Abstract
A DNA construct containing the cholera toxin B subunit (CTB) gene genetically fused to a nucleotide sequence encoding three copies of tandemly repeated diabetes-associated autoantigen, the B chain of human insulin, was produced and transferred into low-nicotine tobaccos by Agrobacterium. Integration of the fusion gene into the plant genome was confirmed by polymerase chain reaction (PCR). The results of immunoblot analysis verified the synthesis and assembly of the fusion protein into pentamers in transgenic tobacco. GM1–ELISA showed that the plant-derived fusion protein retained GM1–ganglioside receptor binding specificity. The fusion protein accounted for 0.11% of the total leaf protein. The production of transgenic plants expressing CTB–InsB3 offers a new opportunity to test plant-based oral antigen therapy against autoimmune diabetes by inducing oral tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral administration of protein antigens can result in diminished peripheral immune responses to a subsequent systemic challenge with the same antigen, in a process known as oral immune tolerance (Weiner et al. 1994; Strobel and Mowat 1998). Oral tolerance induced by autoantigens has recently been applied successfully as a therapeutic approach to treating several experimental autoimmune diseases, including type 1 diabetes in the non-obese diabetic (NOD) mouse (Polanski et al. 1997). Despite the success in animal models, oral tolerance has had only moderate success in treating human autoimmune diseases including type 1 diabetes (McFarland 1996; Chaillous et al. 2000). This difference may be due to a dose-dependent effect, as the amounts of oral insulin and myelin basic protein fed to humans in the respective diabetes and multiple sclerosis prevention trials are relatively low compared with those used in rodent models (Petersen et al. 2003). Taking into consideration the enormously increased intestinal absorptive surface area in humans, and thus the effective use of oral tolerance to treat human disease may critically depend on the use of mucosal adjuvants and/or suitable delivery systems to enhance its efficacy. Another serious limitation in the clinical application of oral tolerance strategies will be the potentially huge cost of producing autoantigens, particularly if repeated regular doses are required to maintain beneficial effects.
Cholera toxin (CT), which is a major pathogenic agent produced by Vibrio cholerae, is a potent mucosal immunogen and adjuvant, and most of these activities are retained by the nontoxic B subunit of CT (CTB). This is in part due to its high binding affinity for the GM1-ganglioside receptor that is present on mucosal epithelium (Cuatrecassas 1973). Because of this, CTB has been widely exploited as a potentially potent adjuvant or carrier molecule for mucosal vaccines. Indeed, oral administration of CTB linked to antigens has been shown to trigger acceptable peripheral and mucosal immune responses that were otherwise unattainable when the antigen was given alone (Dertzbaugh and Elson 1993; Holmgren et al. 1994). Furthermore, CTB has recently been shown to induce oral tolerance for linked antigens. For example, myelin basic protein (MBP) conjugated to CTB prevented or suppressed experimental allergic encephalomyelitis (EAE), when using lower concentrations of the conjugate than is usually necessary to induce tolerance with the protein alone (Sun et al. 2000). Oral administration of insulin or the B chain of insulin conjugated to CTB enhanced oral tolerance, and reduced the dose of antigen and the administration rate necessary for suppression of diabetes in NOD mice (Ploix et al. 1999; Sadeghi et al. 2002). Promising results against type II collagen-induced arthritis have been obtained by intranasal administration of low doses of type II collagen–CTB conjugate (Tarkowski et al. 1999). These results indicate that mucosal administration of autoantigens conjugated to CTB may represent a useful future treatment approach for human autoimmune diseases, such as type I diabetes. At the present time, the mechanism underlying CTB's efficacy as a mucosal adjuvant or carrier molecule remains to be defined. It has been suggested that the utility of CTB as a mucosal adjuvant or carrier molecule is due to its ability to efficiently deliver associated antigens to gut-associated lymphoid tissues (GALT), the major inductive site for mucosal immune responses (Holmgren et al. 1994).
To produce CTB-conjugates, two methods have been commonly used: chemical coupling or a genetic approach to produce a CTB fusion protein using microorganisms such as E. coli as an expression system. A potential limitation of the chemical coupling method is that it could affect the immunogenicity or tolerogenicity by chemical modification of the target antigen or by generating a heterogenous population of CTB-conjugates, where only a fraction retains the ability to form pentamers that can bind to GM1 (McGhee et al. 1992). The recombinant approach offers the advantage of free contamination with the holotoxin CT and more uniform immunogen or tolerogen compositions (Staats et al. 1994). Unfortunately, the E. coli-derived inclusion body-associated CTB fusion proteins have to be redissolved, assembled in vitro into pentameric CTB, and then purified, all of which are laborious and costly.
