Abstract
Tissue inflammation and damage with the abnormal and overactivation of innate immune system results with the development of a hereditary disease group of autoinflammatory diseases. Multiple numbers of DNA damage develop with the continuous exposure to endogenous and exogenous genotoxic effects, and these damages are repaired through the DNA damage response governed by the genes involved in the DNA repair mechanisms, and proteins of these genes. Studies showed that DNA damage might trigger the innate immune response through nuclear DNA accumulation in the cytoplasm, and through chronic DNA damage response which signals itself and/or by micronucleus. The aim of the present review is to identify the effect of mutation that occurred in DNA repair genes on development of DNA damage response and autoinflammatory diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Autoinflammatory diseases
The term autoinflammation was first suggested in 1999, and was used to describe a group of syndromes which is characterized with recurrent fever, and various systemic inflammation forms with no symptoms of typical autoimmune disease such as antigen-specific T lymphocytes or high titer autoantibodies [1]. Then, the rapidly increasing clinical studies concluded with the description of autoinflammatory disease in a separate disease group. Autoinflammatory diseases are a hereditary disease group characterized with the tissue inflammation and damage as a consequence of the abnormal and overactivation of the innate immune system [2]. Both the autoimmune and autoinflammatory diseases develop with the attack of immune system to its own tissues; however, autoinflammatory diseases are differentiated from the autoimmune disease because it mainly stems from the mutations in the genes regulating the innate immune system [3]. Recurrent fever, significant increase in acute phase proteins, urticarial and cutaneous rashes, stomachache, involvement of muscle, joint, skin, gastrointestinal system, and internal organs, serositis (peritonitis, pleuritis, and pericarditis), arthromyalgia, and lymphadenopathy are the most common symptoms of the autoinflammatory diseases [2].
Pathogenesis of the autoinflammatory diseases
In 2002, John Bertin et al. found that an adaptor protein of ASC (apoptosis-associated speck-like protein containing a CARD) binded the NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3)’ to procaspase-1 through homotypic interactions, and this complex was important for the lipopolysaccharide (LPS) stemmed caspase-1 activation, and IL-1β division [4]. Later, Jurg Tschopp completely characterized the components creating this complex, and named the complex as inflammasome [5]. Inflammasomes are a protein complex with the combination of receptor, adaptor, and effector proteins. NLRP1, NLRP2, NLRP3, NLRP6, NLRP7, and NLRC4 proteins from the NOD-like receptor family in this protein complex, and ve AIM2 and pyrin from HIN-200 protein family function as receptors, and ASC as an adaptor protein, and caspase-1 functions as an effector protein [6]. The activation of Caspase-1 results with the proteolytic division and release of proinflammatory cytokines IL-1β, and IL-18, and activates Gasdermin-d protein proteolytically. The divided N-terminal part of Gasdermin-d generates pores on the host cell membrane for mediating the release of the cytoplasmic contents, and thus triggers a hyperinflammatory cell death type of pyroptosis. Pyroptosis causes the occurrence of proinflammatory intracellular components such as the high mobility group box 1 (HMGB1) protein, resulting with the production of more inflammation signal in proximal cells [6, 7]. The biologic activities of IL-1β and IL-18, and pyroptosis are highly useful for host defence in exposure of an infection agent. However, the activation of IL-1β and IL-18 which are induced by the endogenous danger signals also stimulates the sterile inflammation which is a risk factor for the development of autoinflammatory and metabolic diseases. Therefore, inflammasome activation must be strictly controlled for preventing the open tissue damage [8]. Inflammasome activity is regulated in several steps. The activation of NF-κβ pathway initiated by the toll-like receptor (TLR) ligands is the first step for inflammasome activity. This activation stimulates the NLRP3 and IL-1β transcription, and prepares the cell to give a strong response. The anti-inflammatory signals in this stage may block the inflammasome formation by inhibiting the altogether localization of the inflammasome components [9]. In addition, other adaptor and signal molecules such as Caspase 8 may have a role in inflammasome regulation [8]. The final formation and activation of inflammasome requires various signals such as oxidative stress, nucleic acids, and potassium flow [10, 11]. The mutations in these genes encoding all these mechanisms in inflammasome formation and regulation result with the abnormal activation of the innate immune system resulting with inflammation attacks and autoimmune or autoinflammatory diseases [12] (Fig. 1). There is evidence to suggest that inflammation can be directly involved in suppressing the DNA repair mechanism. Recent data support the idea that NLRP3 and IPAF/NLRC4 inflammasome activation may be directly involved in the caspase-1 block of DNA repair. It has been shown that inflammation-mediated activation of Caspase-1 triggers Caspase-7 cleavage, which mediates proteolytic deactivation of poly (ADP-ribose) polymerase-1, a DNA damage repair enzyme [13, 14]. Poly-ADP-ribosylation mediated by PARP-1 causes chromatin decondensation at damage sites, activation of repair mechanisms and accelerates DNA damage repair. These observations suggest that by inactivating poly (ADP-ribose) polymerase-1, NLRP3 inflammasome may play a more direct role in DNA repair suppression [15].
Genetic mutations in autoinflammation pathogenesis: 1. Gain of Function mutations increase cytoplasmic pattern recognition receptors (PRRs), NOD-like receptors (NLRs) or retinoic acid-dependent gene (RIG)-like receptors (RLRs) that enable proinflammatory cytokine production. 2. Loss-of-function mutations in genes encoding enzymes or molecules that are critical in the cell’s homeostatic balance lead to the production of “cell stress molecules” that activate cytoplasmic sensors and proinflammatory cytokines. 3. Loss-of-function mutations in genes encoding negative regulators of the innate immune response prevent the termination or decrease of the innate immune response
Classification of the autoinflammatory diseases
Autoinflammatory diseases are generally investigated under two titles as monogenic and polygenic diseases. These diseases can also be classified in accordance with the pathogenesis and clinical features [16]. Monogenic autoinflammatory diseases consist of a group of rare disorders which are characterized with generally hereditary, childhood period diseases with episodic fever, skin symptoms, and disease-specific organ inflammation models [2]. Polygenic autoinflammatory diseases are characterized with inflammation no significant association with a microbial infection, not causing a specific antibody or T cell activation, and with unknown origin [16].
DNA damage, repair mechanisms, and repair genes
DNA damage may emerge associated with endogenous or exogenous reasons. Endogenous DNA damage develops with insertions, deletions, deamination, methylation, depurination, depyrimidination, replication errors, with the chemical reaction of the natural water and reactive oxygen species and the hydrolytic and oxidative reactions of active DNA [17] and exogenous DNA damage develops with the DNA damage of the environmental, physical, and chemical agents [18]. These endogenous, and exogenous factors may cause damages such as DNA single-strand breaks, double-strand breaks, deletions, insertions, formation of abasic areas, and formation of DNA–protein cross links. The cells in multiple cell organisms develop mechanisms which continuously detect the possible DNA damages, repair, and protect the natural position of the genome to maintain the body functions for longer time and protect themselves against the dangers, and also use the innate and adaptive immune system mechanisms against the bacterial and viral invasions. The close interaction of the DNA damage response (DDR) and defence strategies, and high-degree-coordinated processes are required for these mechanisms to be effective in multicellular organisms [19].
