Abstract
The final enzymatic step in the synthesis of the flavor compound vanillin (4-hydroxy-3-methoxybenzaldehyde) is believed to be methylation of 3,4-dihydroxybenzaldehyde. We have isolated and functionally characterized a cDNA that encodes a multifunctional methyltransferase from Vanilla planifolia tissue cultures that can catalyze the conversion of 3,4-dihydroxybenzaldehyde to vanillin, although 3,4-dihydroxybenzaldehyde is not the preferred substrate. The higher catalytic efficiency of the purified recombinant enzyme with the substrates caffeoyl aldehyde and 5-OH-coniferaldehyde, and its tissue distribution, suggest this methyltransferase may primarily function in lignin biosynthesis. However, since the enzyme characterized here does have 3,4-dihydroxybenzaldehyde-O-methyltransferase activity, it may be useful in engineering strategies for the synthesis of natural vanillin from alternate sources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vanillin (4-hydroxy-3-methoxybenzaldehyde) is the most widely used flavor compound in the world. It is the principal flavor component of the vanilla extract obtained from the cured pods (called beans) of the orchid Vanilla planifolia Andrews. V. planifolia is cultivated in tropical areas, with the largest producers being Madagascar and Indonesia (Dignum et al. 2001; Walton et al. 2003). During pod development glucovanillin accumulates in the region of the placental tissue in the inner core of the pod (Joel et al. 2003). After harvest, the pods are subjected to a curing process during which glucovanillin is hydrolyzed by beta-glucosidase, resulting in the release of vanillin (Havkin-Frenkel et al. 2003). This vanilla extract is valued as a natural flavor but because of its cost and limited availability supplies less than 1% of the world’s yearly demand for vanillin (Walton et al. 2003). Most of the vanillin used by the flavor industry originates from chemical methods using guaiacol, eugenol, or lignin as starting materials (Rao and Ravishankar 2000).
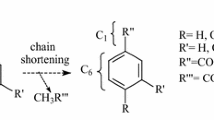
Despite its commercial importance, the biosynthetic pathway for vanillin is still under investigation. Cloning of the enzymes involved in the pathway would be useful in developing alternative strategies for the production of natural vanillin. Vanillin is believed to be synthesized from phenylpropanoid precursors, and different biosynthetic pathways have been proposed. A three-step pathway for vanillin biosynthesis from 4-coumaric acid has been proposed based on precursor accumulation and on feeding cell cultures of V. planifolia with the proposed precursors (Fig. 1) (Havkin-Frenkel et al. 1999; Herz 2000). In this pathway 4-coumaric acid is first converted to 4-hydroxybenzaldehyde through a chain-shortening step. Hydroxylation at position 3 on the ring results in 3,4-dihydroxybenzaldehyde (also called protocatechuic aldehyde). The 3-hydroxyl group is then methylated, producing vanillin. An enzyme from V. planifolia that catalyzes the chain-shortening step, 4-hydroxybenzaldehyde synthase, has been reported (Podstolski et al. 2002). Other vanillin biosynthetic pathways have been proposed (Dignum et al. 2001; Walton et al. 2003), but none of the proposed enzymes have been characterized.
Plant O-methytransferases (OMTs) that use S-adenosylmethionine (SAM) as the methyl donor are involved in the synthesis of a diverse range of secondary products (Ibrahim et al. 1998). Methylation of the 3-OH of caffeic acid and related phenylpropanoid compounds has been widely studied due to its presumed involvement in the synthesis of lignin. Caffeic acid O-methyltransferase (COMT) enzymes have been characterized from numerous species (Ibrahim et al. 1998). The substrate preferences and kinetic properties of recombinant COMT from alfalfa have been compared, resulting in a re-evaluation of the lignin biosynthetic pathway (Parvathi et al. 2001). Caffeic acid was actually found to be the least effective substrate for the enzyme, leading to the proposal that the primary physiological substrate is likely 5-hydroxyconiferaldehyde, which is a key substrate in the production of S lignin (Osakabe et al. 1999; Li et al. 2000; Parvathi et al. 2001). High catalytic efficiency with caffeoyl aldehyde as well as 5-hydroxyconiferaldehyde suggested that COMT may catalyze both the 3-OH and 5-OH methylations in S lignin biosynthesis (Dixon et al. 2001; Parvathi et al. 2001). For historical reasons, even though caffeic acid is not the preferred substrate, these enzymes are still often referred to as COMTs.