We have been investigating the use of transgenic plants expressing specific pancreatic β-cell autoantigen to prevent and treat autoimmune diabetes by inducing oral immune tolerance. Transgenic plants offer several advantages for oral tolerance strategy, not the least of which is its high production potential for relevant autoantigens with near unlimited scale up (Kusnadi et al. 1997). As protein-purification costs can eliminate the economic advantage of any production system, an additional advantage of transgenic plants for oral tolerance is that plants can also become effective delivery systems without extensive purification. Plant expression also largely eliminates concerns regarding potential pathogens that could be transmitted to humans. Lastly, augmented immune responses to plant-produced vaccines may suggest increased stability for plant expressed recombinant proteins to gastrointestinal degradation. Collectively these features make plants an ideal expression and delivery system for oral tolerance. Recently we demonstrated that transgenic plants can be used to produce β-cell autoantigen glutamic acid decarboxylase (GAD) and immunoregulatory cytokines, such as interleukin-4 (IL-4). The experimental NOD mice were protected from diabetes when given murine GAD67 plant tissue or human GAD65 together with IL-4 plant tissue (Ma et al. 1997, 2004, 2005). The present study was aimed at producing a fusion protein comprising CTB and the B chain of human insulin fusion protein in transgenic plants for inducing oral tolerance to prevent autoimmune diabetes. Like GAD, insulin B chain is another major autoantigen in autoimmune diabetes (Polanski et al. 1997; Song et al. 1999). Arakawa et al. (1998) have previously reported the production of transgenic potato plants expressing a fusion protein comprising CTB and the whole human insulin molecule. While oral administration of transgenic potatoes expressing the CTB–insulin fusion protein delayed the development of diabetes in NOD mice, one concern over administering the whole hormone insulin molecule is its risk for causing hypoglycemia (Karounos et al. 1997; Bot et al. 2001). In contrast, the administration of the B chain of insulin is generally safe for both humans and animals, as the B chain itself has no enzymatic activity. Here we demonstrate that a CTB–InsB3 fusion protein consisting of cholera toxin B-subunit and three copies of tandemly repeated human insulin B chain can be efficiently produced in transgenic tobacco plants. Moreover, the plant-derived fusion protein retains important functional characteristics of the native CTB, including pentamerization and GM1-ganglioside receptor binding. Transgenic plants expressing the B chain of human insulin linked to mucosal adjuvant CTB offers a new opportunity to test plant-based oral antigen therapy for treating autoimmune diabetes by inducing oral immune tolerance.
Materials and methods
Construction of plant transformation vector pBIN–CTB–InsB3
Plant expression vector pBIN–CTB–InsB3 was constructed using polymerase chain reaction (PCR) methods together with standard recombinant DNA techniques. Briefly, a single stranded synthetic gene encoding all 30 amino acids of the B chain of human insulin was initially produced as a DNA primer (5′-TTTGTGAACCAACACCTGTGCGGCTCACACCTGGTGGAAGCTCTCTACCTAGT GTGCGGGGAACGAGGCTTCTTCTACACACCCAAG ACC-3′) and used as the DNA template to obtain the double-stranded synthetic insulin B chain gene (insB) by the process of PCR amplification using the following pair primers: 5′-AATTCTCGAGTTTGTGAACCAACAC-3′ (forward primer; a XhoI restriction site (underlined) on the 5′ end of the primer was incorporated to facilitate subsequent ligation of the insulin B chain DNA), and 5′-ATATGTCGACGGTCTTGGGTGTGTA-3′ (reverse primer with a SalI site (underlined) being added) or 5′-ATATGGTACCGGTCTTGGGTGTGTA-3′ (reverse primer with a KpnI site (underlined) being added). To obtain the first insB–insB fusion, the PCR product with a 5′ end XhoI and a 3′ end SalI was digested with SalI and XhoI respectively, to generate compatible ends for tail-to-head ligation. The ligated insB–insB was rescued by blunt-end ligation into pUC19 digested with HincII to produce pUC19/insB–insB. To obtain insB–insB–insB fusion, the PCR product with a 5′ end SalI and a 3′ end KpnI restriction site (underlined) was digested with the same enzymes and ligated into pUC19/insB–insB digested with SalI and KpnI to create pUC19/insB–insB–insB. To create a CTB–insB–insB–insB fusion, the resulting insB–insB–insB DNA was isolated as a single XhoI and KpnI fragment and cloned into the same sites of pUC19–CTB (manuscript in preparation), which contains the bacterial CTB gene (ctxB) that had been modified by replacing the native signal peptide sequence with a plant signal sequence together with the addition of a hinge oligonucleotide encoding a glycine–proline–glycine–proline (GPGP) motif followed by a SalI restriction site engineered to the 3′ end of CTB and is used to produce CTB fusion proteins. The native bacterial ctxB gene was received as a gift from Dr. J. Holmgren (Institute of Medical Microbiology, University of Göteborg, Sweden). The resulting ctxB–insB–insB–insB fusion gene was confirmed by automated sequencing, and then tagged with 6xHis followed by an ER-retention signal motif SEKDEL via the KpnI site located at the 3′ end of the third copy of the insB gene. The complete fusion gene was then isolated as a single Nco1 and Xba1 fragment, and cloned onto pTRL2–GUS (Carrington and Freed 1990) to replace the β-glucuronidase (GUS) gene. The resulting expression cassette comprising the CaMV35S promoter with a double enhancer sequence (2x35S) fused to a 5′ untranslated tobacco etch virus (TEV) leader sequence, ctxB–insB–insB–insB–His/SEKDEL, and the nonpaline synthase (NOS) terminator were excised as a HindIII fragment and cloned onto plant transformation vector pBIN19 (Bevan 1984) to give pBIN–CTB–InsB3. The obtained expression vector was introduced into Agrobacterium tumefaciens strain LBA4404 (pAL4404) by tri-parental mating as described previously (Lige et al. 2001).
