Abstract
Epitopes often require co-delivery with an adjuvant or targeting protein to enable recognition by the immune system. This paper reports the ability of transgenic tomato plants to express a fusion protein consisting of the B subunit of the Escherichia coli heat-labile enterotoxin (LTB) and an immunocontraceptive epitope. The fusion protein was found to assemble into pentamers, as evidenced by its ability to bind to gangliosides, and had an average expression level of 37.8 μg g−1 in freeze-dried transgenic tissues. Processing of selected transgenic fruit resulted in a 16-fold increase in concentration of the antigen with minimal loss in detectable antigen. The species-specific nature of this epitope was shown by the inability of antibodies raised against non-target species to detect the LTB fusion protein. The immunocontraceptive ability of this vaccine will be tested in future pilot mice studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, plants expressing vaccine antigens have proven a promising approach to vaccination (Walmsley and Arntzen 2000). Plant-derived antigens have delayed (Modelska et al. 1998) or prevented (Dalsgaard et al. 1997; Wigdorovitz et al. 1999) the onset of disease in animals under experimental conditions and have proven to be safe and functional in human clinical trials (Tacket et al. 1998, 2000). Fitchen et al. (1995) showed a plant virus capable of expressing an immunocontraceptive epitope in plants as part of a hybrid coat protein. The seven amino acid epitope (amino acids 336–342) from the mouse zona pellucida 3 (ZP3) glycoprotein is well characterized and has previously been proven immunocontraceptive by parenteral administration (Dunbar and Prasad 1997; Millar et al. 1989; Skinner et al. 1994; Tung et al. 1996). Upon fusion of the mouse ZP3 epitope to the coat protein of tobacco mosaic virus and infection of tobacco, high levels accumulated in the inoculated tobacco leaves (Fitchen et al. 1995). Parenteral immunization of mice with the partially purified chimeric virus emulsified with an adjuvant resulted in production of serum antibodies that recognized the ZP3 epitope, yet no effect on fertility was observed (Fitchen et al. 1995).
The heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) has been used as a carrier for subunit vaccines (Bagdasarian et al. 1999; O'Dowd et al. 1999) and has been found immunogenic to mice and humans when expressed by plants (Mason et al. 1998; Tacket et al. 1998). The LT toxin is a strong mucosal immunogen that contains a non-toxic B subunit (LTB) and a toxic A subunit (Sixma et al. 1993). Five B subunits form a donut-shaped pentamer that is essential for binding to GM1 gangliosides (Tsuji et al. 1995). Because the surface of mucosal cells is decorated with GM1 gangliosides, the pentamer targets the holotoxin to the mucosal surface, and subsequent endocytosis of the LT-GM1 complex presents the holotoxin to the mucosal lymphoid tissue.
We aim to produce a successful oral immunocontraceptive vaccine through fusion of an appropriate epitope to LTB, thereby targeting the epitope to antigen-processing cells underlying the mucosal surface of the gut. In this study we investigated the capacity of transgenic tomato plants to express a fusion protein consisting of the LTB of ETEC and a seven amino acid epitope from murine ZP3.
Materials and methods
Fusion of a plant-optimized mouse ZP3 epitope to a plant-optimized synthetic LTB gene
An EcoRV-KpnI fragment from TH210 (Mason et al. 1998) containing a synthetic LTB sequence was inserted into the corresponding sites of pBluescript (Stratagene, La Jolla, Calif.) to make pBlueLTB. The plasmid pLTB-L was created by using a unique BbsI site within the synthetic LTB sequence to fuse the coding region of a seven amino acid linker (translational) to the 3′ end of the LTB coding region in pBlueLTB. The linker was constructed using two commercially synthesized oligomers 5′-AAC TCT GAT CCA CAT GTT CCT and 5′-AGT TAG GAA CAT GTG GAT CAG that had been purified by electrophoresis on a denaturing polyacrylamide gel. The oligomers were phosphorylated with T4 kinase in separate reactions, mixed in equimolar amounts and annealed by heating to 90°C for 5 min before being gradually cooled to 23°C for over an hour. Plasmid pLTB-L was prepared for insertion of the mouse ZP3 epitope by serial digestion with BbsI and KpnI and dephosphorylation with calf intestinal phosphatase.