COMTs have been reported to have activity against 3,4-dihydroxybenzaldehyde. In tobacco, two distinct COMTs with different substrate specificities have been characterized: tobacco class I COMT showed activity against both caffeic acid and 3,4-dihydroxybenzaldehyde with similar efficiencies, whereas class II COMT was active against 3,4-dihyroxybenzaldehyde but not caffeic acid (Maury et al. 1999). COMTs from basil (Gang et al. 2002) and strawberry (Wein et al. 2002) were found to have activity with 3,4-dihydroxybenzaldehyde at 69.4% and 140%, respectively, of their relative activity with caffeic acid. Zubieta et al. (2002) determined the crystal structure of the enzyme from alfalfa, revealing a spacious active site which is consistent with the broad range of substrates acted upon by the enzyme. These results on the broad substrate utilization by COMTs have raised the question of whether methylation of 3,4-dihydroxybenzaldehyde in V. planifolia is mediated by an enzyme specific for this substrate or whether it can occur from a COMT-like enzyme with a broad substrate range. COMT activity is expected to be present in all plant species.
We report here the characterization of a multifunctional O-methyltransferase from V. planifolia that has a broad substrate range, including 3,4-dihydroxybenzaldehyde. The substrate preferences and tissue distribution, however, suggest it may be primarily involved in lignin biosynthesis. This is the first report of a thorough examination of such a broad range of substrates for a purified monocot COMT.
Materials and methods
Plant material
Tissue cultures of Vanilla planifolia were initiated and maintained as previously described (Podstolski et al. 2002). The cultures were transferred to new medium every 2 weeks. The V. planifolia plants used here have been maintained in the greenhouse for 5 years and were the source of stem, leaf, and root tissues. Green V. planifolia pods at different stages of development were obtained from Indonesia (Djasula Wangi).
Enzyme extraction and assay
The preparation of crude protein extracts of the V. planifolia pods and tissue cultures grown in liquid media was modified from that described by Wang et al. (1997). For determining the presence of OMT activity, we homogenized 3 g of tissue in 6 ml of 50 mM BisTris-HCl, pH 6.9, 10 mM 2-mercaptoethanol, 5 mM Na2S2O5, 1% (w/v) PVP-40, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 10% (v/v) glycerol. The homogenate was filtered through cheesecloth and centrifuged 15 min at 10,000 g at 4°C.
Protein concentrations were determined using the Bio-Rad protein assay reagent (Bio-Rad, Richmond, Calif.) with bovine serum albumin as a standard.
O-Methyltransferase assays were as described by Wang et al. (1997). The assays were carried out in 50-μl volumes consisting of 10 μl assay buffer (250 mM Tris-HCl, pH 7.5, 10 mM dithiothreitol), 1 μl 50 mM substrate, 10 μl enzyme (crude extracts or fractions from partial purification), and 1 μl S-[methyl-14C]adenosyl-l-methionine (SAM) (2.18 GBq mmol−1) (Amersham Pharmacia Biotech, Piscataway, N.J.), and 28 μl water. The final concentration of [14C]SAM was 8.4 μM. The samples were incubated at 30°C for 30 min, after which the reactions were stopped by the addition of 2.5 μl 6 M HCl. [14C]SAM was separated from the radiolabeled methylated product by extraction with 100 μl ethyl acetate. Twenty microliters of the organic phase containing the labeled product was used for liquid scintillation counting. The control consisted of all reaction components except the enzyme, and those counts were subtracted from the sample counts. The counts per minute were converted to picomoles of product produced per second (pkat), based on the specific activity of the substrate and the efficiency of the scintillation counter.
For determination of the kinetic parameters for the aromatic substrates, the reaction conditions were modified to include 2 μl [14C]SAM, 100 μM unlabeled SAM, and 3 μg of the purified recombinant protein expressed in Escherichia coli. The final concentration of SAM was 116.8 μM. Substrate concentrations ranged from 0.001 mM to 4 mM. All reactions were done in duplicate. V max and K m were calculated from non-linear regressions of the Michaelis-Menten plots using the prism 4 program (GraphPad Software, San Diego, Calif.).
Thin layer chromatography
The identity of vanillin as the labeled reaction product following methylation of 3,4-dihydroxybenzaldehyde was confirmed by TLC analysis. Twenty-microlitre aliquots of the organic extract were spotted onto a 20×20-cm silica gel 60-precoated TLC plate (EM Industries, Gibbstown, N.J.). Twenty microlitres each of 10 mM vanillin, 10 mM 3,4-dihydroxybenzaldehyde, and a mixture of both were also spotted as standards. The plate was developed in a solvent system of chloroform/acetic acid (9:1, v/v). In this chromatography system the other potential products of 3,4-dihydroxybenzaldehyde methylation—3-hydroxy, 4-methoxybenzaldehyde (isovanillin), and 3,4-dimethoxybenzaldehyde (veratryl aldehyde)—were readily distinguishable from vanillin. The Rfs of vanillin, isovanillin, and veratryl aldehyde were 0.80, 0.74, and 0.91, respectively. The standards were visualized following chromatography by allowing the plate to dry and then examining it under 365 nm UV light. The region of the plate from the reaction product that corresponded to the position of standard vanillin was scraped into scintillation vials and counted.