DNA damage response (DDR)
Lesion-specific sensor proteins initiate a DNA damage response when a DNA damage develops associated with endogenous and exogenous factors. All these mechanisms detecting, informing the DNA damage, and promoting the further repairs are named as the DNA damage response. The DNA damage response in the cell develops in four different ways as DNA repair, interruption of the cell cycle, transcriptional response, and programmed cell death [20]. Any defects in these response pathways may result with genomic instability, genetic disease, cancer, autoimmune, and autoinflammatory diseases [21, 22].
DNA repair mechanisms and associated genes
DNA repair genes include the genes associated with the signaling and regulation of the DNA repair and genes associated with the different repair mechanisms. The mutations developing in these genes are responsible from the development of tumors characterized with complex metabolic changes and various hereditary diseases [23]. DNA repair mechanisms can be classified in three groups as direct repair (photoreactivation repair, methyltransferase repair), excision repair (base excision repair, nucleotide excision repair, and mismatch excision repair), and strand break repair (single-strand break repair and double-strand break repair).
Direct repair
There are two direct repair mechanisms as photoreactivation, and O6-methylguanine-DNA-methyltransferase repair [24].
The UV damage causing the formation of pyrimidine dimers are corrected with the photoreactivation mechanism. Photolyase enzyme in this mechanism performs the repair using the light spectrum energy by breaking the covalent bond holding together the pyrimidine dimers. In O6-methylguanine-DNA-methyltransferase (MGMT) mechanism, the O6-methylguanine-DNA methyl transferase enzyme enables the formation of normal guanine by transferring the CH3 groups of false methylated bases on DNA to its own cysteine residues [25]. MGMT gene is located at 10q26 chromosome region, and the first mammalian DNA repair gene which was shown to be induced by genotoxic stress, and glucocorticoids [26]. Methylation of the gene promoter was associated with various cancer types [27, 28].
Excision repair
Base excision repair (BER)
Repairs the small damages such as a oxidative damage, deamination, alkylation, single base damage forms, and DNA replication errors which emerge spontaneously at DNA spiral or due to the exposure to chemicals. DNA glucosidase, apurinic/apyrimidinic (AP) endonuclease or AP DNA lyase, DNA polymerase, and DNA ligase enzymes have roles in the repair [29]. The changes in BER function were associated with Alzheimer’s disease (AD), Huntington’s disease (HD), other neurologic disorders, inflammation, and leukemia [30, 31].
Nucleotide excision repair (NER)
Repairs the damages which develop in the DNA double-strand normal helix structure such as the occurrence of pyrimidine dimers due to exposure to ultraviolet. XPA, XPB, XPC, XPD, XPE, XPF, XPG, CSA, CSB, ERCC1, RPA, RAD23A, and RAD23B proteins function in the NER repair mechanism, and their deficiencies result with severe diseases such as xeroderma pigmentosum, Cockayne syndrome, segmental progeria (XFE), and Fanconi Anemia (FA) [32].
Mismatch excision repair (MER)
It is responsible from the repair of the errors which develop with the mismatch of normal bases during DNA replication. The proteins which have two different heterodimeric complex such as MSH2–MSH3 (MutSβ), and MSH2–MSH6 (MutSα) in this repair mechanism have roles in the recognition of the mismatch. Other proteins in the repair mechanisms are the PCNA (proliferating cell nuclear antigen), exonucleases (EXO1 gibi), DNA polymerases, replication factors (RPA), and helicases [33]. Mutations in the mismatch excision repair genes are associated with genetic, neurologic, neurodegenerative, and neuromuscular diseases [34].
Single-strand break repair
The DNA single-strand breaks are detected with the poly (ADP-ribose) polymerase protein [35]. Other proteins in the repair mechanism are; XRCC1, polynucleotide kinase, AP endonuclease-1, DNA polymerase β (Pol β), tyrosyl DNA phosphodiesterase-1 (TDP1), Aprataxin, flap endonuclease1 (FEN-1), DNA ligase I (LigI) or DNA ligase IIIa (Lig3a) [36]. The defects in the repair of single-strand breaks were associated with cerebellar ataxia and neurodegeneration [37, 38].
Double-strand break repair
Double-strand breaks may develop after the collapse of the DNA replication fork associated with the factors such as ionized radiation, methyl methane sulfonate, hydroxy urea, oxidative free radicals, topoisomerase errors, and mechanic stress. Repair of these breaks is performed with two different mechanisms as repair homologous recombination (HR) and nonhomologous end joining (NHEJ) [39]. The errors in the repair of double-strand breaks may be associated with radiosensitivity, immune deficiency, developmental abnormalities, microcephalia, and predisposition to cancer [40].
Homologous recombination repair involves the RAD52 protein group (including RAD50, RAD51, RAD52 and RAD54), RPA, XRCC2, XRCC3, and BRCA proteins [41].
NHEJ repair mechanism involves the Ku 70, and Ku 80 complexes, XLF (XRCC4-like factor), APLF (APTX-and-PNK-like factor) TdT (terminal deoxynucleotidyl transferase), and DNA ligase IV-XRCC4 complex [42].
Association of DNA repair genes and immune system in autoinflammatory diseases
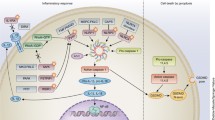
Innate immune system provides a strong defence mechanism through inflammation against infections and tissue damage [43]. Inflammation triggered with the detection of infection by innate immune system cells inactivates the pathogens generally within several hours or days, cleans the tissue which are damaged by the inflammatory process, and enables the hemostatic balance by initiating the tissue repair damage [44]. The organism must preserve the sensitive balance in the activation of the inflammatory responses for protecting the homeostasis. The deterioration of this balance in favor of the inadequate immune response results with the predisposition to infections and tumor development, and excessive immune response or immune response to self-antigens causes allergy, autoimmune or autoinflammatory diseases [44, 45]. The first stage in innate immune response is the recognition of pathogens which can pass the epithelial barrier by the immune response cells. The sensory proteins of these cells recognize the pathogens in the extracellular area and cytoplasmic spaces through pattern recognizing receptors (PRRs), and immune response is triggered with their activation [43]. When innate immune system cells recognize PRRs with one ligand, a signaling cascade is triggered involving generally an adaptor protein, a protein kinase, and phosphorylation of the transcription factors which activate the expression of the immunity genes. PRR signal pathways have negative regulators for preventing the erroneous accumulation of the host ligands which can mimic the microbic PAMPs or can access PRRs with no substrate specificity. Function loss mutations in negative regulators or function gain mutations in PRR signaling activators are associated with autoimmune and autoinflammatory diseases such as lupus, rheumatoid arthritis (RA), inflammatory vasculopathy, and various interferonopathies. In addition, innate immune response may be stimulated in the absence of infection such as the recognition of the self-nucleic acids as foreign factors [46]. Localization of DNA in cytosol which is located normally in the nucleus or mitochondria in normal cell is a sign of cellular damage or infection and results with the activation of the immune response-associated genes. Some studies showed that there were connections between the immune response and self-DNA and DDR. These studies showed that the DNA damage might trigger the innate immune response through nuclear DNA accumulation in the cytoplasm and through chronic DNA damage response which signaled activation with micronucleus and/or with itself [47, 48] (Fig. 2).
