Abstract
Systematic reviews and meta-analyses of randomized trials have long been important synthesis tools for guiding evidence-based medicine. More recently, network meta-analyses, an extension of traditional meta-analyses enabling the comparison of multiple interventions, use new statistical methods to incorporate clinical evidence from both direct and indirect treatment comparisons in a network of treatments and associated trials. There is a need to provide education to ensure that core methodological considerations underlying network meta-analyses are well understood by readers and researchers to maximize their ability to appropriately interpret findings and appraise validity. Network meta-analyses are highly informative for assessing the comparative effects of multiple competing interventions in clinical practice and are a valuable tool for health technology assessment and comparative effectiveness research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meta-analysis is a statistical technique used to synthesize information from a collection of all relevant studies comparing interventions for a medical condition of interest to address a focused research question about effectiveness or safety in the context of a systematic review [1, 2]. Systematic reviews and meta-analyses of randomized controlled trials (RCTs) have been fundamental tools for the practice of evidence-based medicine by clinicians, scientists and policy makers. When done well and transparently reported, systematic reviews and meta-analyses produce information (in which bias has been extensively reduced) that can be helpful for the evaluation of healthcare interventions. However, a potential drawback of traditional meta-analyses is their ability to compare no more than two treatments at a time, a significant obstacle for decision-making when the medical condition under study has many relevant treatment options to be considered in clinical practice [3–8]. Moreover, in situations where there are no studies directly comparing two or more interventions, traditional meta-analysis cannot estimate their comparative benefits and harms. A simple example of this scenario is when information from RCTs is available regarding the effectiveness of two active treatments, generically B and C, in comparison to a common comparator A (commonly placebo or standard practice therapy); an indirect treatment comparison may be used to estimate a comparison of the relative effectiveness of B compared with C. In recent years, advanced approaches to meta-analysis have been increasingly implemented to obtain estimates of the relative effects of multiple treatments, taking into account the “full network” of available trials, and simultaneously incorporating direct and indirect treatment comparisons in a more complete analysis. This is the objective of network meta-analyses, also known as “multiple-treatments meta-analyses” or “mixed-treatment comparisons meta-analyses.”
What is a network meta-analysis ?
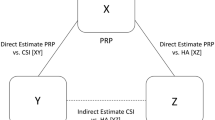
Envision a situation in which a clinician wishes to know the comparative effectiveness of biologic drug therapies used for the treatment of rheumatoid arthritis. Classically, a systematic review of RCTs could only identify studies comparing treatment A (placebo) versus treatment B (or biologic one) and treatment A (placebo) versus treatment C (or biologic two). These sets of studies provide information to compare two sets of treatments (e.g., A vs. B and A vs. C), in what is often termed pairwise or direct comparisons. It can be observed that in this hypothetical systematic review, there are no identified trials that directly compare treatment C versus treatment B, and so direct evidence from these trials is not available to estimate the most clinically relevant comparison between active treatments (B vs. C). Following a traditional approach using pairwise meta-analysis techniques, one cannot establish the relative benefits of treatment B versus C. However, by applying a network meta-analysis (NMA) approach, one can consider many indirect comparisons using information from multiple trials that include a common comparator, A. In this example, an indirect comparison of C versus B can be obtained by synthesizing the results of the trials providing information on the direct comparisons of A versus B and A versus C.