Transformation of tobacco plants
Non-nicotine and low-alkaloid Nicotiana tabacum cv. 81V9 were transformed by co-cultivation of leaf discs with A. tumefaciens LBA4404 containing pBIN–CTB–InsB3 according to the method of Horsch et al. (1985). Primary transgenic plants were selected on MS medium containing 100 mg/l kanamycin. As the regenerated plants matured, they were transferred into the greenhouse and maintained.
PCR and Northern blot analysis of transgenic tobacco plants
Integration of the fusion gene into tobacco genome was confirmed by PCR. Briefly, genomic DNA was isolated from leaves of 6-week-old greenhouse-grown tobacco plants as described previously (Ma et al. 2004), and the presence of the full-length CTB–InsB3 DNA was amplified by PCR using CTB and insulin B chain-specific primers. Transcription of the fusion gene was verified by Northern blot analysis. In brief, total RNA from transgenic tobacco leaf tissue was extracted by using a RNAeasy kit (Qiagen, Mississauga, Ontario, Canada), according to the manufacturer's recommendations. Aliquots (20 μg) of total RNA was separated on a 1.2% agarose/formaldehyde gel and blotted onto the Hybond N+ membrane (Amersham Biosciences, Baie d’Urfe, Quebec, Canada) by vacuum blotting, using 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.2) as transfer buffer, and then cross-linked to the membrane under UV light. A DNA fragment of approximately 600 base pairs spanning the entire coding region of CTB–InsB3 was labeled with [32p]dCTP (deoxycytidine 5′-triphosphate) and used as a hybridization probe. Blots were washed under stringent conditions, and exposed to autoradiographic film (MP-film, Amersham Biosciences).
Detection of CTB–InsB3 fusion protein in transgenic tobacco tissues by Western blot analysis
Expression of CTB–InsB3 fusion protein in transgenic tobacco plants was analyzed by Western blotting. In brief, the uppermost fully expanded leaves of 6-week-old greenhouse-grown tobacco plants were homogenized in liquid nitrogen and resuspended in cold extraction buffer (25 mM Tris pH 7.0, 50 mM NaCl, 2 mM β-mercaptoethanol, and 1 mM phenyl-methylsulfonyl fluoride, 2 μg/ml aprotinin, 2 μg/ml pepstain A, and 2 μg/ml leupeptin). Samples were centrifuged for 10 min at 4°C, and supernatant was collected. Protein concentration was measured according to the method of Bradford using the Bio-Rad reagent with BSA (Sigma, St. Louis, MO) as a standard. Samples were boiled for 15 min in sample buffer or left untreated, proteins were separated by SDS-PAGE and subsequently blotted onto PVDF (polyvinylidene difluoride) membrane (Millipore, Burlington, MA). Blocked for 1 h in 5% skim milk-TBST (20 mM Tris, 150 mM NaCl, 0.02% Tween 20, pH 7.6), the membrane was then incubated for 1 h with a 1:500 dilution (v/v) of a rabbit anti-CTB primary antibody (C-3062, Sigma) or with a 1:1,000 dilution of a mouse anti-human insulin antibody (12018, Sigma). For the primary antibody detection, the second anti-rabbit or anti-mouse peroxidase-linked goat antibody (Amersham Pharmacia Biotech, Piscataway, NJ) was used in combination with the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech).
Quantification of CTB–InsB3 fusion protein levels in transgenic tobacco tissues by enzyme-linked immunosorbent assay (ELISA)
The CTB–InsB3 fusion protein levels in transgenic plants were determined by quantitative ELISA assay as described previously (Ma et al. 2004). In brief, total protein extracts from transgenic leaf tissues were bound to a 96-well microtiter overnight at 4°C. Background was blocked with 1% BSA in PBS, and washed three times with PBST (PBS containing 0.5% Tween). The plate was then incubated with rabbit anti-cholera toxin antibody (Sigma C-3062) at a dilution of 1:8,000 for 2 h at 37°C, followed by three washes with PBST. The plate was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma G-7641) at a dilution of 1:10,000 for 2 h at room temperature. After final washes in PBST, ABTS substrate (Sigma A-1888) was added and allowed to develop for 30 min at room temperature. The OD value was measured at 405 nm in a Dynatech automated plate reader and the OD value from each sample was subtracted from untransformed plant OD value before converting into nanograms CTB–InsB3 per microgram total extracted protein by reference to an ELISA standard curve constructed with purified bacterial CTB (Sigma C-9903). Assays were performed in triplicate unless otherwise indicated and the average expression levels were calculated. Two independent experiments were performed and the data are presented as the mean ± standard deviation.