DNA damage and immune system relationship. A In programmed DNA damage events, error-prone DNA repair mechanisms such as NHEJ, an error-prone DNA repair mechanism, are effective in the development of lymphocytes and increasing antibody diversity during VDJ recombination, while BER and MMR are effective in converting cytosines in Ig genes to uracil. B Random DNA damage events trigger the activation of DDR-induced proinflammatory signals that can lead to chronic inflammation and disease
DNA detection receptors
Three main DNA detection receptors in mammalian cells initiating the immune response against the foreign DNA are AIM2, toll-like receptor9 (TLR9), and cyclic GMP-AMP synthase (cGAS).
TLR9 is localized into endosomal membrane. Recognizes the CpG hypomethylated DNA, and activates the transcription factors. Nuclear factor-B (NF-κβ), and interferon regulatory factor 7 (IRF7), respectively, cause the expression of genes which encode the proinflammatory cytokines, and interferons.
AIM2 is binded to double-stranded DNA in the cytosol, and results with the formation of the multimeric protein complex named the AIM2 inflammasome. AIM2 inflammasome activates the Caspase-1 and causes the maturation of the proinflammatory cytokines IL-1 and IL-18’, and finally to pyroptotic cell death.
cGAS-STING pathway
DNA coming from various sources can enter the cytosol and transfer to cGAS-STING pathway. Binding of cytosolic DNA to cGAS activates the cGAS-STING pathway and causes the production of the secondary messenger endogenous cyclic GMP-AMP (cGAMP). cGAMP is binded to the endoplasmic reticulum (ER) localized adaptor protein of STING (stimulator of the interferon genes) [49]. The stimuli out of the cGAMP such as ER stress, viral liposomes, and cyclic dinucleotides (CDNs) may also activate STING. STING is transferred from ER to golgi, and in this period, binds the transcription factor of interferon regulatory factor 3 (IRF3) which is phosphorylated by TBK1. The activated IRF3 is moved to the nucleus and functions as the transcriptional activator of the type I interferon (IFN) genes [50, 51].
Binding of type I IFN molecules to IFN receptors activates the JAK-STAT pathway and initiates the transcription of hundreds of IFN-stimulated genes (ISG) [52]. In addition, STING gets the kinases such as Iκβ kinase which phosphorylated the NF-κβ inhibitor of Iκβα while transferring to golgi from ER. Phosphorylation of Iκβα results with the translocation of NF-κβ to the nucleus and activates the transcription of genes which encode the proinflammatory cytokines as IL-6 and tumor necrosis factor (TNF). This pathway has a critical role in mediating the immunity defence against the double-stranded DNA viruses [53]. Similarly, RIG-I-like receptor sensors RIG-I and MDA5 bind to dsRNA, and are stimulated by signaling with the adaptor protein MAVS. The common pathway in the signaling of STING and MAVS involve the TBK1 and IRF3 phosphorylation/activation and IFN-β transcription (Fig. 3).
Regulation of the cGAS-STING pathway
cGAS has no sequence specificity for DNA. The host DNA is mainly located in the nucleus and mitochondria, and its separation from cytoplasm prevents the reach of cGAS to self-DNA; however, the resolution of nuclear membrane for sometimes during the cell division may result with the transfer of nuclear DNA to cytosol [54] and may result with the transfer of mitochondrial DNA to cytosol during deterioration in mitophagy [55]. Therefore, there has to be additional mechanisms for limiting the meeting of cGAS-DNA.
The secondary protective mechanisms preventing the binding to self-DNA is the host DNases. DNases are located in the extracellular area [DNase-I and DNase-I-like 3 (DNase IL3)], in phagolysosomal compartment (DNase-II) or cytosol [TREX1 (DNase III)], and prevents the accumulation of self-DNA as much as to bind to cGAS or other sensors [56].
TMEM173 gene and associated autoinflammatory diseases
TMEM173 gene is located on the short arm of the chromosome 5 and has autosomal dominant inheritance and encodes the STING protein important in innate immune system which functions as an adaptor protein with different molecular mechanisms in directly cytosolic DNA sensor (CDS) and type I enterferon signaling. STING-associated vasculopathy with onset in infancy (SAVI) and familial Chilblain lupus disease are the autoinflammatory diseases associated with TMEM173 gene mutation [57, 58] (Table 1).
NEIL1 gene and associated autoinflammatory diseases
NEIL1 gene is localized at 15q24.2 chromosome and encodes a DNA glycosylase endonuclease VIII-like protein 1 (NEIL1) which is important for the initiation of the base excision repair mechanism. The conducted studies detected an association between the NEIL1 gene variant and Behçet’s disease [59, 60] (Table 1).
TREX1 gene and associated autoinflammatory diseases
TREX1 gene is localized at 3p21 chromosome and consists of a single exon, and encodes the TREX1 protein consisting of 314 amino acids. TREX1 gene has a widened C-terminal ‘tail’ area with approximately 70 amino acids which includes a rich strand required for leucine for endoplasmic reticulum (ER) localization. It is a cellular 3’-5’ exonuclease which divides the DNA fragments in the cytoplasm, and consists of 3 exonuclease areas which are important for DNA repair enzyme role [61]. One of the results of its activities is to destroy the viral DNAs before they are detected by cGAS or other PRRs, thus to prevent the IFN induction, and inhibit the innate immune response. The most important function of TREX1 is to protect the innate immune response tolerance against the cytosolic self-DNA by destroying a serious of substrates for preventing the initiation of the autoimmunity [62, 63]. TREX1 mutations result with the accumulation of self-DNA in the cytosols of the cells with TREX1 deficiency, and triggers the systemic inflammation, and uncontrolled autoimmunity with the chronic activation of the cGAS-STING-mediated type I interferon response [56]. In addition, research showed that TREX1 was also associated with the SET complex [64]. SET complex is a DNA repair complex targeted by Gransim A during Caspase-independent, T cell-mediated death. TREX1 is attached to SET complex, moved to nucleus, and deteriorates the 3’ end of DNA during Gransim A-mediated cell. Therefore, cells with TREX1 deficiency are relatively more resistant to apoptosis [64].