Statistical methods for comparing multiple interventions (using a Bayesian or frequentist framework) have been described in detail elsewhere [9–12]. Suppose that treatment B is judged better than treatment A, demonstrating a higher response of clinical relevance in five additional patients with rheumatoid arthritis and that treatment C is judged better than treatment A with a higher response in three additional patients. An indirect estimate of the comparative effectiveness of treatment B versus C would result in a relative benefit for two additional patients (B is considered to be better than C). Doing so, it would be possible to estimate the potential benefits of the various relative comparisons of treatments A, B and C. In addition, NMA also allows for the combination of direct and indirect information in the estimation of a single treatment effect, which has come to be known as a mixed comparison. In this example, the mixed comparison between B and C incorporates the results of the direct comparison of the two treatments (i.e., outcome data from RCTs of B vs. C) with indirect comparison results obtained from the information of other related direct comparisons (i.e., outcome data from RCTs of B vs. A and C vs. A). In practice, mixed comparisons can be estimated as a weighted average of the direct and indirect estimates of treatment effects, and they can complement information for those comparisons in which there is scarce direct information. When both direct and indirect information can be combined, precision of the estimated treatment effect is improved.
Network geometry
Generally, the graphical representation of a network showing the multiple competing treatments being compared for a medical condition is useful to understand and assess the strength of the clinical evidence for each of the various comparisons under study. Reviewing network geometry also improves the transparency of the results for the critical appraisal of a NMA from the perspective of determining the degree of confidence one may place in interpreting particular comparisons.
Network geometry addresses what the shape of the treatment network looks like in terms of the number of included interventions (i.e., “treatment nodes”), the extent to which there are trials comparing different pairs of these interventions (i.e., the adjoining lines or “edges”), and the numbers of patients associated with different comparisons [5, 13]. By studying and presenting the network geometry, one can develop an understanding of how strong the evidence is for some treatment comparisons and whether specific comparisons are selected, under-represented or even avoided by trialists (comparator preference bias) [5, 7]. Generally, the edges between treatment nodes in the network indicate the comparisons made within eligible randomized trials identified during the process of study identification. The widths of these edges are commonly sized to proportionally reflect the numbers of studies evaluating each pair of treatments, and the sizes of each treatment node are typically sized to proportionally reflect the numbers of subjects randomized to each treatment. If there is no link between a given pair of treatment nodes, it suggests there are no identified trials comparing those particular treatments. Examples of possible network geometries are presented in Fig. 1 and show scenarios which range from a sparse network with a complete lack of direct information for most of the comparisons in the network to a more heavily connected network. For example, if all of the treatments have been compared against a common comparator (e.g., placebo), but not among active treatment options themselves, the network geometry looks like a star (Fig. 1a). If all of the active treatments have been compared with each other, the network plot can be represented as a complex polygon with all treatment nodes connected to each other (Fig. 1b) [5]. For a published survey of network geometries encountered in a range of clinical disciplines, we refer readers to Salanti et al. [13].
Examples of treatment networks. Circles represent treatment nodes in the network; lines represent direct comparisons for which data are available from RCTs. Line thickness is proportionally weighted according to the number of studies evaluating each comparison, while nodes are proportionally weighted according to the number of patients that have received each treatment relative to the total number of participants across all studies
Assumptions and validity considerations: transitivity and consistency
The validity of a NMA depends on a set of assumptions. Particularly, indirect and mixed comparisons are generally considered observational in nature and are based on the assumptions of transitivity (e.g., if treatment B is better than A and A is better than C, it is assumed that B is better than C) [14] and consistency or coherence (which refers to the level of agreement between the evidence from direct and indirect comparisons) [15]. These core assumptions for NMAs can be verified conceptually and epidemiologically but are, however, subject to substantial uncertainty. Clinicians and researchers must assess the appropriateness of the transitivity assumption based on careful review of included studies to judge their comparability from both clinical and methodologic viewpoints. In practice, transitivity and consistency require similarity, which means that the sets of trials used to establish the indirect or mixed comparison are sufficiently alike in terms of important clinical characteristics of the patients, settings and study methods (often referred to as effect modifiers) [14, 16].