GM1-ganglioside binding assay
The GM1–ELISA assay was performed to determine the binding capacity of plant-derived CTB–InsB3 fusion protein to GM1-ganglioside. Briefly, the microtiter plate was coated with monosialoganglioside–GM1 (Sigma G-7641), 3 μg/ml dissolved in bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3), pH 9.6 (100 μl per well) at 4°C overnight. Wells coated with bovine serum albumin (BSA) were used as controls. All wells were blocked with 200 μl of 3% fat-free milk in PBS and incubated at room temperature for 2 h. After washing with PBST, 100 μl of total soluble protein extracts from transgenic or wild-type plants was added to the wells coated with monosialoganglioside–GM1 or BSA. The wells added with bacterial CTB were used as a positive control. Plates were incubated overnight at 4°C. After the washing, binding of plant-derived CTB–InsB3 to GM1-ganglioside was visualized by the addition of 100 μl of rabbit anti-cholera toxin antibody (Sigma C-3062) diluted 1:2,000 in PBS containing 1% BSA for 1 h at 37°C, followed by the addition of 100 μl of enzyme-conjugated anti-rabbit IgG and enzyme substrate as for an ordinary ELISA as described above. Horse radish peroxidase-conjugated antibodies were diluted 1:5,000 before use in the assay. As the substrate, 100 μl TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase substrate (Sigma) was used. After 10 min of incubation at room temperature, OD value at 450 nm was measured as described above. A commercial pentameric CTB (sigma) was used as a positive control in the GM1 ELISA
Results
Construction of expression vector and production of transgenic plants
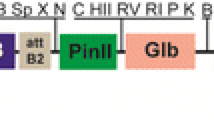
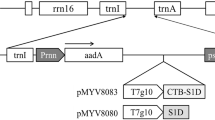
The plant expression vector pBIN–CTB–InsB3, in which the ctxB gene fused in frame to nucleotide sequences encoding three copies of the B chain of human insulin (insB) is illustrated in Fig. 1. Construction of pBIN–CTB–InsB3 has been described in detail in Materials and methods section. Each copy encodes all 30 amino acid residues of the human insulin B chain, was joined together via a two amino acid linker, valine–glutamic acid, that was created from SalI/XhoI pair ligation. A C-terminal 6XHis-tag was introduced to facilitate enrichment and purification of the recombinant protein if necessary, using the Ni–NTA resin that had been used for purification of proteins tagged with six consecutive histidine residues. The inclusion of an ER-retention signal motif SEKDEL in pBIN–CTB–InsB3 was attempted to optimize the expression level of the fusion protein.
Schematic diagram of the plant expression vector pBIN–CTB–InsB3. The T-DNA region inserted into the plant genome contains the nopaline synthase expression cassette (KanR), which confers kanamycin resistance on transformed cells, and the CTB–human insulin B chain fusion protein expression cassette consisting of the CaMV 35S promoter fused to a tobacco etch virus 5′-untranslated region (TEV) and nopalin synthase terminater (Nos-ter). The nucleotide and amino acid sequences of the B chain of human insulin as well as the sequence surrounding the fusion site are shown at the bottom. The underlined sequences correspond to the flexible hinge region. The boxed sequences represent the linking site with CTB or other copy of the B chain of the fused gene
Detection of CTB–InsB3 fusion gene in genomic DNA of transgenic tobacco plants. The CTB–InsB3 fusion gene was amplified from transgenic plant DNA by PCR and the amplification products were separated by agarose gel electrophoresis. M, 1-kb DNA ladder. Lane 1, PCR product obtained from pBIN–CTB–InsB3 plasmid DNA as template. Lanes 2–7, PCR from transgenic tobacco lines. Lane 8, PCR from wild-type tobacco DNA
Following the leaf disc transformation with Agrobacterium containing pBIN–CTB–InsB3, more than 20 independent transgenic tobacco lines were produced. There were no morphological differences between transgenic and untransformed control plants. The presence of the fusion gene in the plant genomic DNA was verified by PCR using CTB and human insulin B chain-specific primers (Fig. 2).
Expression of CTB–InsB3 in transgenic tobacco plants
PCR-positive tobacco plants were initially evaluated for CTB–InsB3 expression at the transcriptional level by Northern blot analysis with a DNA probe specific for both CTB and human insulin B chain sequences. As expected, CTB–InsB3 fusion mRNA of the correct size was detected in selected transgenic tobacco lines. Representative data are shown in Fig. 3. No amplification product was detected in wild-type plants or plants transformed with the control empty vector. These results indicate that the fusion gene was actively transcribed.