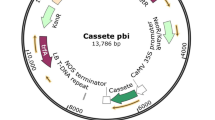
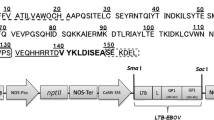
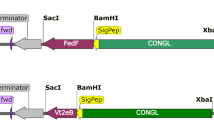
The coding sequence of the mouse ZP3336–342 epitope was analyzed for its codon usage compared with plant genes (Wada et al. 1990). A plant-optimized epitope was designed maintaining the amino acid sequence of the native epitope with added nucleotides at the 5′ and 3′ ends that, on assembly of the oligonucleotides, created a BbsI and KpnI compatible fragment. The oligonucleotides MZepA 5′-AAC TTC CAA ATT CAT GGA CCA AGA AAC TAA GTC TTC GGT AC and MzepB 5′-CGA AGA CTT AGT TTC TTG GTC CAT GAA TTT GGA were synthesized and purified by electrophoresis on a denaturing polyacrylamide gel. The epitope was assembled as described for the linker in the construction of pLTB-L. The epitope was placed on ice before ligation into BbsI+KpnI-digested pLTB-L, making pLTBL7. The plasmids pLTB-L and pLTB7 were sequenced by the dideoxy chain termination method. An NcoI-KpnI fragment of pLTB7 was inserted into pIBT210 (Haq et al. 1995) digested with NcoI and KpnI to make pAW7. The expression cassette was purified from pAW7 after digestion with HindIII and EcoRI and ligated with pGPTV.kan (Becker et al. 1992) digested with HindIII and EcoRI to give pAWBin7 (Fig. 1).
Schematic diagram of the T-DNA region of the binary vector pAWBin7 for expression of the B subunit of Escherichia coli heat labile enterotoxin (LTB)-mouse zona pellucida 3 (ZP3) epitope fusion protein. This region is based on the T-DNA region of pGPTV.kan (Becker et al. 1992). RB Right hand border repeat, LB left hand border repeat, 5′NOS nopaline synthase promoter, NPT2 neomycin phophotransferase gene, Ag7 Agrobacterium gene 7 polyA signal, CaMV 35S cauliflower mosaic virus 35S promoter, TEV 5′-UTR of tobacco etch virus, sLTB coding region of a synthetic LTB gene, L linker, Zep murine ZP3 epitope (amino acids 336–342), VSP 3′-UTR of soybean vegetative storage protein gene
Tomato transformation and processing
In this paper, subscript text used to describe a transgenic plant line, for example T1, indicates the number of sexual cycles that have occurred after the transformation event. The plasmid pAWBin7 was prepared from cultures of E. coli DH5α and electroporated into Agrobacterium tumefaciens EHA105 (Hood et al. 1993). Agrobacterium-mediated transformation of tomato cotyledons (variety Tanksley TA234TM2R) was performed according to Frary and Earle (1996) except that seeds were sterilized by soaking in 70% ethanol for 2 min before rinsing in sterile water and washing in a mixture of 10% bleach and 1% Tween-20 for 2 h. The seeds were rinsed three times in sterile distilled water before plating on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog 1962) (half-strength MS: 50 mg l−1 myo-inositol, 2 mg l−1 thiamine HCl, 0.5 mg l−1 pyridoxine HCl, 0.5 mg l−1 nicotinic acid, 10 g l−1 sucrose and 8 g l−1 Difco bacto agar, pH 5.8). Plantlets were regenerated on medium containing kanamycin at 300 mg l−1. Individual lines were screened by ganglioside-dependent ELISA for LTB expression in leaves and fruit. The lines with best fruit expression were self-pollinated and the resulting seeds germinated on MS medium supplemented with 300 mg l−1 kanamycin. Surviving seedlings were transferred to soil and the glasshouse and self-pollinated. Fresh fruit samples from each T1 tomato plant were taken and freeze-dried in a 100 SRC Virtis freeze-drier for a minimum of 72 h with a maximum shelf temperature of 20°C. The dried fruit were then powdered, pooled, and stored in a dry environment at 4°C.