Takuya Miyakazi et al. showed in their study that TREX1 interacted with a nuclear enzyme of poly (ADP-ribose) polymerase-1 (PARP-1) which have role in DNA damage response. Although PARP-1 is detected in many dimensions of cellular response against DNA damage, it is required for the repair of ssDNA breaks mainly by BER mechanisms [65]. TREX1 gene mutations were found associated with familial Chilblain lupus, and have Aicardi–Goutières syndrome (AGS) diseases.
RNaseH2A, RNaseH2B, and RNaseH2C genes and associated autoinflammatory diseases
DNA–RNA hybrids may develop during many cellular processes such as DNA replication, and transcription. The presence of ribonucleotides in genomic DNA is an undesired condition which makes the DNA more sensitive to strand break. RNase H enzymes hydrolyze the RNA strand of RNA/DNA hybrids. Eukaryotes have two types of RNase H with different biochemical features, and substrate specificity RNase1 and Rnase2. The source of the dominant RNase H activity in mammalian cells is RNase H2. In Eukaryotes, RNase H2 is a multimeric complex which consist of three subtypes as RNaseH2A, RNaseH2B, and RNASEH2C. In this complex, RNASEH2A/B/C are the nucleases which target the DNA–RNA hybrids. RNaseH2A subunit includes a catalytic center, the intertwined helper RNaseH2B, and C subunits are possibly interacting with the other proteins. One PIP box motif in the C-terminal of the RNaseH2B subunit C directs the RNase and PCNA interaction, and localization to replication focuses in DNA replication and/or repair [66]. The mutations in the genes encoding the RNaseH2A, B, and C enzymes were associated with the AGS2, AGS3, and AGS4 diseases [67].
SAMHD1 gene and associated autoinflammatory diseases
Human SAMHD1 protein is a dNTP triphosphohydrolase which hydrolyzes the dNTPs to deoxyribonucleotide (dN)s and triphosphates. SAMHD1 protein has been divided into three areas as N-T SAM area, middle HD area, and C-T phosphorylation/regulatory area. The function of SAM area has not completely been clarified. HD area involves the dNTPase active region consisting of histidine and aspartic acid residues. SAMHD1 keeps the dNTP pools in the cell in appropriate levels for DNA replication and repair, however, potentially below the mutation threshold through dNTPase activity [68]. Mutations detected on SAMHD1 gene were associated with an autoinflammatory disease of Aicardi–Goutières syndrome (AGS). Most proteins of SAMHD1 with this mutation demonstrate various defects in tetramerization, dNTPase activity loss, and nuclear localization [69]. Lupus-like chronic brain inflammation characterized with hyperactivation and irregularity of type 1 interferon responses, and early start of severe neurodevelopmental disorders are detected in AGS patients with mutated SAMHD1 genes [70]. In addition, function loss mutations in SAMHD1 gene were also found associated with another autoinflammatory disease of familial chilblain lupus [71].
ADAR1 gene and associated autoinflammatory diseases
The cytosolic PRRs such as MDA5 and RIG-I detect the double-strand RNA (dsRNA) which is commonly produced during many viral infection, and results with the type I IFN production, and induce the hundreds of IFN-stimulated gene with antiviral activity [72]. ADAR1 gene encodes the enzyme responsible from the RNA regulation through the region-specific deamination of adenosines. The deamination activity of this enzyme converts adenosine to iosine in the double-strand regions of RNA molecules, and thus damages the dsRNA stability. Therefore, ADAR1 has a critical role in the regulation of the innate immune system activation through suppressing the endogenous RNA perception [73]. The mutations in ADAR1 gene results with an autoinflammatory disease of AGS associated with the spontaneous IFN production in the absence of virus infection [74].
IFIH1 gene and associated autoinflammatory diseases
IFIH1 encodes MDA5 which is a cytosolic double-strand RNA receptor and important in the innate immune response against pathogens. The function gain mutations in helicase area of IFIH1 cause the increase of the type I interferon levels and results with the occurrence of the systemic symptoms of an autoinflammatory disorder of AGS [75].
General characteristics of autoinflammatory diseases that are suggested to be related to DNA repair genes are summarized below.
-
STING-associated vasculopathy with onset in infancy (SAVI): it is a rare autoinflammatory disease characterized by severe skin lesions and interstitial lung disease. Gain-of-function mutations in TMEM173 gene which encodes a protein called STING is with associated. STING activation stimulates the induction of type I interferons that activate interferon responses. Gain-of-function in STING leads to autoactivation without ligand binding [58].
-
Familial chilblain lupus disease: it is an autoinflammatory disease of childhood onset, characterized by typical skin manifestations and acral ischemia. In these patients, a gain-of-function mutation in the TMEM173 gene causes structural type I IFN activation [76]. In addition, loss of function mutations in the TREX1 or SAMHD1 gene also cause the activation of antiviral type I interferon (IFN) induced by the recognition of non-metabolized nucleic acids by the immune system [62].
-
Behcet’s disease: it is a chronic, recurrent multisystem disease of unknown etiology characterized by recurrent oral and genital ulcers, skin lesions and uveitis. It has been associated with many genes, particularly those related to immune system regulation and inflammation. The inflammation that occurs during Behçet’s disease exposes cells to abnormal ROS levels, and the DNA damage repair mechanism is necessary to manage the effects of this oxidative stress. NEIL1 gene variant (rs5745908) is thought to provide information about the role of oxidative stress and increased leukocyte tissue infiltration in patients with Behçet’s disease [60].
-
Aicardi–Goutières syndrome: it is a rare disease characterized by bilateral basal ganglia calcification, cerebralatrophy, lymphocytosis in the cerebrospinal fluid and increased interferon levels (IFN-α), which occurs as a result of defects in the DNA repair mechanism. AGS may be caused by seven different genes (AGS1-7), TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAHMD1, IFIH1, and ADAR (DRADA), involved in interferon type 1 metabolism [77]. Loss-of-function mutations in the TREX1 gene cause accumulation of ssDNA derived from an endogenous retroelement, leading to STING-dependent type I IFN transcription (AGS1). Similarly, loss of function mutations in RNASEH2B, RNASEH2C, RNASEH2A, SAMHD1, and ADAR1 result in type I IFN transcription through still unknown signaling processes (AGS2-6). MDA5, encoded by IFIH1, is one of the dsRNA sensors. Gain-of-function mutation in MDA5 causes constitutive or enhanced IFN-β transcription. Although the disease generally shows autosomal recessive inheritance, there are also cases in which specific heterozygous loss of function mutations are observed in TREX1 and ADAR1. In addition, it has been determined that all AGS-related mutations in IFIH1 show autosomal dominant inheritance [16].