Northern blot analysis of CTB–insulin B chain fusion mRNA expression in transgenic plants. Total RNA was extracted from the uppermost fully expended green leaves of 6-week-old greenhouse grown transgenic or wild-type tobacco plants. Twenty micrograms of RNA was separated on agarose/formaldehyde gels, blotted onto Hybond N+ membrane, and hybridized with a 32P-labeled DNA fragment spanning the entire coding region of the fusion gene. The data only from those transgenic lines (T1, T2, T3, T4, T6, and T7) that were selected for further study are presented here. WT, wild-type plants. Numbers on the left indicate positions of RNA size markers in kb
Transgenic lines expressing the highest levels of CTB–InsB3 mRNA were further analyzed for fusion protein production by Western blotting. When probed with anti-CTB polyclonal antibody, two prominent protein bands running closely in the range of 35–39 kDa (as indicated by the large arrow in Fig. 4a) was detected in boiled extracts from transgenic lines T1, T4, T6, and T7. Moreover, a weak band of approximately 19 kDa (as indicated by the small arrow in Fig. 4a) was also detected in boiled extracts from transgenic line T7. The 19-kDa protein corresponds in size to the expected monomeric form of CTB–InsB3, whereas the two larger bands at 35–39 kDa represent in size the dimeric form of the fusion protein. As expected, none of these protein bands were detected in boiled extracts from untransformed control plants. To further demonstrate that the plant-derived CTB–InsB3 was assembled into pentameric structures, Western blot analysis of unboiled extracts was also conducted. As shown in Fig. 4b, a single band running above the 93-kDa marker was recognized by anti-CTB antibody. This band most likely represents the CTB–InsB3 pentamer, as the predicted size of CTB–InsB3 pentamer is in the range of 95–100 kDa. The corresponding band could not be detected in unboiled extracts from untransformed plants. The integrity and assembly of the CTB–InsB3 fusion protein were additionally analyzed by Western blot using anti-human insulin antibody. As expected, anti-human insulin antibody also recognized the same two bands at 35–39 kDa on the Western blot of boiled transgenic extracts (as indicated by arrow in Fig. 4c), as revealed by the anti-CTB antibody described above. The anti-human insulin antibody could not bind the 19-kDa band corresponding to the CTB–InsB3 monomer (Fig. 4c). Moreover, the anti-human insulin antibody also recognized the band migrating above the 93-kDa marker in Western blot of unboiled transgenic extracts (Fig. 4d). Together, these results confirmed the identity of the fusion protein and showed cross-reactivity of CTB–InsB3 with both antibodies, demonstrating that transgenic plants are not only able to synthesize the full-length CTB–InsB3 fusion protein, but are also able to assemble into large oligomeric complexes essential for its biological function.
Western blot analysis of CTB–InsB3 fusion protein expression in transgenic tobacco plants. Total protein extracts (40 μg/lane) from leaf tissues of transgenic plants were fractionated by SDS-PAGE gel, blotted onto PVDF membrane, and probed with anti-CTB (a and b) or anti-insulin antibody (c and d). B, bacterial CTB standard (Sigma); Ins, recombinant insulin standard (Sigma); T1–T8, individual transgenic tobacco lines; WT, wild-type tobacco. Denatured proteins in panel were boiled for 15 min before loading onto the gel. CTB–InsB3 monomer is indicated by a small arrow whereas the CTB–InsB3 dimeric form is indicated by a large arrow. Numbers on the left indicate positions of protein size markers in kDa
Quantification of CTB–InsB3 fusion protein in transgenic tobacco leaf tissues
The expression levels of CTB–InsB3 fusion protein in transgenic leaf tissues were determined by ELISA. Using this method, the amount of plant CTB–InsB3 fusion protein was calculated by comparison of the optical density (OD) values obtained for transgenic extracts with the OD values for a known amount of bacterial CTB standard. The amount of CTB–InsB fusion protein was then expressed as a percentage of the total soluble plant protein (TSP). As shown in Fig. 5A, variable levels of CTB–InsB3 fusion protein were seen in leaf extracts from individual tobacco lines, with transgenic lines T1, T6, and T7 being the top producers expressing CTB–InsB3 at levels ranging from 0.08 to 0.11% of TSP.
(A) ELISA quantification of the CTB–InsB3 fusion protein. The amount of the CTB–InsB3 in total soluble protein (TSP) of transgenic tobacco leaf tissues was estimated by using ELISA as described in detail in the Materials and methods section. The CTB–InsB3 fusion protein concentration was expressed as a percentage of TSP from transgenic plants. Data shown here represent averages of three experiments. The numbers on the bottom of the figure represent the different tobacco lines. WT: wild-type tobacco used as a negative control. The error bar represents the standard deviation. (B) GM1 binding analysis of the CTB–InsB3 fusion protein by ELISA. Microtiter plates, coated with bovine serum albumin (BSA) or GM1-ganglioside, were incubated with total transgenic extracts containing the CTB–InsB3 fusion protein or purified bacterial CTB. The amount of total protein from transgenic lines T1, T4, T6, or T7 that was added to each well was adjusted to obtain the concentration of CTB at approximately 7.9, 7.2, 8.9, or 8.0 ng/ml, respectively. The concentration of bacterial CTB used was 10 ng/ml. The absorbance of the GM1-ganglioside–CTB–InsB3 antibody complex in each case was measured. The values represent the averages of three experiments. The error bar represents the standard deviation
GM1 receptor-binding assays of plant-derived CTB–InsB3 fusion protein
Biological functions of CTB, such as the ability to bind to GM1-ganglioside, depends on the formation of a pentameric structure composed of identical monomers (Hardy et al. 1988). To demonstrate that the plant-derived CTB–InsB3 binds to GM1-gangliosides, a GM1–ELISA was performed. The results, as shown in Fig. 5B, show that both plant fusion protein and commercial CTB bind gangliosides efficiently. No binding activity was detected when the plate was coated with irrelevant bovine serum albumin (BSA). These results further suggest that the CTB–InsB3 pentamer is required for biological activity. Thus, the addition of the insulin B chain to the C-terminus of CTB protein did not affect pentamerization or GM1 receptor binding of the protein.