Arabidopsis thaliana transformation
The plasmid TH110 containing the LTB gene (Haq et al. 1995) was prepared from cultures of E. coli DH5α and electroporated into Agrobacterium tumefaciens LBA4404 (Hoekema et al. 1983). Floral dip Agrobacterium-mediated transformation of Arabidopsis thaliana was performed according to Clough and Bent (1998). This method involved the growth of Arabidopsis thaliana to flowering stage and dipping the developing floral tissue into a solution containing Agrobacterium tumefaciens, 5% sucrose and 500 μl l−1 of the surfactant Silwet L-77. The resulting seeds were collected and selected for transgenic lines by germination and growth on medium containing 50 μg ml−l kanamycin.
Protein extraction and antigen analysis
The LTB fusion protein and possible cleavage products were purified from the pooled, freeze-dried, transgenic tomato fruit sample by galactose affinity chromatography (Clements and Finkelstein 1979). Briefly, 1 g Sephadex G75 (Sigma, St. Louis, Mo.) was activated with 100 ml 1 mM HCl in a glass sintered funnel. The gel was allowed to swell for 15 min before applying a vacuum. The gel was flushed with 100 ml 1 mM HCl, and then washed with 100 ml phosphate-buffered saline (PBS). A gel slurry was made in a 15 ml falcon tube by adding 3 ml ice-cold PBS to 7 g activated Sephadex. Pooled, freeze-dried, transgenic tomato fruit powder was vortexed with ice-cold PBS +0.1% Triton X-100 +10μg ml−1 leupeptin at a ratio of 1:4 (powder:PBS) and centrifuged at 3,750 rpm for 5 min at 4°C. Tomato fruit extract supernatant (3 ml) was added to 2 ml Sephadex slurry in a 15 ml Falcon tube and incubated overnight on a bench rocker at 4°C. The tomato extract/Sephadex gel was added to a sintered glass funnel and a vacuum was applied. The gel was flushed with 50 ml ice-cold PBS then made up to 5 ml with ice-cold PBS. The slurry was degassed using a vacuum desiccator then poured into a 10 ml disposable column (Bio-Rad; Hercules, Calif.) being careful not to create air bubbles. The bottom of the column was snapped off and the PBS allowed to flow through. The column was washed with 20 ml ice-cold PBS; LTB products were then eluted with 4 ml 0.2 M galactose in PBS. Absorbance at 280 nm of the collected fractions was read. The aliquot with the highest absorbance was used in Western blot analysis. Freeze-dried, wild-type tomato fruit powder was also subjected to galactose affinity chromatography before use in Western analysis.
An aliquot (30 μl) of each sample was added to 6 μl 6× SDS gel loading buffer (300 mM Tris-HCl, pH 6.8, 600 mM dithiothreitol, 12% SDS, 0.6% Bromophenol Blue, 60% glycerol), then boiled for 10 min and placed on ice. The samples were centrifuged at 12,000 rpm for 5 min at 4°C in an Eppendorf 5415C microcentrifuge before being subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was run in Tris-glycine buffer (25 mM Tris, 250 mM glycine, pH 8.3, 0.1% SDS) at 30 mA/gel until the dye front ran about 5 mm from the gel bottom. The separated proteins were transferred from the gel to a polyvinylidenedifluoride membrane using a Bio-Rad Trans Blot Cell (50 V for 2 h). The following membrane incubations were performed at room temperature while membrane washes were in PBST (PBS +0.1% Tween-20). The membrane was blocked with PBST containing 1% dry milk (PBSTM) for 1 h using slow rotation in a Hybaid (Ashford, Middlesex, UK) oven then briefly washed twice before incubating with the primary antibody (anti-LTB, Benchmark Biolabs, Lincoln, Neb.) diluted 1:1,000 in PBSTM. The membrane was briefly rinsed in PBST before a 15 min wash and 3×5 min washes. The membrane was then incubated in a 1:12,000 dilution of an anti-rabbit IgG horseradish peroxidase conjugate (Sigma) for 1 h with slow rotation. The membrane was rinsed in PBST, and then subjected to a 15 min wash and 3×5 min washes. Detection was performed using the Amersham ECL+ kit according to the manufacturer's instructions.