Conclusions and recommendations
Many gene mutations triggering the IFN-α expression and pathogenesis have been detected in highly heterogenous autoinflammatory disorders, and some of these genes were also associated with the DNA repair [78]. Cells are continuously exposed to endogenous metabolic byproducts or environmental genotoxic effects and multiple number of DNA lesions develop. These DNA damages are repaired by the DNA repair mechanisms. The excision of short ssDNA byproducts develop as a consequence of the activities of some DNA repair mechanisms destroying the DNA lesions [79]. RPA and Rad51 are ssDNA-binding proteins highly expressed in the nucleus which have significant roles in DNA replication, recombination, and repair pathways [80, 81]. Short ssDNA was reported to pass the nuclear membrane, however, was withdrawn to nucleus by binding to DNA replication and repair factors of RPA and Rad51 in a study by Christine Wolf and his colleagues. In addition, the study showed that the function disorder occurring on RPA and Rad51 increased the cytosolic leakage of ssDNA, and resulted with the cGAS-dependent type I IFN activation [82]. Researchers reported that TREX1 was fixed to outer nuclear membrane for enabling the immediate degradation of ssDNA leaking to cytosol, the accumulated ssDNA in fibroblasts with TREX deficiency resulted with the RPA and Rad51 consumption and cause the replication stress and p52 and type 1 IFN activation [82].
Morita and his colleagues reported in their study that inflammatory myocarditis developed in TREX1 null mice owing to the interferon-dependent autoimmune response, and the first association possibility of immunity activation and TREX1 was revealed [83] TREX1 mutations were detected in AGS patients in the prospective studies on human [84], and recent studies revealed that the function loss mutations on TREX1 were found associated with various autoimmune and inflammatory diseases as familial chilblain lupus, systemic lupus erythematosus, and cerebral leukodystrophy, and retinal vasculopathy in addition to AGS [85]. Mutations on TMEM173 gene which is an encoder of the STING protein stimulators were found associated with autoinflammatory diseases such as STING-associated vasculopathy (SAVI) and familial chilblain lupus [57, 58]. Both hereditary and de novo TMEM173 mutations were detected in SAVI patients in the conducted studies. STING-associated vasculopathy in patients with de novo TMEM173 mutations showed an early onset (< 8 weeks) and severe phenotype [57, 86]. However, the patients with familial TMEM173 mutations were shown to have late-onset (teenage or adult) disease and more mild clinical symptoms [76, 87].
Yan et al. showed that SAVI-associated STING has some missense mutations (V147L, N154S, and V155M) [56]. In addition, Dobbs et al. showed that N154S and V155M mutants structurally activated the STING-mediated IFN response independent of cGAMP binding [51].
NEIL1 gen encodes a DNA glucosidase which has a role in the initiation of BER pathway. Mikhail Ognenovsk et al. reported in their study that the association of NEIL1 gene variant (rs5745908) and Behçet’s disease was detected. This variant was suggested to provide data on the increase of leukocyte tissue infiltration and the role of oxidative stress in Behçet’s disease patients [60]. Recent studies showed that PNPT1 deficiency caused the accumulation of double-strand mtRNA on cytoplasm, and resulted with the abnormal type I interferon activation [88, 89].
Studies up to present associated the AGS disease with TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1 genes [90]. Daniel Bamborschke et al. reported in their current study that they have identified homozygous PNPT1 mutations (c.1399C > T, p.Pro467Ser) in a child with AGS phenotype. Researchers showed that PNPT1 deficiency due to these mutations resulted with type I interferonopathy, and suggested that PNPT1 mutations might be demonstrated among the reasons of AGS disease [91].
SAMHD1 holds the dNTP pools in the cell in appropriate levels for DNA replication and repair through dNTPase activity, however, below the potentially mutagenic threshold. Daddacha et al. showed that SAMHD1 was localized to double-strand break regions and supported the homologous recombination owing to its complex forming ability with endonuclease CtBP-interacted protein (CtIP) [92]. In addition, Beloglazova et al. reported that SAMHD1 demonstrated exonuclease activity on ssRNA and ssDNA [93]. Franzolin and colleagues in their current study reported that SAMHD1 gene had a genome stability protective effect similar to genes in DNA repair mechanisms [94]. Most proteins of the mutated SAMHD1 gene demonstrate various defects in tetramerization, dNTPase activity loss, and nuclear localization. The function gain mutations in helicase area of IFIH1 cause the increase of the levels of the type I interferon levels. Amari et al. indicated that they have found an IFIH1 gene variant which caused AGS7 in their study, however, prospective studies were required for revealing the function of this mutation [95]. Although AGS7 is accepted as a relatively mild subtype compared with other Aicardi–Goutières syndromes, the c.2439A > T variant of AGS7 was shown to be fetal in early infancy in the studies of Amari et al. [95].
In general, the irregulatory of innate immune system causes “autoinflammatory” diseases. The mutations in the genes encoding the proteins which regulate DNA damage response, innate immune response receptors, and in DNA repair mechanisms were found associated with the autoinflammatory diseases such as AGS, SAVI, familial chilblain lupus, RVCL, and Behçet’s disease in the studies conducted so far. In addition, a defect repairing the oxidative DNA damage in DNA repair mechanisms in familial Mediterranean fever (FMF) patients was suggested to cause the accumulation of the DNA lesions, however, whether the pyrin encoded by MEFV gene was included in the DNA repair has not been clarified yet. Further studies are required for clarifying the possible role of pyrin and the association of DNA gene variants with autoinflammatory diseases.
Search strategy
We conducted a comprehensive review of the literature for English articles published between 1996 and 2020, using PubMed, Scopus, Springer Link, Science Direct and Web of Science as databases. We seek for articles and abstracts published with the following keywords: “Autoinflammatory diseases”, “DNA repair genes”, “DNA damage”, “DNA damage response,” ‘’DNA repair mechanisms’ and “Immune system”. We screened and reviewed titles and abstracts to identify studies examining DNA damage and DNA repair genes in autoinflammatory diseases. We also manually searched the references of the selected articles for any relevant reference that we might have missed. All these articles were subsequently filtered by the two co-authors (DK and SD) to select only those that met the objectives of this review, resulting in 95 articles.