Discussion
We report here the production of a fusion protein containing CTB fused to three tandem copies of the B chain of human insulin in transgenic tobacco plants. The B chain was chosen because it bears major type 1 diabetes-associated epitopes of significance for disease in humans as well as in NOD mice. Feeding of B chain or B chain peptides such as amino acid residues 9–23 has been shown to prevent the development of diabetes, while feeding of A chain showed no protective effect (Song et al. 1999; Polanski et al. 1997). The design of three tandem copies of human insB-encoding sequence in the fusion gene construct was used in attempt to increase the expression amount of the target antigen in plants, as each CTB–InsB3 hybrid protein molecule synthesized contained two extra B chain molecules compared to CTB conjugated to a single B chain. Previously we have shown that induction of oral tolerance to GAD using transgenic plant systems is critically dependent on the amount of GAD delivered by plant tissues (Ma et al. 1997, 2004). In theory, it is possible to further increase the expression amount of B chain antigen in plants simply by incorporating additional copies of insB gene to CTB. However, when a CTB fusion construct containing six tandem copies of insB gene was tested, the expression level of the fusion protein was found to be reduced (data not shown).
The capacity of CTB as a mucosal adjuvant or carrier molecule for conjugated antigens is dependant on its ability to bind GM1-ganglioside in its pentameric form. It is therefore critical to ensure that fusion of foreign proteins or peptides to CTB will not impair the ability or efficiency of CTB to form pentamers. The results from Western blot analysis demonstrated the synthesis and assembly of the full-length CTB–InsB3 fusion protein into oligomeric structures of pentamer size (Fig. 4). Moreover, GM1–ELISA showed that plant-derived CTB–InsB3 binds to GM1-receptors as efficiently as the native CTB (Fig. 5). This suggests that the B chain of insulin carried by the fusion protein molecule had no demonstrable adverse effect on assembly, stability, or GM1 binding of the parental CTB molecule. It has been reported that pentamerization and GM1 binding capacity of CTB linked with proteins or peptides can be affected by the length of the partner protein or peptide, or by the conformational changes induced by the fusion partner (Liljeqvist et al. 1997). To minimize any negative perturbation effects of the fusion partner on the pentamer formation and ganglioside-binding capacity of CTB, the B chain of insulin was purposely fused to the C-terminal end of the CTB and expressed as a C-terminal fusion protein because it has been suggested that the N-terminal of CTB is in closed proximity to the GM1-binding pocket (Zhang et al. 1995). Furthermore, a flexible hinge tetrapeptide (GPGP) was introduced between the CTB and insulin B chain moieties to reduce potential sterical hindrance and permit high intramolecular flexibility between the CTB and insulin B chain proteins. An endoplasmic reticulum (ER)-retention signal sequence SEKDEL was also added to the C-terminal of the fusion protein combined with substitution of the native signal peptide of CTB with the signal sequence from the plant protein peanut peroxidase. Both SEKDEL and KDEL have been shown to function as an effective ER-retention signal in plant cells (Fiedler et al. 1997). Haq et al. (1995) have shown that addition of SEKDEL to the C-terminal of the B subunit of the CTB-related bacterial heat-labile enterotoxin (LTB) is efficient at retaining the recombinant protein in the ER compartment. This compartmentalization facilitates oligomerization of LTB into pentamers and increased protein expression levels in plants. The use of a plant signal was intended to ensure the correct signal peptide processing of CTB, a step required for importing the fusion protein into the ER for a higher level accumulation. There have been inferences from previous studies that transgenic plants may fail to process the native signal peptide of CTB (Arakawa et al. 1997; Jani et al. 2002). Nevertheless, a recent study by Wang et al. (2001) has demonstrated a dramatic increase in the expression levels of CTB in transgenic tobacco plants when its native signal peptide was substituted with a plant signal peptide sequence, even without the further addition of an ER-retention signal. ELISA quantification of CTB–InsB3 expression levels in transgenic tobacco leaf tissues revealed that the fusion protein accumulated up to 0.11% of total soluble protein. Although higher levels of CTB expression (0.3% of TSP) has been achieved in transgenic potato plants (Arakawa et al. 1997), the expression level of CTB achieved in tobacco plants was relatively low, ranging from 0.02 to 0.09% of TSP (Wang et al. 2001; Jani et al. 2004). The higher expression of the CTB–InsB3 fusion protein achieved in tobacco plants in this study may be partly due to a synergistic effect of the combination of a plant signal peptide and the ER-targeting signal SEKDEL.
As expected, Western blot analysis of unboiled transgenic extracts revealed a single prominent band of the expected size for CTB–InsB3 pentamer (Fig. 4), suggesting a high degree of oligomerization of the fusion protein. Interestingly, Western blot analysis of heat-treated extracts (boiled for 15 min before loading on to SDS-PAGE) showed that CTB–InsB3 still exists predominantly in the dimeric form, rather than a monomeric form. This may suggest that the fusion protein is very heat stable. The presence of two forms of the dimer with slightly different molecular mass is interesting. This could not be due to different N-glycosylation, as CTB and the B chain of human insulin are both non-glycoproteins. One possibility is that incomplete processing of the engineered plant signal peptide may have occurred in some of the fusion proteins. Nevertheless, a strong interaction of unboiled CTB–InsB3 with gangliosides revealed by GM1-binding ELISA (Fig. 5) suggests that the plant-made CTB–InsB3 pentamers are fully biologically active. Taken together, these findings suggest that transgenic plants can be a valuable system for the production of diabetes-associated chimeric insulin B chain antigen, and perhaps many other recombinant pharmaceutical proteins.