For ELISA analysis, leaf samples and fresh fruit from greenhouse-grown plants or dried fruit samples were homogenized in 2 ml g−1 (fresh samples) or 50 ml g−1 (dried fruit) ice-cold extraction buffer (50 mM sodium phosphate, pH 6.6, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10 μg ml−1 leupeptin, 1 mM phenylmethylsulfonylfluoride) in a QBiogene (Carlsbad, Calif.) Fast Prep machine. Insoluble materials were pelleted by centrifugation at 14,000 rpm in an Eppendorf 5415C microcentrifuge at 4°C for 5 min. The supernatant was kept on ice during analysis then stored at −80°C.
Ganglioside-dependent ELISA was performed on two samples of 1:100 dilution of each extract as described (Haq et al. 1995) using a primary antibody of either 1:1,000 anti-LTB antiserum, 1:100,000 anti-mouse ZP3 epitope antiserum or 1:100,000 anti-brushtail possum (Trichosurus vulpecula) ZP3 epitope antiserum dilution in PBSTM.
Although ganglioside-dependent ELISA was used for detecting the epitope in the fusion protein in tomato plant samples, the standards were directly bound to the plates. Standards for mouse ZP3 epitope detection were the seven amino acid epitope or the seven amino acid epitope conjugated to BSA. The epitope was chemically conjugated to BSA using the Pierce (Rockford, Ill.) Imject (R) Maleimide Activated BSA kit according to the manufacturer's instructions.
Isolation of nucleic acids from plant material
Nucleic acid was isolated from young leaf tissue of T1 tomato plants by grinding 200–250 mg material in a 1.5 ml microcentrifuge tube using liquid nitrogen and a nail. The powder was resuspended in 500 μl extraction buffer (420 g l−1 urea, 312.5 mM NaCl, 50 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1% sarcocine) and incubated for 30 min with 500 μl phenol:chloroform:isoamyl alcohol (25:24:1) on a sample rotator. The resulting slurry was centrifuged for 15 min at 14,000 rpm, 4°C. The upper aqueous phase was collected and total RNA precipitated by addition of an equal volume of 4 M LiCl and incubation at −20°C for at least 8 h. Samples were centrifuged at 15,000 rpm for 15 min at 4°C and the supernatant removed to separate tubes. The RNA pellet was resuspended in 50 μl RNase-free sterile water. Genomic DNA was precipitated from the saved supernatant through addition of a 10% volume 7.5 M ammonium acetate, 1 volume ice-cold isopropanol and incubation at −20°C for at least 8 h. The DNA was washed with 70% ice-cold ethanol, allowed to air-dry for 5 min and resuspended in 50 μl sterile water + RNase (50 μg ml−1).
Southern analysis
Samples containing 15 μg DNA were prepared for Southern analysis according to Sambrook et al. (1989).
A labeled probe was made by PCR using pAW7 as a template with the primer set TEV (5′-GCA TTC TAC TTC TAT TGC AGC) and VSP (5′-GTG CAT ATC AGC ATA C). Digoxigenin (DIG)-labeled dCTP was incorporated into the 588 bp amplicon according to the manufacturer's instructions (PCR DIG Probe Synthesis kit, Boehringer Mannheim, Germany). The amplifications were performed over 32 cycles using a Hybaid PCR Express Thermo-Cycler. The template was first melted at 94°C for 4 min followed by 30 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 90 s. A final extension step was performed at 72°C for 5 min before soaking at 4°C.
Hybridization and membrane detection was performed as per the manufacturer's instructions (Boehringer Mannheim: DIG wash and block buffer set and DIG Luminescent Detection Kit) using a probe concentration of 5 μl ml−1 DIG Easy Hyb. Labeled membranes were visualized after exposure to film.
Northern analysis
Northern analysis was performed as described in Sambrook et al. (1989). Briefly, 18 μl total RNA from selected tomato plant lines was denatured with formaldehyde/formamide and run for 2 h in a 1% agarose, MOPS-acetate-EDTA gel. RNA was then transferred to Zeta Probe membrane (Bio-Rad) by upward capillary action and fixed by UV cross-linkage. Membrane hybridization and detection were performed as with Southern analysis except that hybridization steps were performed at 42°C.