NMA methodology is not limited strictly to the calculation of “numerical estimates” based on the synthesis of indirect and mixed comparisons for multiple competing interventions. It may also be used to examine the totality of underlying evidence available for relevant treatments to manage a medical condition in order to detect current research gaps in the existing comparisons and to explore potential sources which may place findings of an NMA at risk of biases [17–19]. One can use network analyses to undertake an examination of the geometry of the clinical evidence (i.e., to identify the totality of all the randomized comparisons that have been performed in terms of comparisons present and missing, the numbers of patients studied, and so forth). Identifying these geometry patterns is informative for detecting research gaps in the existing evidence and designing future research agendas that could potentially improve evidence-based practice. It is possible to consider exploring potential sources of bias (e.g., reporting bias) [18] and also to calculate estimates adjusted for important study and patient characteristics that can be considered as effect modifiers, or even providing subgroup analyses [20]. In addition to the estimation of summary outcome measures of a familiar nature (e.g., odds ratios, mean differences), NMAs can also produce supporting information of considerable appeal to researchers in the form of treatment rankings (e.g., generation of a ranking of different treatments according to the probability of being the “best” treatment, second best, third best and so forth; these values are often summarized by a parameter for each treatment commonly called the surface under the cumulative ranking curve [SUCRA]) [21]. It has been suggested in many published network meta-analyses that these rankings may facilitate interpretation of the results from indirect and mixed comparisons and that they can be especially important for clinicians who wish to know what is the best treatment for a particular clinical condition. However, there are risks of focusing one’s primary interpretation and conclusions on treatment rankings, especially when the network structure may be considered weak (e.g., there exists few or no trials for some of the included comparisons, the trials are of small sample size, and/or are of high risk of bias) or where the systematic review may not have appropriately considered all relevant treatments [7, 19]. Furthermore, treatment rankings may exaggerate small differences in benefits. For this reason, it is generally recommended that clinicians not focus their interpretation of the data solely on the treatment ranks, but rather consider them alongside relative and absolute effect measures.
Example: Results from a published NMA of randomized controlled trials of biologics for rheumatoid arthritis
The number of published NMAs has increased substantially in recent years [22, 23] and has been conducted to evaluate a wide range of healthcare interventions in a diverse array of clinical disciplines. The last few years have also seen a considerable increase in the use of NMA to assess the comparative effectiveness and safety of interventions in the field of clinical rheumatology [22, 23]. One example of a frequently studied condition using NMA methods has been the assessment of the efficacy and safety of treatments such as biologics for rheumatoid arthritis [24–30]. Below, we summarize and comment on a published example of the use of Bayesian NMA methods (see Box 1) based on a study that compared the efficacy of biologics (abatacept, adalimumab, certolizumab pegol, etanercept, golimumab, infliximab and rituximab, tocilizumab) used in combination with a non-biologic disease-modifying antirheumatic drug (DMARD) in patients with rheumatoid arthritis previously treated with methotrexate or other DMARD [24].
