967
Tepfer D (1990) Genetic transformation using Agrobacterium rhizogenes. Physiol Plant 79:140–146
Acknowledgments
We wish to thank Paul Hooykaas for the generous gift of A. rhizogenes strain LBA9402, John Balsevich for supplying seed of S. vaccaria varieties, Prakash Venglat and Darwin Reed for technical assistance, and Paul Arnison, John Balsevich, Joe Hammerlindl, Jon Page and Don Palmer for useful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Petersen.
NRCC Publication No. 48435.
Rights and permissions
About this article
Cite this article
Schmidt, J.F., Moore, M.D., Pelcher, L.E. et al. High efficiency Agrobacterium rhizogenes-mediated transformation of Saponaria vaccaria L. (Caryophyllaceae) using fluorescence selection. Plant Cell Rep 26, 1547–1554 (2007). https://doi.org/10.1007/s00299-007-0369-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0369-9