Partial purification of V. planifolia OMT
For protein purification, we homogenized a crude extract of the tissue culture in ten volumes fresh weight of extraction buffer in an Ultra-Turrax T25 tissue homogenizer (IKA Works, Wilmington, N.C.). Partial purification of DOMT activity from the crude extract on an adenosine-agarose affinity column was modified from that described by Wang and Pichersky (1998). A 1-ml adenosine-agarose (Sigma, St Louis, Mo.) column was prepared as described by Attieh et al. (1995). Ten milliliters of tissue culture crude extract was applied to the adenosine-agarose column, and the column was then washed with 6 ml 50 mM Bis-Tris, pH 6.9, 10 mM 2-mercaptoethanol, 10% glycerol followed by elution with 10 ml wash buffer containing 2.5 mM adenosine. One-milliliter fractions were collected and assayed for DOMT and COMT activities. Fractions containing activity were combined and concentrated using Microcon YM30 devices (Amicon, Beverly, Mass.).
PCR amplification of the OMT cDNA fragment
Degenerate oligonucleotide primers for PCR were designed based on conserved sequences in COMTs from other plant species. The sequences of the degenerate primers were: primer A, 5′-GTIGTIATGGARWSNTGGTAY-3′ and primer B, 5′-RAACATRACICCNCCNACRTG-3′. The symbols used for the mixed bases are I=deoxyinosine, N=A, C, T, G; R=A, G; S=C, G; W=A, T; Y=C, T. The amino acid sequences encoded by primers A and B are VLMESWY and HVGGDMF, respectively.
The degenerate oligonucleotide primers were used in PCR amplification of the cDNA library prepared from the V. planifolia tissue cultures, and the reactions were carried out using the Elongase Amplification System (Invitrogen, Carlsbad, Calif.). The 100-μl reactions contained 60 mM Tris-SO4, pH 9.1, 18 mM (NH4)2SO4, 1.5 mM MgSO4, 200 μM each dNTP, 3 μg of each oligonucleotide, and 2 μl Elongase enzyme mix. PCR analyses were carried out in a GeneAmp 9600 thermocycler (Perkin-Elmer Life Sciences, Boston, Mass.). Touchdown PCR (Don et al. 1991) cycling parameters were used. The initial denaturation was conducted at 94°C for 30 s. Cycle 1 consisted of a 30-s denaturation at 94°C, a 30-s annealing at 66°C, and a 2-min extension at 68°C. At each two subsequent cycles, the annealing temperature was decreased by 1°C until 56°C was reached. An additional 30 cycles at an annealing temperature of 56°C were performed, followed by a final extension at 68°C for 10 min. PCR products were resolved on a 1.2% (w/v) agarose gel, and a single band of about 350 bp was detected. The DNA band was excised and purified using a commercial kit (QIAquick Gel Extraction kit, Qiagen, Valenica, Calif.). The purified band was ligated into the pGEM-T Easy vector (Promega, Madison, Wis.) and transformed into JM109 E. coli competent cells. Plasmids were purified from E. coli transformants using a commercial kit (QIAprep Spin Miniprep kit, Qiagen) and sequenced using SP6 and T7 primers.
cDNA library screening
A cDNA library was constructed by Stratagene (LaJolla, Calif.) in the λ ZAP-Express vector using poly(A+) RNA from V. planifolia tissue culture. Using the 350-bp PCR clone as probe, we screened 450,000 plaque-forming units. The cloned 350-bp fragment was labeled with [α32P]dCTP using a commercial kit (Prime-It II Random Primer Labeling kit, Stratagene).
The plaque lifts were prehybridized at 42°C in 50% (v/v) formamide, 5× SSC, 5× Denhardt’s solution [1× Denhardt’s solution is 0.02% (w/v) Ficoll, 0.02% (w/v) PVP, 0.02% (w/v) BSA], 50 mM sodium phosphate, pH 6.8, 1% (w/v) sodium dodecyl sulfate (SDS), 100 μg ml−1 calf thymus DNA, and 2.5% (w/v) dextran sulfate. The hybridization solution was 5×105 cpm ml−1 of [32P]-labeled fragment, 50% (v/v) formamide, 5× SSC, 1× Denhardt’s solution, 20 mM sodium phosphate, pH 6.8, 1% (w/v) SDS, 100 μg ml−1 calf thymus DNA, and 5% (w/v) dextran sulfate. Hybridized membranes were washed with 2× SSPE (20× SSPE is 20 mM disodium EDTA, 160 mM sodium hydroxide, 200 mM monobasic sodium phosphate, and 3.6 M sodium chloride), 0.5% (w/v) SDS for 15 min at room temperature, 2× SSPE, 0.5% (w/v) SDS for 15 min at 65°C, and 0.2× SSPE, 0.2% (w/v) SDS for 15 min at 65°C. The washed filters were exposed to X-Ray film (XOMAT-AR, Kodak, Rochester, N.Y.) with an intensifying screen. Positive plaques were subjected to two additional rounds of screening to isolate single positive plaques. The cDNA inserts from positive plaques were excised from the λ-vector as recombinant pBK-CMV phagemids (Short et al. 1988). A full-length clone was completely sequenced by primer walking.