References
McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, O’Shea JJ, Kastner DL (1999) Germ line mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 97:133–144. https://doi.org/10.1016/s0092-8674(00)80721-7
Zen M, Gatto M, Domeneghetti M, Palma L, Borella E, Iaccarino L, Punzi L, Doria A (2013) Clinical guidelines and definitions of autoinflammatory diseases: contrasts and comparisons with autoimmunity—a comprehensive review. Clin Rev Allergy Immunol 45(2):227–235. https://doi.org/10.1007/s12016-013-8355-1
Brydges S, Kastner DL (2006) The systemic autoinflammatory diseases: inborn errors of the innate immune system. Curr Top Microbiol Immunol 305:127–160. https://doi.org/10.1007/3-540-29714-6_7
Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J (2002) PYPAF7, a novel PYRIN containing Apaf1-like protein that regulates activation of NF-κB and caspase-1-dependent cytokine processing. J Biol Chem 277(33):29874–29880. https://doi.org/10.1074/jbc.M203915200
Martinon F, Burns K, Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 2:417–426. https://doi.org/10.1016/s1097-2765(02)00599-3
Sharma D, Kanneganti T-D (2006) The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J Cell Biol 213(6):617–629. https://doi.org/10.1083/jcb.201602089
Man SM, Karki R, Kanneganti TD (2017) Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev 277(1):61–75. https://doi.org/10.1111/imr.12534
Man SM, Kanneganti TD (2015) Regulation of inflammasome activation. Immunol Rev 265(1):6–21. https://doi.org/10.1111/imr.12296
Hwang I, Yang J, Hong S, Lee JE, Lee SH, Alnemri TF, Alnemri ES, WookYu J (2015) Non-transcriptional regulation of NLRP3 inflammasome signaling by IL-4. Immunol Cell Biol 93(6):591–599. https://doi.org/10.1038/icb.2014.125
Kesavardhana S, Kanneganti TD (2017) Mechanisms governing inflammasome activation, assembly and pyroptosis induction. Int Immunol 29(5):201–210. https://doi.org/10.1093/intimm/dxx018
Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G (2013) K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38(6):1142–1153. https://doi.org/10.1016/j.immuni.2013.05.016
Georgin-Lavialle S, Fayand A, Rodrigues F, Bachmeyer C, Savey L, Grateau G (2019) Autoinflammatory diseases: state of the art. Presse Med 48(1):e25–e48. https://doi.org/10.1016/j.lpm.2018.12.003 (Pt 2)
Malireddi RK, Ippagunta S, Lamkanfi M, Kanneganti TD (2010) Cutting edge: proteolytic inactivation of poly (ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J Immunol 185:3127–3130
Erener S, Petrilli V, Kassner I, Minotti R, Castillo R, Santoro R, HassaTschopp POJ, Hottigeret MO (2012) Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-kappa B target genes. Mol Cell 46:200–211
Licandro G, Khor HL, Beretta O, Lai J, Derks H, Laudisi F, Conforti-Andreoni C, Qian HL, Teng GG, Ricciardi-Castagnoli P, Mortellaro A (2013) The NLRP3 inflammasome affects DNA damage responses after oxidative and genotoxic stress in dendritic cells. Eur J Immunol 43(8):2126–2137. https://doi.org/10.1002/eji.201242918
de Jesus AA, Scott W, Yin LC, Goldbach-Mansky R (2015) Molecular mechanisms in genetically defined autoinflammatory diseases: disorders of amplified danger signaling. Annu Rev Immunol 33:823–874. https://doi.org/10.1146/annurev-immunol-032414-112227
Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB (2010) Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 49(11):1603–1616. https://doi.org/10.1016/j.freeradbiomed.2010.09.006
Cadet J, Wagner JR (2013) DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol 5(2):a012559. https://doi.org/10.1101/cshperspect.a012559
Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K, Kotsinas A, Orsolic I, Bursac S, Cokaric-Brdovcak M, Zoumpourlis V, Kletsas D, Papafotiou G, Klinakis A, Volarevic S, Gu W, Bartek J, Halazonetis TD, Gorgoulis VG (2013) Functional inter play between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol 15(8):967–977. https://doi.org/10.1038/ncb2795
Mari GG, Zotter A, Vermeulen W (2011) DNA damage response. Cold Spring Harb Perspect Biol 3(1):a000745. https://doi.org/10.1101/cshperspect.a000745
Jeggo PA, Pearl LH, Carr AM (2016) DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 16(1):35–42. https://doi.org/10.1038/nrc.2015.4
De Cauwer A, Mariotte A, Sibilia J, Bahram S, Georgel P (2018) DICER1: a key player in rheumatoid arthritis, at the crossroads of cellular stress, ınnate ımmunity, and chronic ınflammation in aging. Front Immunol 9:1647. https://doi.org/10.3389/fimmu.2018.01647
Christmann M, Tomicic MT, Roos WP, Kaina B (2013) Mechanisms of human DNA repair: an update. Toxicology 193(1–2):3–34. https://doi.org/10.1016/s0300-483x(03)00287-7
Sancar A, Lindsey-Boltz LA, Kaçmaz KU, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. https://doi.org/10.1146/annurev.biochem.73.011303.073723
Jin B, Robertson KD (2013) DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol 754:3–29. https://doi.org/10.1007/978-1-4419-9967-2_1
Grombacher T, Mitra S, Kaina B (1996) Induction of the alkyltransferase (MGMT) gene by DNA damaging agents and the glucocorticoid dexamethasone and comparison with the response of base excision repair genes. Carcinogenesis 17(11):2329–2336. https://doi.org/10.1093/carcin/17.11.2329
Wani HA, Majid S, Bhat AA, Amin S, Farooq R, Bhat SA, Naikoo NA, Beigh MA, Kadla SA (2019) Impact of catechol-O-methyltransferase gene variants on methylation status of P16 and MGMT genes and their downregulation in colorectal cancer. Eur J Cancer Prev 28(2):68–75. https://doi.org/10.1097/CEJ.0000000000000485
Martínez-Ramírez OC, Pérez-Morales R, Castro-Hernández C, Gonsebatt ME, Casas-Ávila L, Valdés-Flores M, Petrosyan P, de León-Suárez VP, Rubio J (2019) Association of the promoter methylation and the rs12917 polymorphism of MGMT with formation of DNA bulky adducts and the risk of lung cancer in Mexican Mestizo population. DNA Cell Biol 38(4):307–313. https://doi.org/10.1089/dna.2018.4526
Robertson AB, Klungland A, Rognes T, Leiros I (2009) DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci 66(6):981–993. https://doi.org/10.1007/s00018-009-8736-z
Sliwinska A, Kwiatkowski D, Czarny P, Toma M, Wigner P, Drzewoski J, Fabianowska-Majewska K, Szemraj J, Maes M, Galecki P, Sliwinski T (2016) The levels of 7,8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1)—a potential diagnostic biomarkers of Alzheimer’s disease. J Neurol Sci 368:155–159. https://doi.org/10.1016/j.jns.2016.07.008
Ba X, Aguilera-Aguirre L, Rashid QT, Bacsi A, Radak Z, Sur S, Hosoki K, Hegde ML, Boldogh I (2014) The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int J Mol Sci 15(9):16975–16997. https://doi.org/10.3390/ijms150916975