Results
Antigen expression in T0 and T1 tomato plants
Phenotypes were indistinguishable among control tomato plants and T0 tomato plants with low or high antigen expression levels. Because the fruit were the target organs for animal feed trials, tomato plant lines with high fruit expression were prioritized. Tomato plants 16.1, 17.4, 17.8, and 17.9 showed the highest LTB expression in fruit, thus progeny from these lines were further analyzed.
Of 21 T1 tomato plants germinated on kanamycin, LTB expression was detected in leaf and/or dried fruit samples of 17 plants using GM1 ganglioside-dependent ELISA (Table 1). Expression of LTB in leaf samples varied from 0 to 10 μg g−1 fresh weight (FW) while in the concentrated processed fruit samples, expression ranged from 0 to 64.7 μg g−1 dry weight (DW). To obtain an average expression level for the LTB fusion protein, dried fruit samples from tomato plants expressing LTB above 8 μg g−1 DW were pooled and mixed well. The concentration of LTB in the pooled, freeze-dried transgenic tomato fruit powder was 37.8 μg g−1 DW.
As previously observed in the T0 generation, T1 tomato plants showing high antigen expression in fruit or leaves had indistinguishable phenotypes from control plants or plants with low antigen expression levels. Tomato plants were ranked in descending order with regard to LTB leaf and fruit expression. Regression analysis of the leaf and fruit LTB expression rankings showed only a weak positive correlation (r 2 =0.246).
To assess the integrity of the LTB fusion protein, anti-LTB Western analysis was performed. Anti-LTB Western blot analysis of T1 tomato materials showed a band roughly centered at 14 kDa in the purified extract from the pooled, freeze-dried T1 transgenic tomato fruit sample. A band roughly centered at 12 kDa was detected in the bacterial and A. thaliana LTB positive controls (Fig. 2, lanes B and P, respectively) while no bands were detected in the wild-type tomato, negative control lane.
Anti-LTB Western blot analysis of T1 tomato plant material. Lanes: P Crude protein extract from Arabidopsis plant expressing LTB, B 30 ng bacterial LTB, N wild-type, freeze-dried tomato extract purified by galactose affinity chromatography, TT transgenic, freeze-dried tomato extract purified by galactose affinity chromatography. Sizes (kDa) are indicated on the left of the gel
An accurate ELISA reading for the mouse ZP3 epitope standard could not be developed. When the epitope alone was used as a standard, OD readings were low and inconsistent (data not shown). This most likely resulted from strategic antibody recognition sites being concealed upon binding of such a small epitope to the plate. The concentration of the BSA-epitope conjugate was based on a BSA standard curve. Because conjugation to BSA resulted in multiple chemical linkages of the epitope to one BSA molecule, the conjugate standard gave an inflated OD reading of epitope concentration. Instead, epitope-specific ELISA was performed and the test samples were compared with negative control samples. The BSA-epitope conjugate was used as a positive control and not as a means of determining epitope concentration. Although the OD readings were low, processed fruit samples positive for LTB consistently gave epitope-specific OD readings significantly higher than those obtained for the negative control dried samples (α =0.05) (Fig. 3). In contrast, ELISA assays to detect the mouse epitope using the anti-possum ZP3 epitope antiserum did not return OD readings above background (data not shown).
Effect of processing on antigen content
To determine the effect of freeze-drying on mass and antigen content, two fruit each from five T2 tomato plants (offspring of 161.1 and 174.8) were weighed and assayed for LTB content before and after processing. Freeze-drying reduced fruit mass to 6% of its initial value (data not shown). The antigen content on a per gram basis showed a 16-fold increase, suggesting no appreciable loss of antigen by the processing method employed (Fig. 4).
Southern and northern analysis of T1 tomato plants
All 21 tomato plants germinated on kanamycin possessed at least one copy of the transgene of interest (Fig. 5a). The maximum number of transgene inserts detected was at least four in tomato plants 174.5 and 174.7. However, the number of gene insertions did not necessarily reflect the LTB expression level (Table 1). With the exception of the negative control, all tomato plants tested transcribed the foreign gene of interest (Fig. 5b).