Expression of the V. planifolia OMT in E. coli
The coding sequence of the OMT was amplified by PCR using oligonucleotides that introduced XhoI sites at the 5′ and 3′ ends. The sequences of the oligonucleotides used for amplification were 5′-CATATGCTCGAGATGGCTACATGGGTGGAGCAC-3′ and 5′-CGGATCCTCGAGCTATTTGTTGAATTCCAT-3′. The PCR amplification product was separated on a 1% (w/v) agarose gel, and the DNA band was excised from the gel and extracted using a commercial kit (QIAquick Gel Extraction kit, Qiagen). The PCR product was digested with XhoI and again gel-purified. The digested PCR product was then ligated to the XhoI-digested dephosphorylated pET-15b expression vector (Novagen, Madison, Wis.) and transformed into ElectroMAX DH10B cells (Invitrogen) via electroporation. Plasmids from positive transformants were completely sequenced to confirm that no errors had been introduced through the PCR process. A plasmid containing the perfect OMT sequence, as well as an empty vector control, were then transformed in BL21(DE3) cells (Novagen) for protein expression.
For purification of the recombinant protein, a BL21(DE3) OMT transformant was grown at 37°C in Luria-Bertoni medium supplemented with 50 μg ml−1 ampicillin to OD600=0.5. Protein expression was then induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.05 mM. Additional ampicillin to a concentration of 50 μg ml−1 was also added and the cells grown overnight at 20°C. The cells were collected by centrifugation at 12,000 g for 15 min, lysed using BugBuster Protein Extraction reagent (Novogen), and treated with Benzonase Nuclease (Novogen) according to the manufacturer’s instructions. Cell debris was removed by centrifugation at 12,000 g for 20 min, the clarified lysate applied to a His-Bind column (Novogen), and the recombinant OMT protein eluted according to the manufacturer’s instructions. The eluted protein was passed through a PD10 column (Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with the OMT assay buffer and concentrated threefold using Ultrafree-4 centrifugal filter units (Millipore, Bedford, Mass.). The concentrated protein was used for enzyme activity assays.
Antibody production and immunoblot analysis
The purified recombinant protein was used for preparation of V. planifolia OMT-specific antiserum. It was mixed with an equal volume of Freund’s complete (first injection) or incomplete (subsequent injections) adjuvant and injected into the subscapular space of a rabbit. Three injections of about 100 μg of protein each were given at 4-week intervals.
For immunoblot analysis, proteins from leaves, stems, roots, pods, and the tissue culture were extracted by homogenizing tissue samples using a mortar and pestle in phosphate-buffered saline (1.5 mM NaH2PO4, 8.1 mM Na2HPO4, and 145.5 mM NaCl) in a ratio of 0.4 g tissue: 800 μl−1 buffer. The extracts were centrifuged to remove debris and the protein concentrations of the supernatants determined using the Bio-Rad protein assay reagent. Protein samples (20 μl) were mixed with an equal volume of 2× SDS sample buffer [2×: 125 mM Tris, pH 6.8, 4.6% (w/v) SDS, 10% (v/v) 2-mercaptoethanol, 20% (v/v) glycerol and 0.002% bromophenol blue (w/v) (Laemmli 1970)], then boiled for 5 min and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 10% polyacrylamide gels. The proteins were transferred to nitrocellulose membranes (NitroPure, Osmonics, Westborough, Mass.) in 10 mM 3-(cyclohexylamino)-1-propane-sulfonic acid (CAPS), pH 11, 10% methanol (v/v). Processing and detection by chemiluminescence (Western Lightening Chemiluminescence kit; Perkin-Elmer Life Science, Foster City, Calif.) was according to the manufacturer’s instructions.
Results and discussion
DOMT activity in V. planifolia pods and tissue culture
The proposed three-step vanillin biosynthetic pathway postulates a DOMT activity as the final step, resulting in the production of vanillin. We obtained green V. planifolia pods at different stages of development from Indonesia and assayed the crude extracts of the inner region of the pods where vanillin is synthesized (Joel et al. 2003) for DOMT activity by following the transfer of [14C] from radiolabeled SAM to 3,4-dihydroxybenzaldehyde. DOMT activity doubled between 3 months and 5 months after pollination and was maintained at a constant level for 11 months after pollination (Table 1). The increase in DOMT activity at 5 months after pollination corresponds to the developmental stage at which glucovanillin accumulation in the pods begins (Havkin-Frenkel et al. 1999).