Orme et al. [24] reviewed 29 RCTs of eight biologics forming a star-shaped network (Fig. 2). This network geometry shows that there were no head-to-head comparisons between the active treatments and that all biologics were compared only with placebo in the corresponding set of RCTs. The major outcomes reviewed were benefits defined as a 25, 50, 75 % improvement in patient- and physician-reported criteria of the American College of Rheumatology; here, we shall focus on the results for the 50 % improvement (ACR50). Using a conventional approach to meta-analysis, findings are limited to summary estimates of the direct (pairwise) comparisons for which information is available (namely comparison of each biologic to placebo). Compared with DMARD, all biologics were associated with a clinically important difference in ACR50 response rate (Table 1). From the perspective of selecting a treatment for patients, analyzing the data and presenting results in this manner makes it difficult to draw an overall conclusion about which biologic is best. Furthermore, it is difficult to adjust for differences in patient and study-level characteristics among included studies when analyses are conducted in a disparate fashion, and it is also limiting in terms of assessing whether findings are internally consistent. The NMA approach can provide a set of relative effects for comparing biologics against each other, considering indirect evidence for each comparison and can also provide estimates of the probability each treatment is best. For example, Table 2 presents estimates of comparative effectiveness of combination therapy of etanercept + DMARD versus other active combination therapies (as reported by Orme et al. [24]) using random-effects models and fixed-effect models. Random-effects NMA differs from fixed-effect NMA in that it allows the true treatment effect (odds ratios between two treatments) to vary between studies due to heterogeneity. One could similarly estimate comparisons of adalimumab + DMARD versus other active combinations, and so forth. Table 3 presents estimates of the probability each treatment is best for ACR50 and absolute efficacy of each treatment. Further, NMA allows one to formally assess inconsistency [31] and adjust for differences in patient and study-level characteristics [32]. Finally, network meta-analytic methods have evolved considerably in recent years and there are now a suite of diagnostic tools available to help choose appropriate statistical models [31–35]. For example, in this analysis [24], the authors chose the random-effects model (vs. the fixed-effects) for the base-case because model diagnostics indicated the random-effects model provided a better fit to the data. When interpreting these findings, the authors recognized that evidence syntheses considering indirect comparisons need to account for differences among trials in terms of patient and study-level characteristics disease duration and duration of follow-up. Others have noted the considerable heterogeneity among patient and study-level characteristics of previous NMAs comparing biologics for rheumatoid arthritis [36–38]. Accordingly, Orme et al. [24] conducted numerous sensitivity analyses to adjust for patient and study-level characteristics, which are crucial when conducting and appraising NMAs.
Example of network geometry: RCTs evaluating biologics in patients with rheumatoid arthritis [24]. Treatment network diagram based on data from [24] for standard regimens. Note: Circles represent the biologic as a treatment node in the network, while lines represent direct comparisons using placebo RCTs. DMARD disease-modifying antirheumatic drug, kg kilogram, mg milligram, wk(s) week(s)
Box 2 provides a short summary of issues relevant to evaluating and interpreting systematic reviews incorporating NMAs [6, 39].
Final considerations
NMA, an extension of traditional pairwise meta-analysis, is a novel statistical method that incorporates clinical evidence from both direct and indirect treatment comparisons in a complete network of trials to assess the benefits and risks of multiple interventions. Efforts to educate readers and researchers alike about the core methodological concepts and assumptions underlying NMAs are important to provide tools to enable proper interpretation of findings. NMAs should be carried out as part of a rigorous systematic review process to ensure high methodological quality. In order to realize the full potential of these tools, methods and results of NMAs should be transparent and completely reported. It is also necessary to know their inherent limitations. Apart from the transitivity and consistency assumptions, NMA may suffer from potential biases affecting studies, which provide direct and indirect evidence. The assessment of the impact of such potential biases on the results of meta-analyses of indirect and mixed-treatment comparisons is still limited, and the methods to adjust for these biases are still under development. It is important to continue working on the development and validation of the methods for NMAs and, for the time being, interpreting reports of NMAs in light of these potential limitations [7, 35].