Southern and northern analysis of T1 tomato plant nucleic acids. A Southern analysis of T1 tomato lines. Lanes: 1 Wild-type tomato control, 2 wild-type tomato control spiked with pAW7 HindIII digest, 3 transgenic tomato line 161.1, 4–10 transgenic tomato lines from parent 17.4, 11–15 transgenic tomato lines from parent 17.8, 16–23 transgenic tomato lines from parent 17.9. Size of DNA fragments is shown in base pairs. B Northern analysis of T1 tomato lines. Lanes: 1 Wild-type tomato negative control, 2–4 transgenic tomato lines from parent 17.4, 5–7 transgenic tomato lines from parent 17.8, 8 transgenic tomato line 179.1, 9 wild-type tomato control, 10–17 transgenic tomato lines from parent 17.9. Size of RNA bands is shown in bases
Discussion
Previous studies have successfully produced LTB fusions in bacterial expression systems (Cardenas and Clements 1993; Pillai et al. 1996; Schodel et al. 1991; Sewani et al. 1998) and cholera toxin B fusions in plants (Arakawa et al. 1998, 2001); however, this is the first report of a plant-expressed LTB fusion. The ability to detect both components of the LTB fusion protein in processed fruit by GM1 ganglioside-dependent ELISA indicated the addition of a linker and epitope (15 amino acids) to the carboxy terminus of LTB did not prevent LTB pentamer formation. Anti-LTB Western blot analysis showed a band centered about 14 kDa in the crude protein extract from the pooled, freeze-dried T1 transgenic tomato fruit sample (Fig. 2, lane TT), a band roughly centered at 12 kDa in the Arabidopsis and bacterial-derived LTB positive controls (Fig. 2, lanes P and B) and no band in the negative control of wild-type tomato (Fig. 2, lane N). LTB is expected to be directed to the endoplasmic reticulum in plant cells via its cleavable amino-terminal signal peptide, which targets LTB to the periplasmic space in E. coli (Mason et al. 1998). Since LTB has a potential glycosylation site at position 90, Arabidopsis-derived LTB was compared to the LTB fusion protein using Western analysis to ensure the shift in size of the LTB product in the fusion protein lane was due to the presence of the epitope and not due to glycosylation. Because the Arabidopsis-derived LTB was the same size as the bacterial LTB, it was not glycosylated (Fig. 2, lanes P and B). The positive mouse ZP3 ELISAs using the transgenic tomato materials supports the view that the shift in size of the LTB product in lane TT (from 12 to 14 kDa) was caused by the seven amino acid linker and the eight amino acid mouse ZP3 epitope (1.7 kDa). No cleavage of the fusion protein was observed, therefore the fusion protein concentration was taken as the LTB content of the sample.
Harvesting and processing of T2 tomato fruit was performed as described for the T1 generation yet the LTB expression was approximately 10-fold higher at 354.7 μg g−1 DW. Since the choice of tomato lines for further propagation and analysis was based on high LTB expression in fruit, it was thought that the 9-fold increase in LTB expression was caused by the selfing of elite tomato lines. The inability to detect the mouse epitope component of the fusion protein by ELISA using antibodies raised against the epitope from a different species (Trichosurus vulpecula) lends evidence to the species-specific nature of this epitope. As species specificity will be an important characteristic for the final application of this vaccine, future small animal trials have been planned to confirm this aspect. This study found the fusion protein to accumulate in the fruit of tomato plants without antigen expression resulting in stunting of plant growth as reported by Mason et al. (1998). This suggests either that the murine ZP3 epitope decreases the reported toxicity of LTB in potato cells or that tomato is more tolerant of LTB accumulation than potato.
Analysis of leaf and fruit LTB expression levels showed only a weak positive correlation (r 2 =0.246). Leaf expression levels could therefore not be used as an early screen for high fruit LTB expression. Plant lines for northern analysis were selected to include plants that had high (lines 174.1 and 174.2), medium (line 179.2), low (lines 178.5, 178.6, and 179.6) or no detectable (lines 178.3, 179.3, 179.5, 179.7 and 179.8) fusion protein expression in the fruit. The Northern analysis performed was not quantitative; however, it did reveal that RNA of the expected size for the transgene of interest (approximately 1,134 bases) was detected in all tested transgenic plants and not in the wild-type control (Fig. 5B). Thus total gene silencing can be eliminated as a possible reason for the lack of correlation and general variation in antigen expression in this study. Southern analysis revealed several plant lines to contain labeled DNA fragments of the same size after HindIII digestion, for example plant lines 178.1, 178.2, 178.3, and 178.5 (Fig. 5A). However, we observed that LTB expression levels varied widely between these plants (Table 1). Since it was demonstrated that total transgene silencing was not occurring, we believe the variation in antigen expression was due to either mutation of the gene leading to a different protein conformation, and thus decreased or loss of recognition of the antigen by the polyclonal antibody in ELISA detection, or partial transgene silencing.