Tissue cultures of V. planifolia have been established that accumulate glucovanillin (0.17% of dry weight) and its proposed precursors, including 3,4-dihydroxybenzaldehyde (0.11% of dry weight) (Havkin-Frenkel et al. 1996; Knorr et al. 1993). Crude extracts of the tissue cultures were found to have both DOMT and COMT activities (Table 1). Using 3,4-dihydroxybenzaldehyde as the substrate, we identified [14C]vanillin as the product on the basis of co-migration with the unlabeled standard vanillin on a TLC plate: 78% of the radioactivity present in the crude reaction product was recovered from the TLC plate at the position of authentic vanillin.
Partial purification of DOMT from V. planifolia tissue culture
Since the V. planifolia tissue cultures demonstrated DOMT activity at levels similar to those found in the pods and since they were a convenient source of plant material, our first approach to characterizing the enzyme was to purify it from the tissue cultures. Affinity purification by binding to adenosine-conjugated agarose has been successfully applied for the purification of some OMTs (Attieh et al. 1995; Wang and Pichersky 1998). DOMT and COMT activities were partially co-purified 18.2-fold and 16.7-fold, respectively, from the tissue culture crude protein extract by chromatography on an adenosine-agarose column (Table 1). The yield of partially purified protein from the adenosine-agarose column was 1.3%. SDS gel analysis of the active fractions revealed a major band at approximately 42 kDa and a minor band at approximately 27 kDa (data not shown). COMTs from other species are in the range of 37.6–42.3 kDa (Ibrahim et al. 1998). The 42- kDa band found in the SDS gel of the active fractions appeared to be a single band and was likely the source of the OMT activities. Peptide sequencing of the 42-kDa band, however, revealed that it was heterogeneous, and no sequences similar to COMTs were obtained.
Additional purification attempts were made using hydroxyapatite or Q-Sepharose column chromatography, but neither of these was successful in separating the DOMT and COMT activities from each other (data not shown). These results raised the question of whether there may be two enzymes with similar properties that cannot be separated or whether a single methyltransferase enzyme is present in the crude extracts that has activity against both substrates.
V. planifolia OMT cDNA clone
To test whether the DOMT activity detected in V. planifolia tissue cultures originated from a multifunctional methyltransferase that could methylate both 3,4-dihydroxybenzaldehyde and caffeic acid, we isolated a cDNA clone based on conserved sequences in COMTs from other species for expression in E. coli. Degenerate oligonucleotides based on the peptide sequences VLMESWY and HVGGDMF were used in PCR analyses of a cDNA library prepared from the V. planifolia tissue culture. A 350-bp amplified band was cloned whose sequence was similar to COMTs from other plants. The PCR clone was used to screen the cDNA library, and a full-length clone was obtained. A 365-amino acid protein with a molecular weight of 40,659 Da was predicted from the cDNA sequence.
Similarity of the V. planifolia OMT to other sequences
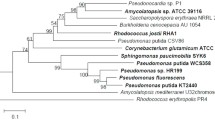
The V. planifolia OMT amino acid sequence is similar to that of COMTs reported from other plant species. COMT sequences previously reported to be from V. planifolia (Xue and Brodelius 1998) have been withdrawn from the NCBI database and now appear to be actually from Catharanthus roseus (Schroder et al. 2002). Phylogenetic analysis comparing 18 similar methyltransferase sequences illustrates the relationship of the V. planifolia OMT sequence to methyltransferases reported from other species (Fig. 2). The amino acid sequence of the V. planifolia OMT shows a similar level of divergence from the other monocot OMTs as from the dicot OMTs, perhaps reflecting its phylogenetic distance from the other reported monocot COMTs. The V. planifolia OMT amino acid sequence is 56% identical to that of Medicago sativa (alfalfa) and 60% identical to that of Triticum aestivum (wheat). V. planifolia is classified in the order Asparagales, whereas the other monocot species in the COMT sequence comparison are in the order Poales (Angiosperm Phylogeny Group 1998). A recent evaluation of expressed sequence tags (ESTs) from onion (Allium cepa), also in the order Asparagales, revealed genomic differences with the order Poales and similarities with the Eudicots (Kuhl et al. 2004). An apparently full-length onion EST with a similarity to COMTs, including all of the substrate binding residues, also groups between the Eudicot and Poales monocot sequences (Fig. 2). The onion sequence, however, shows no more similarity to the V. planifolia sequence than to the other sequences in the comparison. The onion sequence is 64% and 58% identical to that of alfalfa and V. planifolia, respectively.