References
Higgins JPT, Green S (eds) (2008) Cochrane handbook for systematic reviews of interventions. Wiley, Chichester
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151:264–269
Caldwell DM, Ades AE, Higgins JP (2005) Simultaneous comparison of multiple treatments: combining direct and indirect evidence. BMJ 331:897–900
Salanti G (2012) Indirect and mixed-treatment comparison, network, or multipletreatments meta-analysis: many names, many benefits, many concerns for the next generation evidence synthesis tool. Res Synth Methods 3:80–97
Ioannidis JP (2009) Integration of evidence from multiple meta-analyses: a primer on umbrella reviews, treatment networks and multiple treatments meta-analyses. CMAJ 181:488–493
Mills EJ, Ioannidis JP, Thorlund K, Schünemann HJ, Puhan MA, Guyatt GH (2012) How to use an article reporting a multiple treatment comparison meta-analysis. JAMA 308:1246–1253
Mills EJ, Thorlund K, Ioannidis JP (2013) Demystifying trial networks and network meta-analysis. BMJ 346:f2914
Cipriani A, Higgins JP, Geddes JR, Salanti G (2013) Conceptual and technical challenges in network meta-analysis. Ann Intern Med 159:130–137
Higgins JP, Whitehead A (1996) Borrowing strength from external trials in a meta-analysis. Stat Med 15:2733–2749
Bucher HC, Guyatt GH, Griffith LE, Walter SD (1997) The results of direct and indirect treatment comparisons in meta-analysis of randomized controlled trials. J Clin Epidemiol 50:683–691
Lumley T (2002) Network meta-analysis for indirect treatment comparisons. Stat Med 21:2313–2324
Lu G, Ades AE (2004) Combination of direct and indirect evidence in mixed treatment comparisons. Stat Med 23:3105–3124
Salanti G, Kavvoura FK, Ioannidis JP (2008) Exploring the geometry of treatment networks. Ann Intern Med 148:544–553
Catalá-López F, Hutton B, Moher D (2014) The transitivity property across randomized controlled trials: if B is better than A, and C is better than B, will C be better than A? Rev Esp Cardiol. doi:10.1016/j.recesp.2013.11.016
Song F, Xiong T, Parekh-Bhurke S, Loke YK, Sutton AJ, Eastwood AJ et al (2011) Inconsistency between direct and indirect comparisons of competing interventions: meta-epidemiological study. BMJ 343:d4909
Jansen JP, Naci H (2013) Is network meta-analysis as valid as standard pairwise meta-analysis? It all depends on the distribution of effect modifiers. BMC Med 11:159
Trinquart L, Abbé A, Ravaud P (2012) Impact of reporting bias in network meta-analysis of antidepressant placebo-controlled trials. PLoS One 7:e35219
Trinquart L, Chatellier G, Ravaud P (2012) Adjustment for reporting bias in network meta-analysis of antidepressant trials. BMC Med Res Methodol 12:150
Mills EJ, Kanters S, Thorlund K, Chaimani A, Veroniki AA, Ioannidis JP (2013) The effects of excluding treatments from network meta-analyses: survey. BMJ 347:f5195
Salanti G, Marinho V, Higgins JP (2009) A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin Epidemiol 62:857–864
Salanti G, Ades AE, Ioannidis JP (2011) Graphical methods and numerical summaries for presenting results from multiple-treatment meta-analysis: an overview and tutorial. J Clin Epidemiol 64:163–171
Bafeta A, Trinquart L, Seror R, Ravaud P (2013) Analysis of the systematic reviews process in reports of network meta-analyses: methodological systematic review. BMJ 347:f3675
Lee AW (2014) Review of mixed treatment comparisons in published systematic reviews shows marked increase since 2009. J Clin Epidemiol 67:138–143
Orme ME, Macgilchrist KS, Mitchell S, Spurden D, Bird A (2012) Systematic review and network meta-analysis of combination and monotherapy treatments in disease-modifying antirheumatic drug-experienced patients with rheumatoid arthritis: analysis of American College of Rheumatology criteria scores 20, 50, and 70. Biologics 6:429–464
Singh JA, Christensen R, Wells GA, Suarez-Almazor ME, Buchbinder R, Lopez-Olivo MA et al (2009) A network meta-analysis of randomized controlled trials of biologics for rheumatoid arthritis: a cochrane overview. CMAJ 181:787–796. Erratum in: CMAJ 2010 182(8):806