Marín M, Ramírez MJ, Carmona MA, Jia N, Ogi T, Bogliolo M, Surrales J (2019) Functional comparison of XPF missense mutations associated to multiple DNA repair disorders. Genes (Basel) 10(1):60. https://doi.org/10.3390/genes10010060
Li Z, Pearlman AH, Hsieh P (2016) DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38:94–101. https://doi.org/10.1016/j.dnarep.2015.11.019
Schmidt MHM, Pearson CE (2016) Disease-associated repeat instability and mismatch repair. DNA Repair (Amst) 38:117–126. https://doi.org/10.1016/j.dnarep.2015.11.008
D’Amours D, Desnoyers S, D’Silva I, Poirier GG (1999) Poly (ADP-ribosyl) ation reactions in the regulation of nuclear functions. Biochem J 342:249–268 (Pt 2 PMC1220459)
Caldecott KW (2003) XRCC1 and DNA strand break repair. DNA Repair (Amst) 2(9):955–969. https://doi.org/10.1016/s1568-7864(03)00118-6
Yoon G, Caldecott KW (2018) Nonsyndromic cerebellar ataxias associated with disorders of DNA single-strand break repair. Handb Clin Neurol 155:105–115. https://doi.org/10.1016/B978-0-444-64189-2.00007-X
Madabhushi R, Pan L, Tsai LH (2014) DNA damage and its links to neurodegeneration. Neuron 83(2):266–282. https://doi.org/10.1016/j.neuron.2014.06.034
Kowalczykowski SC (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol 7(11):a016410. https://doi.org/10.1101/cshperspect.a016410
McKinnon PJ, Caldecott KW (2007) DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet 8:37–55. https://doi.org/10.1146/annurev.genom.7.080505.115648
West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4(6):435–445. https://doi.org/10.1038/nrm1127
Chang HHY, Pannunzio NR, Adachi N, Lieber MR (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18(8):495–506. https://doi.org/10.1038/nrm.2017.48
Medzhitov R (2008) Origin and physiological roles of inflammation. Nature 454(7203):428–435. https://doi.org/10.1038/nature07201
Newton K, Dixit VM (2012) Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4(3):a006049. https://doi.org/10.1101/cshperspect.a006049
Cao X (2016) Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol 16(1):35–50. https://doi.org/10.1038/nri.2015.8
Roers A, Hiller B, Hornung V (2016) Recognition of endogenous nucleic acids by the ınnate ımmune system. Immunity 44(4):739–754. https://doi.org/10.1016/j.immuni.2016.04.002
Gasser S, Raulet DH (2006) The DNA damage response arouses the immune system. Cancer Res 66(8):3959–3962. https://doi.org/10.1158/0008-5472.CAN-05-4603
Li T, Chen ZJ (2018) The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215(5):1287–1299
Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339(6121):826–830. https://doi.org/10.1126/science.1229963
Barber GN (2011) STING-dependent signaling. Nat Immunol 12(10):929–930. https://doi.org/10.1038/ni.2118
Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N (2015) STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18(2):157–168. https://doi.org/10.1016/j.chom.2015.07.001
Oda H, Kastner DL (2017) Genomics, biology, and human illness: advances in the monogenic autoinflammatory diseases. Rheum Dis Clin North Am 43(3):327–345. https://doi.org/10.1016/j.rdc.2017.04.011
Gao D, Wu J, Wu Y-T, Du F, Aroh C, Yan N, Sun L, Chen ZJ (2013) Cyclic GMP–AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341(6148):903–906. https://doi.org/10.1126/science.1240933
Motwani M, Fitzgerald KA (2017) cGAS micro-manages genotoxic stress. Immunity 47(4):616–617. https://doi.org/10.1016/j.immuni.2017.09.020
Rongvaux A (2018) Innate immunity and tolerance toward mitochondria. Mitochondrion 41:14–20. https://doi.org/10.1016/j.mito.2017.10.007
Yan N (2017) Immune diseases associated with TREX1 and STING dysfunction. J Interferon Cytokine Res 37(5):198–206. https://doi.org/10.1089/jir.2016.0086
Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GAM, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CCR, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R (2014) Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371(6):507–518. https://doi.org/10.1056/NEJMoa1312625
Crow YJ, Casanova J-L (2014) STING-associated vasculopathy with onset in infancy—a new interferonopathy. N Engl J Med 371(6):568–571. https://doi.org/10.1056/NEJMe1407246
Edmonds MJ, Carter RJ, Nickson CM, Williams SC, Parsons JL (2017) Ubiquitylation-dependent regulation of NEIL1 by mule and TRIM26 is required for the cellular DNA damage response. Nucleic Acids Res 45(2):726–738. https://doi.org/10.1093/nar/gkw959
Ognenovski M, Renauer P, Gensterblum E, Kötter I, Xenitidis T, Henes JC, Casali B, Salvarani C, Direskeneli H, Kaufman KM, Sawalha AH (2016) Whole exome sequencing identifies rare protein-coding variants in Behçet’s disease. Arthritis Rheumatol 68(5):1272–1280. https://doi.org/10.1002/art.39545
deSilva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T (2007) The crystal structure of TREX1 explains the 3’ nucleotide specificity and reveals a polyproline II helixfor protein partnering. J Biol Chem 282(14):10537–10543. https://doi.org/10.1074/jbc.M700039200
Yang YG, Lindahl T, Barnes DE (2007) Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131(5):873–886. https://doi.org/10.1016/j.cell.2007.10.017
Stetson DB, Ko JS, Heidmann T, Medzhitov R (2008) Trex1 prevents cell-intrinsicinitiation of autoimmunity. Cell 134(4):587–598. https://doi.org/10.1016/j.cell.2008.06.032
Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J (2006) The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell 23(1):133–142. https://doi.org/10.1016/j.molcel.2006.06.005
Miyazaki T, Kim YS, Yoon J, Wang H, Suzuki T, Morse HC (2014) The 3’-5’ DNA exonuclease TREX1 directly ınteracts with poly (ADP-ribose) polymerase-1 (PARP1) during the DNA damage response. J Biol Chem 289(47):32548–32558. https://doi.org/10.1074/jbc.M114.547331
Hyjek M, Figiel M, Nowotny M (2019) RNases H: structure and mechanism. DNA Repair (Amst) 84:102672. https://doi.org/10.1016/j.dnarep.2019.102672
Livingston JH, Crow YJ (2016) Neurologic phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1: Aicardi–Goutières syndrome and beyond. Neuropediatrics 47(6):355–360. https://doi.org/10.1055/s-0036-1592307
Maehigashi T, Kim DH, Schinazi RF, Kim B (2018) SAMHD1-mediated negative regulation of cellular dNTP levels: HIV-1, ınnate ımmunity, and cancers. In: Fernandez-Lucas J (ed) Enzymatic and chemical synthesis of nucleic acid derivatives, chapter 12, pp 313–325
White TE, Brandariz-Nunez A, Martinez-Lopez A, Knowlton C, Lenzi G, Kim B, Ivanov D, Diaz-Griffero F (2017) A SAMHD1 mutation associated with Aicardi–Goutieres syndrome uncouples the ability of SAMHD1 to restrict HIV-1 from its ability to downmodulate type I interferon in humans. Hum Mutat 38(6):658–668. https://doi.org/10.1002/humu.23201
Crow YJ (2015) Type I interferonopathies: Mendelian type I interferon up-regulation. Curr Opin Immunol 32:7–12. https://doi.org/10.1016/j.coi.2014.10.005