Unrooted neighbor-joining tree comparing the V. planifolia OMT amino acid sequence with COMTs reported from other species. The tree was created using the clustal x and treeview programs. GenBank accession numbers for the corresponding DNA sequences are: Allium cepa, CF442066 (Kuhl et al. 2004); Catharanthus roseus, AY028439 (Schroder et al. 2002); Clarkia breweri, AF006009 (Wang and Pichersky 1997); Coffea canephora, AF454631 (unpublished); Eucalyptus gunnii, X74814 (Poeydomenge et al. 1994); Festuca arundinacea, AF153825 (unpublished); Lolium perenne, AF010291 (McAlister et al. 1998); Medicago sativa, M63853 (Gowri et al. 1991); Nicotiana tabacum class I, X74452 (Jaeck et al. 1996); Ocimum basilicum, AF154918 (Wang et al. 1999); Populus tremuloides, X62096 (Bugos et al. 1991); Prunus amygdalus, X83217 (Garcia-Mas et al. 1995); Saccharum officinarum, AJ231133 (Selman-Housein et al. 1999); Sorghum bicolor, AY217766 (Bout and Vermerris 2003); Thalictrum tuberosum, AF064696 (Frick and Kutchan 1999); Triticum aestivum, AY226581 (Jang et al. 2003); Vanilla planifolia, AY555144; Zea mays, M73235 (Collazo et al. 1992)
Although there is considerable overall amino acid sequence variability among the monocot and dicot COMTs, all of the residues identified from the crystal structure of the alfalfa enzyme as being involved in substrate binding or positioning (Zubieta et al. 2002) are generally well-conserved among all of the enzymes, including the V. planifolia OMT. A comparison of the deduced amino acid sequence of the V. planifolia OMT with that of alfalfa and the monocot wheat is shown in Fig. 3. The one non-conserved substrate binding residue in the V. planifolia enzyme is N185, which is H183 at the corresponding position of the alfalfa enzyme. In other COMT sequences, leucine and valine are often found at the relative position of the alfalfa I316; the V. planifolia sequence is a leucine at that position.
Comparison of the deduced amino acid sequences of the V. planifolia OMT with COMTs from alfalfa and wheat. The alignment was done using the geneworks program. Boxes enclose identical amino acids, gaps were inserted to maximize alignment. The experimentally determined substrate binding/positioning residues in the alfalfa enzyme are indicated by asterisks, the SAM binding residues are indicated by overlines, and the catalytic residues indicated by arrowheads
Expression of V. planifolia OMT in E. coli
The protein encoded by the V. planifolia OMT cDNA was expressed as an N-terminal polyhistidine-tagged fusion in E. coli based on the expression vector pET-15b. To confirm that the transformant E. coli cells had OMT activity, we measured the activity of the crude cell lysates. COMT and DOMT activity in crude lysates of the empty vector control cells after IPTG induction was only 20% and 28%, respectively, of that of the induced cells containing the vector with the V. planifolia OMT. These results confirmed that the V. planifolia cDNA did encode an OMT. For determination of the kinetic characteristics of the enzyme, we purified the recombinant protein using nickel-agarose affinity chromatography (Fig. 4). The expressed protein tended to rapidly accumulate in insoluble inclusion bodies, so conditions were developed using a low concentration of IPTG and a low incubation temperature to allow accumulation of soluble OMT protein.
Purification of the V. planifolia recombinant His-tagged fusion protein from Escherichia coli. Lane 1 20 μg of crude extract of E. coli transformant cells containing the expression vector for the V. planifolia OMT, lane 2 2 μg of the purified V. planifolia recombinant OMT. The positions of protein standards, in kiloDaltons, are indicated on the left
The kinetic parameters of the purified recombinant protein were determined with several phenolic and phenylpropanoid substrates (Table 2). This represents the first examination of a purified COMT from a monocot for such a broad range of substrates. The substrates 5-OH-ferulic acid ethyl ester and caffeic acid ethyl ester were included in the analysis since they were available, although they are not naturally occurring and therefore unlikely to serve as substrates in vivo. Surprisingly, they were the preferred substrates for the enzyme. Of the physiological substrates, caffeoyl aldehyde and 5-OH-coniferaldehyde were preferred over 5-OH-ferulic acid, 3,4-dihyroxybenzaldehyde, or caffeic acid. In general, the relative substrate preferences for the V. planifolia enzyme were similar to those reported for alfalfa COMT (Parvathi et al. 2001), which has been confirmed by down-regulation to be involved in S lignin biosynthesis (Guo et al. 2001). This suggests that the V. planifolia enzyme characterized here also may function primarily in the synthesis of lignin.
Kinetic characterizations of COMTs from other species have rarely included 3,4-dihydroxybenzaldehyde as a substrate. The tobacco (Maury et al. 1999), basil (Gang et al. 2002), and strawberry (Wein et al. 2002) COMTs are the only ones reported to have this activity. That the V. planifolia OMT characterized here also has this activity suggests that this feature may be common among other COMT enzymes.