Bergman GJ, Hochberg MC, Boers M, Wintfeld N, Kielhorn A, Jansen JP (2010) Indirect comparison of tocilizumab and other biologic agents in patients with rheumatoid arthritis and inadequate response to disease-modifying antirheumatic drugs. Semin Arthritis Rheum 39:425–441
Launois R, Avouac B, Berenbaum F, Blin O, Bru I, Fautrel B et al (2011) Comparison of certolizumab pegol with other anticytokine agents for treatment of rheumatoid arthritis: a multiple-treatment Bayesian metaanalysis. J Rheumatol 38:835–845
Turkstra E, Ng SK, Scuffham PA (2011) A mixed treatment comparison of the short term efficacy of biologic disease modifying anti-rheumatic drugs in established rheumatoid arthritis. Curr Med Res Opin 27:1885–1897
Schmitz S, Adams R, Walsh CD, Barry M, FitzGerald O (2012) A mixed treatment comparison of the efficacy of anti-TNF agents in rheumatoid arthritis for methotrexate non-responders demonstrates differences between treatments: a Bayesian approach. Ann Rheum Dis 71:225–230
Ghogomu EA, Maxwell LJ, Buchbinder R, Rader T, Pardo Pardo J, Johnston RV et al (2011) Adverse effects of biologics: a network meta-analysis and cochrane overview. Cochrane Database Syst Rev 2:CD008794
Dias S, Welton NJ, Sutton AJ, Caldwell DM, Lu G, Ades AE (2013) Evidence synthesis for decision making 4: inconsistency in networks of evidence based on randomized controlled trials. Med Decis Mak 33:641–656
Dias S, Sutton AJ, Welton NJ, Ades AE (2013) Evidence synthesis for decision making 3: heterogeneity–subgroups, meta-regression, bias, and bias-adjustment. Med Decis Mak 33:618–640
Dias S, Sutton AJ, Ades AE, Welton NJ (2013) Evidence synthesis for decision making 2: a generalized linear modeling framework for pairwise and network meta-analysis of randomized controlled trials. Med Decis Mak 33:607–617
Dias S, Welton NJ, Sutton AJ, Ades AE (2013) Evidence synthesis for decision making 1: introduction. Med Decis Mak 33:597–606
Li T, Puhan MA, Vedula SS, Singh S, Dickersin K, Ad hoc network meta-analysis methods meeting working group (2011) Network meta-analysis-highly attractive but more methodological research is needed. BMC Med 9:79
Ioannidis JP, Karassa FB, Druyts E, Thorlund K, Mills EJ (2013) Biologic agents in rheumatology: unmet issues after 200 trials and $200 billion sales. Nat Rev Rheumatol 9:665–673
Thorlund K, Druyts E, Aviña-Zubieta JA, Wu P, Mills EJ (2013) Why the findings of published multiple treatment comparison meta-analyses of biologic treatments for rheumatoid arthritis are different: an overview of recurrent methodological shortcomings. Ann Rheum Dis 72:1524–1535
Ades AE, Madan J, Welton NJ (2011) Indirect and mixed treatment comparisons in arthritis research. Rheumatology (Oxford) 50(suppl 4):iv5–iv9
Ades AE, Caldwell DM, Reken S, Welton NJ, Sutton AJ, Dias S (2013) Evidence synthesis for decision making 7: a reviewer’s checklist. Med Decis Mak 33:679–691
Nikolakopoulou A, Chaimani A, Veroniki AA, Vasiliadis HS, Schmid CH, Salanti G (2014) Characteristics of networks of interventions: a description of a database of 186 published networks. PLoS One 9:e86754
Sutton AJ, Abrams KR (2001) Bayesian methods in meta-analysis and evidence synthesis. Stat Methods Med Res 10:277–303
Acknowledgments
D.M. is funded by a University Research Chair, University of Ottawa; B.H. is a Canadian Institutes of Health Research DSEN (Drug Safety and Effectiveness Network) New Investigator; and C.C. is supported by the Canadian Institutes of Health Research Vanier Canada Graduate Scholarship Program.
Conflict of interest
The authors do not have any conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Catalá-López, F., Tobías, A., Cameron, C. et al. Network meta-analysis for comparing treatment effects of multiple interventions: an introduction. Rheumatol Int 34, 1489–1496 (2014). https://doi.org/10.1007/s00296-014-2994-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-014-2994-2