The observed variability in antigen expression from plant to plant in this report is also often found also from fruit to fruit. This is inherent in transgenic plants and is an important problem to solve with regards to plant-derived vaccines as it affects the dosage in any single administration. Although palatable raw, a tomato fruit-derived vaccine has the limit of being perishable. The pooling of processed tomato fruit samples in this study provided a batch of plant material with concentrated, antigenically active fusion protein of uniform concentration that displayed a shelf-life of more than 5 months if kept dry (data not shown). Freeze-drying is a well established technology that is relatively inexpensive. It should thus prove useful for the development of plant-derived edible vaccines as it provides a means to standardize the antigen concentration in batches and provides an elongated shelf-life without making the cost of the vaccine prohibitive to either veterinary medicine or human vaccine programs, even in developing countries.
Abbreviations
- DW :
-
Dry weight
- ELISA :
-
Enzyme-linked immunosorbant assay
- ETEC :
-
Entertoxigenic Escherichia coli
- FW :
-
Fresh weight
- LTB :
-
B subunit of Escherichia coli heat-labile enterotoxin
- ZP3 :
-
Zona pellucida glycoprotein 3
References
Arakawa T, Yu J, Chong DK, Hough J, Engen PC, Langridge WH (1998) A plant-based cholera toxin B subunit-insulin fusion protein protects against the development of autoimmune diabetes. Nat Biotechnol 16:934–938
Arakawa T, Yu J, Langridge WH (2001) Synthesis of a cholera toxin B subunit-rotavirus NSP4 fusion protein in potato. Plant Cell Rep 20:343–348
Bagdasarian MM, Nagai M, Frey J, Bagdasarian M (1999) Immunogenicity of Actinobacillus ApxIA toxin epitopes fused to the E. coli heat-labile enterotoxin B subunit. Vaccine 17:441–447
Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20:1195–1197
Cardenas L, Clements JD (1993) Development of mucosal protection against the heat-stable enterotoxin (ST) of Escherichia coli by oral immunization with a genetic fusion delivered by a bacterial vector. Infect Immun 61:4629–4636
Clements JD, Finkelstein RA (1979) Isolation and characterization of homogeneous heat-labile enterotoxins with high specific activity from Escherichia coli cultures. Infect Immun 24:760–769
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dalsgaard K, Uttenthal A, Jones TD, Xu F, Merryweather A, Hamilton WD, Langeveld JP, Boshuizen RS, Kamstrup S, Lomonossoff GP, Porta C, Vela C, Casal JI, Meloen RH, Rodgers PB (1997) Plant-derived vaccine protects target animals against a viral disease. Nat Biotechnol 15:248–252
Dunbar BS, Prasad SV (1997) Contraceptive vaccine comprising a glycosylated 55KD zona pellucida protein immunogen and method of use of the same in contraception. Off Gaz US Patent Trademark Office Patents 1199:1077
Fitchen J, Beachy RN, Hein MB (1995) Plant virus expressing hybrid coat protein with added murine epitope elicits autoantibody response. Vaccine 13:1051–1057
Frary A, Earle ED (1996) An examination of factors affecting the efficiency of Agrobacterium-mediated transformation of tomato. Plant Cell Rep 16:235–240
Haq TA, Mason HS, Clements JD, Arntzen CJ (1995) Oral immunization with a recombinant bacterial antigen produced in transgenic plants. Science 268:714–716
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti Plasmid. Nature 303:179–180
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218
Mason HS, Haq TA, Clements JD, Arntzen CJ (1998) Edible vaccine protects mice against Escherichia coli heat-labile enterotoxin (LT): potatoes expressing a synthetic LT-B gene. Vaccine 16:1336–1343