V. planifolia OMT expression in different tissues
Expression of the V. planifolia OMT in different tissues was evaluated by immunoblot analysis (Fig. 5a). Strong immunoreactive bands were detected in the root, stem, and tissue culture samples at approximately 41 kDa, the size expected for the V. planifolia OMT. A weak immunoreactive band at that position was seen in the leaf sample, and no immunoreactive band at the size of the OMT was detected in the pod samples. The origin of the higher molecular-weight bands observed in the stem and leaf samples is not known. A gel of the same samples run in parallel and stained with Coomassie Blue is shown in Fig. 5b, confirming the presence of adequate protein in the pod sample. The pod samples were from the 8-months-after-pollination pods obtained from Indonesia, as used in Table 1. The same results were obtained in other blots using other pod extracts (data not shown). These results were unexpected and surprising, since both the pods and tissue cultures synthesize vanillin (Havkin-Frenkel et al. 1999) and both had DOMT activity at similar levels (Table 1), and they suggest that the DOMT activities detected in these tissues originate from distinct enzymes that do not exhibit antibody cross-reactivity. If this OMT is involved in the synthesis of vanillin it must be present in the pods at low levels that are not detectable by immunoblot analysis of proteins from crude extracts. Since DOMT activity was detectable in the pods, however, the absence of an immunoreactive protein band suggests that this OMT is not the main contributor to the observed activity. Although the V. planifolia OMT characterized here can convert 3,4-dihydroxybenzaldehyde to vanillin in vitro, the kinetic parameters and the tissue distribution suggest its primary function is likely to be in lignin biosynthesis. Whether it also functions in vanillin biosynthesis in the tissue cultures in vivo is not yet known. Such a dual role in lignification and secondary metabolite synthesis has been proposed for a strawberry COMT (Wein et al. 2002; Schwab 2003).
a Immunoblot analysis of V. planifolia OMT in different tissues. Protein extracts were subjected to SDS-PAGE and immunoblot analysis using the OMT antiserum. b. Coomassie Blue-stained gel of the same extracts used for the immunoblot in a. The positions of protein standards, in kiloDaltons, are indicated on the left
Overall, the results presented here can be considered to be circumstantial evidence that suggests the existence of an additional distinct OMT present in the pods that is also capable of catalyzing the conversion of 3,4-dihydroxybenzaldehyde to vanillin. Confirmation of this will require the isolation of the putative enzyme. Since the OMT characterized here does have DOMT activity, however, it may be useful in engineering strategies for the synthesis of natural vanillin from alternate sources. The functional characterization of the V. planifolia OMT presented here reveals the evolutionary conservation of the substrate binding amino acid residues and relative substrate specificities of a COMT from a less well-studied order of monocots, the Asparagales.
Abbreviations
- COMT :
-
Caffeic acid O-methyltransferase
- DOMT :
-
3,4-Dihydroxybenzaldehyde-O-methyltransferase
- OMTs :
-
O-Methyltransferases
- SAM :
-
S-adenosyl-l-methionine
References
Angiosperm Phylogeny Group (1998) An ordinal classification for the families of flowering plants. Ann Mo Bot Gard 85:531–553
Attieh JM, Hanson AD, Saini HS (1995) Characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J Biol Chem 270:9250–9257
Bout S, Vermerris W (2003) A candidate-gene approach to clone the sorghum brown midrib gene encoding caffeic acid O-methltransferase. Mol Genet Genomics 269:205–214
Bugos RC, Chiang VL, Campbell WH (1991) cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol 17:1203–1215
Collazo P, Montoliu L, Puigdomenech P, Rigau J (1992) Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol 20:857–867
Dignum MJW, Kerler J, Verpoorte R (2001) Vanilla production: technological, chemical, and biosynthetic aspects. Food Rev Int 17:199–219
Dixon RA, Chen F, Guo D, Parvathi K (2001) The biosynthesis of monolignols: a “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57:1069–1084
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Frick S, Kutchan TM (1999) Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J 17:329–339
Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E (2002) Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14:505–519
Garcia-Mas J, Messeguer R, Arus P, Puigdomenech P (1995) The caffeic acid O-methyltransferase from Prunus amygdalus (GenBank X83217). Plant Physiol 108:1341
Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA (1991) Stress responses in alfalfa (Medicago sativa L.) X. Molecular cloning and expression of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol 97:7–14
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88
Havkin-Frenkel D, Podstolski A, Knorr D (1996) Effect of light on vanillin precursors formation by in vitro cultures of Vanilla planifolia. Plant Cell Tissue Organ Cult 46:169–170
Havkin-Frenkel D, Podstolski A, Witkowska E, Molecki P, Mlkolajczyk M (1999) Vanillin biosynthetic pathways: an overview. In: Fu TJ, Singh G, Curtis WR (eds) Plant cell and tissue culture for the production of food ingredients. Kluwer/Plenum, Dordrecht/New York, pp 35–43
Havkin-Frenkel D, French JC, Graft NM, Joel DM, Pak FE, Frenkel C (2003) Interrelation of curing and botany in vanilla (Vanilla planifolia) bean. Acta Hortic 629:93–102
Herz LE (2000) Dynamics and regulatory control of the vanillin biosynthetic pathway in V. planifolia embryo cultures. PhD thesis, Rutgers University, N.J.
Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36:1–10
Jaeck E, Martz F, Stiefel V, Fritig B, Legrand M (1996) Expression of class I O-methyltransferase in healthy and TMV-infected tobacco. Mol Plant Microbe Interact 9:681–688
Jang CS, Kim JY, Haam JW, Lee MS, Kim DS, Li YW, Seo YW (2003) Expressed sequence tags from a wheat-rye translocation line (2BS/2RL) infested by larvae of Hessian fly [Mayetiola destructor (Say)]. Plant Cell Rep 22:150–158
Joel DM, French JC, Graft N, Kourteva G, Dixon RA, Havkin-Frenkel D (2003) A hairy tissue produces vanillin. Isr J Plant Sci 51:157–159
Knorr D, Caster C, Dorneburg H, Dorn R, Graf S, Havkin-Frenkel D, Podstolski A, Werrman T (1993) Biosynthesis and yield improvement of food ingredients from plant cell and tissue cultures. Food Technol 47:57–63
Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, McCallum J, Catanach A, Rutherford P, Sink KC, Jenderek M, Prince J, Town CD, Havey MJ (2004) A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16:114–125
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li L, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275:6537–6545
Maury S, Geoffroy P, Legrand M (1999) Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methltransferse and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol 121:215–223
McAlister FM, Jenkins CLD, Watson JM (1998) Sequence and expression of a stem-abundant caffeic acid O-methyltransferase cDNA from perennial ryegrass (Lolium perenne). Aust J Plant Physiol 25:225–235
Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA 96:8955–8960
Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25:193–202
Podstolski A, Havkin-Frenkel D, Malinowski J, Blount JW, Kourteva G, Dixon RA (2002) Unusual 4-hydroxybenzaldehyde synthase activity from tissue cultures of the vanilla orchid Vanilla planifolia. Phytochemistry 61:611–620
Poeydomenge O, Boudet AM, Grima-Pettenati J (1994) A cDNA encoding S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase from Eucalyptus. Plant Physiol 105:749–750
Rao SR, Ravishankar GA (2000) Vanilla flavour: production by conventional and biotechnological routes. J Sci Food Agric 80:289–304
Schroder G, Wehinger E, Schroder J (2002) Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry 59:1–8
Schwab W (2003) Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62:837–849
Selman-Housein G, Lopez M, Hernandez D, Civardi L, Miranda F, Rigau J, Puigdomenech P (1999) Molecular cloning of cDNAs coding for three sugarcane enzymes involved in lignification. Plant Sci 143:163–171
Short U, Fernandez JM, Sorge JA, Huse WD (1988) λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res 16:7583–7599
Walton NJ, Mayer MJ, Narbad A (2003) Vanillin. Phytochemistry 63:505–515
Wang J, Pichersky E (1997) Nucleotide sequence of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase from Clarkia breweri (accession no. AF006009). Plant Physiol 114:1567
Wang J, Pichersky E (1998) Characterization of S-adenosyl-l-methionine:(iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch Biochem Biophys 349:153–160
Wang J, Dudareva N, Bhakta S, Raguso RA, Pichersky E (1997) Floral scent production in Clarkia breweri (Onagraceae) II. Localization and developmental modulation of the enzyme S-adenosyl-l-methionine:(iso)eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiol 114:213–221
Wang J, Dudareva N, Kish CM, Simon JE, Lewinsohn E, Pichersky E (1999) Nucleotide sequences of two cDNAs encoding caffeic acid O-methyltransferases (accession nos. AF154917 and AF154918) from sweet basil (Ocimum basilicum). Plant Physiol 120:1205
Wein M, Lavid N, Lunkenbein S, Lewinsohn E, Schwab W, Kaldenhoff R (2002) Isolation, cloning and expression of a multifunctional O-methyltransferase capable of forming 2,5-dimethyl-4-methoxy-3(2H)-furanone, one of the key aroma compounds in strawberry fruits. Plant J 32:755–765
Xue Z-T, Brodelius PE (1998) Kinetin-induced caffeic acid O-methyltransferases in cell suspension cultures of Vanilla planifolia Andr. and isolation of caffeic acid O-methyltransferase cDNAs. Plant Physiol Biochem 36:779–788
Zubieta C, Kota P, Ferrer J-L, Dixon RA, Noel JP (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14:1265–1277
Acknowledgements
We thank Dr. Richard Dixon, Noble Foundation, for the sequencing and Dr. Eran Pichersky, University of Michigan, for advice on the affinity chromatography. This work was supported by a grant from David Michael & Company.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Barz
Rights and permissions
About this article
Cite this article
Pak, F.E., Gropper, S., Dai, W.D. et al. Characterization of a multifunctional methyltransferase from the orchid Vanilla planifolia . Plant Cell Rep 22, 959–966 (2004). https://doi.org/10.1007/s00299-004-0795-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-004-0795-x