Ravenscroft JC, Suri M, Rice GI, Szynkiewicz M, Crow YJ (2011) Autosomal dominant inheritance of a heterozygous mutation in SAMHD1 causing familial chilblain lupus. Am J Med Genet A 155A(1):235–237. https://doi.org/10.1002/ajmg.a.33778
Schneider WM, Chevillotte MD, Rice CM (2014) Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. https://doi.org/10.1146/annurev-immunol-032713-120231
Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349(6252):1115–1120
Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JPSM, Lourenco CM, MaleJrMignot AMWMC, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O’Connell MA, Lovell SC, Crow YJ (2012) Mutations in ADAR1 cause Aicardi–Goutières syndrome associated with a type I interferon signature. Nat Genet 44(11):1243–1248. https://doi.org/10.1038/ng.2414
Rice GI, Del Toro DY, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Jv Cordeiro N, Dale RC, Davidson JE, De Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenço CM, Mancardi MM, Masurel-Paulet A, McInnes IB, Menezes MP, Mignot C, O’Sullivan J, Orcesi S, Picco SP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ (2014) Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46(5):503–509. https://doi.org/10.1038/ng.2933
König N, Fiehn C, Wolf C, Schuster M, Costa EC, Tüngler V, Alvarez HA, Chara O, Engel K, Goldbach-Mansky R, Günther C, Lee-Kirsch MA (2017) Familial chilblain lupus due to a gain-of-function mutation in STING. Ann Rheum Dis 76(2):468–472. https://doi.org/10.1136/annrheumdis-2016-209841
Lanzi G, Fazzi E, D’Arrigo S, Orcesi S, Maraucci I, Uggetti C, Bertini E, Lebon P (2005) Thenaturalhistory of Aicardi–Goutie`ressyndrome: follow-up of 11 Italian patients. Neurology 64(9):1621–1624 (Goutie`res F. Aicardi-Goutie`ressyndrome. Brain Dev 27(3):201–06)
Davidson S, Steiner A, Harapas CR, Masters SL (2018) An update on autoinflammatory diseases: interferonopathies. Curr Rheumatol Reports 20(7):38. https://doi.org/10.1007/s11926-018-0748-y
Guo J, Hanawalt PC, Spivak G (2013) Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoguanine in human cells. Nucleic Acids Res 41(16):7700–7712. https://doi.org/10.1093/nar/gkt524
Bochkarev A, Bochkareva E (2004) From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol 14(1):36–42. https://doi.org/10.1016/j.sbi.2004.01.001
Mine J, Disseau L, Takahashi M, Cappello G, Dutreix M, Louis-Viovy J (2007) Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res 35(21):7171–7187. https://doi.org/10.1093/nar/gkm752
Wolf C, Rapp A, Berndt N, Staroske W, Schuster M, Dobrick-Mattheuer M, Kretschmer S, König N, Kurth T, Wieczorek D, Kast K, Cardoso MC, Günther C, Lee-Kirsch MA (2016) RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat Commun 2016(7):11752. https://doi.org/10.1038/ncomms11752
Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE (2004) Gene-targeted mice lacking the Trex1 (DNase III) 3’–>5’ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 24(15):6719–6727. https://doi.org/10.1128/MCB.24.15.6719-6727.2004
Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T (2006) Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 cause Aicardi–Goutières syndrome at the AGS1 locus. Nat Genet 38(8):917–920. https://doi.org/10.1038/ng1845
Rice GI, Rodero MP, Crow YJ (2015) Human disease phenotypes associated with mutations in TREX1. J Clin Immunol 35(3):235–243. https://doi.org/10.1007/s10875-015-0147-3
Picard C, Thouvenin G, Kannengiesser C, Dubus JC, Jeremiah N, Rieux-Laucat F, Crestani B, Belot A, Thivolet-Béjui F, Secq V, Ménard C, Reynaud-Gaubert M, Reix P (2016) Severe pulmonary fibrosis as the first manifestation of interferonopathy (TMEM173 mutation). Chest 150(3):e65-71. https://doi.org/10.1016/j.chest.2016.02.682
Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC, Goudin N, Frémond ML, Nitschke P, Molina TJ, Blanche S, Picard C, Rice GI, Crow YJ, Manel N, Fischer A, Bader-Meunier B, Rieux-Laucat F (2014) Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest 124(12):5516–5520. https://doi.org/10.1172/JCI79100
Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ (2018) Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560(7717):238–242. https://doi.org/10.1038/s41586-018-0363-0
Pajak A, Laine I, Clemente P, El-Fissi N, Schober FA, Maffezzini C, Calvo-Garrido J, Wibom R, Filograna R, Dhir A, Wedell A, Freyer C, Wredenberg A (2019) Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo. PLoS Genet 15(7):e1008240. https://doi.org/10.1371/journal.pgen.1008240
Harapas CR, Steiner A, Davidson S, Masters SL (2018) An update on autoinflammatory diseases: ınflammasomopathies. Curr Rheumatol Reports 20(7):40. https://doi.org/10.1007/s11926-018-0750-4
Bamborschke D, Kreutzer M, Koy A, Koerber F, Lucas N, Huenseler C, Herkenrath P, Lee-Kirsch MA, Cirak S (2020) PNPT1 mutations may cause Aicardi–Goutie’res-syndrome. Brain Dev S0387–7604(20):30283–30287. https://doi.org/10.1016/j.braindev.2020.10.005
Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, Minten EV, Whelan DR, Schlafstein AJ, Zhang H, Anand R, Doronio C, Withers AE, Shepard C, Sundaram RK, Deng X, Dynan WS, Wang Y, Bindra RS, Cejka P, Rothenberg E, Doetsch PW, Kim B, Yu DS (2017) SSAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep 20(8):1921–1935. https://doi.org/10.1016/j.celrep.2017.08.008
Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi–Goutieres syndrome and HIV-1 restriction. J Biol Chem 288(12):8101–8110. https://doi.org/10.1074/jbc.M112.431148
Franzolin E, Coletta S, Ferraro P, Pontarin G, D’Aronco G, Stevanoni M, Palumbo E, Cagnin S, Bertoldi L, Feltrin E, Valle G, Russo A, Bianchi V, Rampazzo C (2020) SAMHD1-deficient fibroblasts from Aicardi–Goutières syndrome patients can escape senescence and accumulate mutations. FASEB J 34(1):631–647. https://doi.org/10.1096/fj.201902508R
Amari S, Tsukamoto K, Ishiguro A, Yanagi K, Kaname T, Ito Y (2019) An extremely severe case of Aicardi–Goutières syndrome 7 with a novel variant in IFIH1. Eur J Med Genet 63(2):103646. https://doi.org/10.1016/j.ejmg.2019.04.003
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
The idea of the article belongs to SD. Literature search was performed by DK. Data analysis was performed by DK and SD. The first draft of the manuscript was written by DK, critically revised the work by SD.
Corresponding author
Ethics declarations
Conflict of interest
The authors of this paper have no conflicts of interest.
Disclaimer
No part of this review, including figures, is copied or published elsewhere in whole or in part.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kivanc, D., Dasdemir, S. The relationship between defects in DNA repair genes and autoinflammatory diseases. Rheumatol Int 42, 1–13 (2022). https://doi.org/10.1007/s00296-021-04906-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-021-04906-3