No expression of the immunodominant B chain of insulin has so far been achieved in transgenic plants, although Arakawa et al. (1998) have previously reported the production of transgenic potato plants expressing a CTB–insulin fusion protein. The production of transgenic plants expressing a fusion protein comprising CTB and the B chain of insulin rather than CTB and the whole insulin molecule, as reported here, offers at least two important advantages. Firstly, as CTB and the B chain of insulin were present in a 1:3 ratio in the CTB–InsB3 fusion protein, the amount of the immunodominant B chain antigen delivered would be tripled when compared to similar amounts of the CTB–insulin fusion protein being delivered. This may be critical to the success of using transgenic plants to induce oral tolerance to treat type 1 diabetes in humans, as large amounts of relevant antoantigen are required for the tolerizing effect. Secondly, administration of transgenic plants expressing the B chain of insulin is safer compared to administration of those plants expressing the whole insulin molecule. To date administration of the whole insulin molecule, including dosing studies, has been tempered by the fear of hypoglycemia (Karounos et al. 1997; Bot et al. 2001). The CTB–human insulin B chain fusion protein expressed in E. coli has been shown, after in vitro assembly into pentameric forms, to induce efficient immunosuppression when fed to mice (Sadeghi et al. 2002). The limitation of E. coli expression systems for production of CTB–insulin B chain fusion protein is that it is very laborious and costly.
In summary, we have demonstrated the feasibility of using transgenic tobacco plants for the production of a hybrid pancreatic β cell specific autoantigen, the B chain of human insulin coupled to CTB. Functional analysis of the plant-derived fusion protein showed that it retains the biological and immunological characteristics of the native CTB including its ability to pentamerize and to bind to GM1-ganglioside receptor. Production of transgenic plants expressing CTB–InsB3 fusion protein may provide a highly effective, low-cost approach for preventing and treating autoimmune diabetes by inducing oral tolerance. Animal feeding studies with CTB–InsB3 transgenic plants have already begun to assess its therapeutic efficiency.
References
Arakawa T, Chong DK, Merritt JL, Langridge WH (1997) Expression of cholera toxin B subunit oligomers in transgenic potato plants. Transgenic Res 6:403–413
Arakawa T, Yu J, Chong DK, Hough J, Engen PC, Langridge WH (1998) A plant-based cholera toxin B subunit–insulin fusion protein protects against the development of autoimmune diabetes. Nat Biotechnol 16:934–938
Bao LG, Ma SW, van Huystee RB (2001) Effect of the site-directed removal of N-glycosylation from cationic peanut peroxidase on its function. Arch Biochem Biophys 386:17–24
Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucl Acids Res 12:8711–8721
Bot A, Smith D, Bot S, Hughes A, Wolfe T, Wang L, Woods C, von Herrath M (2001) Plasmid vaccination with insulin B chain prevents autoimmune diabetes in nonobese diabetic mice. J Immunol 167:2950–2955
Carrington JC, Freed DD (1990) Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64:1590–1597
Chaillous L, Lefevre H, Thivolet C, Boitard C, Lahlou N, Atlan-Gepner C, Bouhanick B, Mogenet A, Nicolino M, Carel JC, Lecomte P, Marechaud R, Bougneres P, Charbonnel B, Sai P (2000) Oral insulin administration and residual beta-cell function in recent-onset type 1 diabetes: A multicentre randomised controlled trial. Diabete Insuline Orale Group. Lancet 356:545–549
Cuatrecassas P (1973) Gangliosides and membrane receptors for cholera toxin. Biochemistry 12:3558–3566
Dertzbaugh MT, Elson CO (1993) Comparative effectiveness of the cholera toxin B subunit and alkaline phosphatase as carriers for oral vaccines. Infect Immun 61:48–55
Fiedler U, Phiillips J, Artsaenko O, Conrad U (1997) Optimization of scFv antibody production in transgenic plants. Immunotechnology 3:205–216
Haq TA, Mason HS, Clements JD, Arntzen CJ (1995) Oral immunization with a recombinant bacterial antigen produced in transgenic plants. Science 268:714–716
Holmgren J, Czerkinsky C, Lycke N, Svennerholm AM (1994) Strategies for the induction of immune responses at mucosal surfaces making use of cholera toxin B subunit as immunogen, carrier, and adjuvant. Am J Trop Med Hyg 50:42–54
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Jani D, Meena LS, Rizwan-ul-Haq QM, Singh Y, Sharma AK, Tyagi AK (2002) Expression of cholera toxin B subunit in transgenic tomato plants. Transgenic Res 11:447–454
Jani D, Singh NK, Bhattacharya S, Meena LS, Singh Y, Upadhyay SN, Sharma AK, Tyagi AK (2004) Studies on the immunogenic potential of plant-expressed cholera toxin B subunit. Plant Cell Rep 22:471–477