Millar SE, Chamow SM, Baur AW, Oliver C, Robey F, Dean J (1989) Vaccination with a synthetic zona pellucida peptide produces long-term contraception in female mice. Science 246:935–938
Modelska A, Dietzschold B, Sleysh N, Fu ZF, Steplewski K, Hooper DC, Koprowski H, Yusibov V (1998) Immunization against rabies with a plant-derived antigen. Proc Natl Acad Sci USA 95:2481–2485
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Planta 15:473–497
O'Dowd AM, Botting CH, Precious B, Shawcross R, Randall RE (1999) Novel modifications to the C-terminus of LTB that facilitate site-directed chemical coupling of antigens and the development of LTB as a carrier for mucosal vaccines. Vaccine 17:1442–1453
Pillai D, Dixit A, Alok D, Garg LC (1996) Translational fusion of heat labile enterotoxin chain B and beta-subunit of human chorionic gonadotropin: periplasmic expression in Escherichia coli and its immunogenicity. FEBS Lett 387:23–26
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Schodel F, Will H, Johansson S, Sanchez J, Holmgren J (1991) Synthesis in Vibrio cholerae and secretion of hepatitis B virus antigens fused to Escherichia coli heat-labile enterotoxin subunit B. Gene 99:255–259
Sewani CR, Bagdasarian MM, Ireland JJ, Bagdasarian M (1998) Display of an inhibin epitope in a surface-exposed loop of the E. coli heat-labile enterotoxin B subunit. Vaccine 16:1611–1619
Sixma TK, Kalk KH, van Zanten BAM, Dauter Z, Kingma J, Witholt B, Hol WGJ (1993) Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. J Mol Biol 230:890–918
Skinner SM, Killian GJ, Miller LA, Dunbar BS (1994) Characterization of antigenicity and immunogenicity patterns of native and recombinant zona pellucida proteins in the white-tailed deer (Oidocoileus virginianus). J Reprod Fertil 101:295–303
Tacket CO, Mason HS, Losonsky G, Clements JD, Levine MM, Arntzen CJ (1998) Immunogenicity in humans of a recombinant bacterial antigen delivered in a transgenic potato. Nat Med 4:607–609
Tacket CO, Mason HS, Losonsky G, Estes MK, Levine MM, Arntzen CJ (2000) Human immune responses to a novel norwalk virus vaccine delivered in transgenic potatoes. J Infect Dis 182:302–305
Tsuji T, Watanabe K, Miyama A (1995) Monomer of the B subunit of heat-labile enterotoxin from enterotoxigenic Escherichia coli has little ability to bind to GM1 ganglioside compared to its coligenoid. Microbiol Immunol 39: 817–819.
Tung KSK, Ang J, Lou Y (1996) ZP3 peptide vaccine that induces antibody and reversible infertility without autoimmune oophoritis. Am J Reprod Immunol 35:181–183
Wada K, Aota S, Tsuchiya R, Ishibashi F, Gojobori T, Ikemura T (1990) Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res 18:2367–2411
Walmsley AM, Arntzen CJ (2000) Plants for delivery of edible vaccines. Curr Opin Biotechnol 11:126–129
Wigdorovitz A, Perez Filgueira DM, Robertson N, Carrillo C, Sadir AM, Morris TJ, Borca MV (1999) Protection of mice against challenge with foot and mouth disease virus (FMDV) by immunization with foliar extracts from plants infected with recombinant tobacco mosaic virus expressing the FMDV structural protein VP1. Virology 264:85–91
Acknowledgements
For valuable technical assistance, the investigators would like to thank Que Quiang, Tracey Cranston and Joan Lenz. This work was supported by the Co-operative Research Centre for Conservation and Management of Marsupials, Axis Genetics, and Dow AgroSciences LLC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.D. Earle
Rights and permissions
About this article
Cite this article
Walmsley, A.M., Alvarez, M.L., Jin, Y. et al. Expression of the B subunit of Escherichia coli heat-labile enterotoxin as a fusion protein in transgenic tomato. Plant Cell Rep 21, 1020–1026 (2003). https://doi.org/10.1007/s00299-003-0619-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-003-0619-4