Karounos DG, Bryson JS, Cohen DA (1997) Metabolically inactive insulin analog prevents type I diabetes in prediabetic NOD mice. J Clin Invest 100:1344–1348
Kusnadi AR, Ann R, Nikolov ZL, Howard JA (1997) Production of recombinant proteins in transgenic plants: Practical considerations. Biotechnol Bioeng 56:473–485
Liljeqvist S, Stahl S, Andreoni C, Binz H, Uhlen M, Murby M (1997) Fusions to the cholera toxin B subunit: Influence on pentamerization and GM1 binding. J Immunol Methods 210:125–135
Ma SW, Zhao DL, Yin ZQ, Mukherjee R, Singh B, Qin HY, Stiller CR, Jevnikar AM (1997) Transgenic plants expressing autoantigens fed to mice to induce oral immune tolerance. Nat Med 3:793–796
Ma SW, Huang Y, Yin Z, Menassa R, Brandle JE, Jevnikar AM (2004) Induction of oral tolerance to prevent diabetes with transgenic plants requires glutamic acid decarboxylase (GAD) and IL-4. Proc Natl Acad Sci USA 101:5680–5685
Ma SW, Huang Y, Antonelle D, Yin Z, Mi Q, Menassa R, Brandle JE, Jevnikar AM (2005) Production of biologically active human interleukin-4 in transgenic tobacco and potato. Plant Biotech J 3:309–318
McFarland HF (1996) Complexities in the treatment of autoimmune disease. Science 274:2037–2038
McGhee JR, Mestecky J, Dertzbaugh MT, Eldridge JH, Hirasawa M, Kiyono H (1992) The mucosal immune system: From fundamental concepts to vaccine development. Vaccine 10:75–88
Petersen JS, Bregenholt S, Apostolopolous V, Homann D, Wolfe T, Hughes A, De Jongh K, Wang M, Dyrberg T, von Herrath MG (2003) Coupling of oral human or porcine insulin to the B subunit of cholera toxin (CTB) overcomes critical antigenic differences for prevention of type I diabetes. Clin Exp Immunol 134:38–45
Ploix C, Bergerot I, Durand A, Czerkinsky C, Holmgren J, Thivolet C (1999) Oral administration of cholera toxin B–insulin conjugates protects NOD mice from autoimmune diabetes by inducing CD4+ regulatory T-cells. Diabetes 48:2150–2156
Polanski M, Melican NS, Zhang J, Weiner HL (1997) Oral administration of the immunodominant B-chain of insulin reduces diabetes in a co-transfer model of diabetes in the NOD mouse and is associated with a switch from Th1 to Th2 cytokines. J Autoimmun 10:339–346
Sadeghi H, Bregenholt S, Wegmann D, Petersen JS, Holmgren J, Lebens M (2002) Genetic fusion of human insulin B-chain to the B-subunit of cholera toxin enhances in vitro antigen presentation and induction of bystander suppression in vivo. Immunology 106:237–245
Song HY, Abad MM, Mahoney CP, McEvoy RC (1999) Human insulin B chain but not A chain decreases the rate of diabetes in BB rats. Diab Res Clin Pract 46:109–114
Staats HF, Jackson RJ, Marinaro M, Takahashi I, Kiyono H, McGhee JR (1994) Mucosal immunity to infection with implications for vaccine development. Curr Opin Immunol 6:572–583
Strobel S, Mowat AM (1998) Immune responses to dietary antigens: Oral tolerance. Immunol Today 19:173–181
Sun JB, Xiao BG, Lindblad M, Li BL, Link H, Czerkinsky C, Holmgren J (2000) Oral administration of cholera toxin B subunit conjugated to myelin basic protein protects against experimental autoimmune encephalomyelitis by inducing transforming growth factor-beta-secreting cells and suppressing chemokine expression. Int Immunol 12:1449–1157
Tarkowski A, Sun JB, Holmdahl R, Holmgren J, Czerkinsky C (1999) Treatment of experimental autoimmune arthritis by nasal administration of a type II collagen–cholera toxoid conjugate vaccine. Arthritis Rheum 42:1628–1634
Wang XG, Zhang GH, Liu CX, Zhang YH, Xiao CZ, Fang RX (2001) Purified cholera toxin B subunit from transgenic tobacco plants possesses authentic antigenicity. Biotechnol Bioeng 72:490–494
Weiner HL, Friedman A, Miller A, Khoury SJ, al-Sabbagh A, Santos L, Sayegh M, Nussenblatt RB, Trentham DE, Hafler DA (1994) Oral tolerance: Immunologic mechanisms and treatment of animal and human organ-specific autoimmune diseases by oral administration of autoantigens. Annu Rev Immunol 12:809–837
Zhang RG, Westbrook ML, Westbrook EM, Scott DL, Otwinowski Z, Maulik PR, Reed RA, Shipley GG (1995) The 2.4 A crystal structure of cholera toxin B subunit pentamer: Choleragenoid. J Mol Biol 251:550–562
Acknowledgements
The authors thank Dr. J. Holmgren, Institute of Medical Microbiology, University of Göteborg, Sweden, for providing bacterial CTB DNA clone. This research was supported by the Natural Sciences and Engineering Research Council (NSERC) and the London Health Sciences Centre MOTS Research Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Ebinuma
Rights and permissions
About this article
Cite this article
Li, D., O’Leary, J., Huang, Y. et al. Expression of cholera toxin B subunit and the B chain of human insulin as a fusion protein in transgenic tobacco plants. Plant Cell Rep 25, 417–424 (2006). https://doi.org/10.1007/s00299-005-0069-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0069-2