Abstract
The aim of this study was to investigate the association of single-nucleotide polymorphisms (SNPs) in IL23R with ankylosing spondylitis (AS) in Chinese Han population. Six SNPs were selected for analysis in AS patients and controls. The IL23R mRNA expression was detected using RT-PCR. The differences in the genotypes of rs11209032 and the differences in the genotypes and allele frequencies of rs6677188 between cases and controls were significant. The two SNPs rs11209032 and rs6677188 were in strong linkage disequilibrium. Haplotype analysis noted a higher proportion of GAC in cases and a higher proportion of GTC in controls. The patients with AS showed an elevated level of IL23R mRNA in PBMCs. This study suggested that IL23R polymorphisms were associated with susceptibility to AS in the Chinese population and that IL23R may be involved in the development of AS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ankylosing spondylitis (AS) is a kind of seronegative spondyloarthropathy, which is characterized by sacroiliitis and a variety of extra-articular manifestations, with an estimated prevalence of 0.1–0.9%. Genetic factors have been strongly implicated in its etiology, and heritability as assessed by twin studies has been estimated to be >90%. Although HLA-B27 is present in >95% with AS and is the major gene associated with this disease, only 1–5% of random B27 carriers develop the disease, and B27 contributes to only 20–30% of the overall genetic predisposition [1]. In addition to B27 allele contribution, several case–control studies have noted that some non-HLA genes such as IL-1 family gene cluster, CYP450 and the ANKH gene are associated with susceptibility to AS [2–4].
IL23R is the receptor for interleukin-23 (IL-23), a pro-inflammatory cytokine, which is composed of a unique p19 subunit in addition to a p40 subunit shared with IL-12, and is involved in memory T-cell regulation [5]. IL23R was identified as a novel member of the hemopoietin receptor family as a subunit of the receptor for IL-23 and associated constitutively with Jak2 and in a ligand-dependent manner with stat3. IL23R expression on T cells, NK cells, monocytes, and DCs corresponds with the ability of those cells to respond to IL-23 [6, 7]. The IL23R gene is located on chromosome 1p31 and forms a receptor for IL-23, together with the beta 1 subunit of IL-12 (IL-12RB1) [6]. At least six alternatively spliced mRNAs of IL23R gene is expressed to generate diverse isoforms of the receptor protein [8]. IL23R polymorphisms have recently been reported to be strongly associated with both IBD and psoriasis [9–11]. AS is clinically related to both inflammatory bowel disease (IBD) and psoriasis, with an increased risk of AS in both conditions and an increased prevalence of IBD and psoriasis in AS.
A recent study genotyped 14,436 nonsynonymous SNPs from independent cases of AS and observed strong association between IL23R polymorphisms and AS [12]. The SNPs rs11209026, rs1343151 and rs11209032 lying in IL23R have been observed to be associated significantly with AS in the UK and North American populations [12]. Thus, we performed genotyping of the three SNPs and another three SNPs near these, based on their physical distances, to examine the association of IL23R polymorphisms in the AS population of China.
On the basis of previous data, we enrolled 138 cases and 129 ethnically matched controls from a Chinese population. In addition to analyzing the genotypes and allele frequencies of the SNPs, we also performed linkage disequilibrium (LD) analysis and haplotype analysis to seek further evidence of association with AS in the region. Furthermore, we examined the IL23R mRNA expression in peripheral blood mononuclear cells (PBMCs) to find out whether there were abnormalities in AS patients.
Materials and methods
Patients
This study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University. Informed consent was obtained from all patients. All the subjects were of Chinese Han population and satisfied the Modified New York Criteria for the Classification of AS. Information was collected systematically and included age of onset, HLA-B27, CRP, ESR, standardized measures of disease activity and function for AS [Bath AS Disease Activity Index (BASDAI) and Bath AS Functional Index (BASFI)]. Control subjects were recruited from medical students and volunteers from the Chinese Han population and had no history of autoimmune diseases.
As much as 138 AS sporadic cases and 129 ethnically matched controls were assessed for genotype assay. The mean age of patients with AS was 29.2 years (SD 10.2); 89.1% of AS were male and 87.6% were HLA-B27-positive. The mean age of control was 35.4 years (SD 12.1) and 51% of controls were men. The mean BASDAI score for AS patients was 3.7 years (SD 2.0) and the mean BASFI was 2.3 (SD 2.2).
Thirty patients with AS (26 men and 4 women) with an average age of 29.7 years (SD 7.2) and 30 healthy individuals (20 men and 10 women) with an average age of 26.9 years (SD 7.9) were included in IL23R expression study. The mean BASDAI, BASFI, ESR and CRP for AS patients were 5.4 (SD 1.7), 4.7 (SD 2.5), 56.97 mm/h (SD 11.45) and 38.33 mg/L (SD 27.03), respectively.
Polymerase chain reaction and sequencing
Fasting venous blood samples were obtained from the study participants and genomic DNA was extracted from blood lymphocytes by a standard salting out procedure. The SNPs rs11209026, rs1343151 and rs11209032 were selected based on previous studies and another three SNPs nearby were also included. SNP genotyping was performed by PCR direct sequencing method. The reaction system contained 1 μl of 100 ng/ul genomic DNA, 0.25 μl of Taq polymerase, 0.5 μl of 2.5 mM dNTP mixture, 2.5 μl of buffer (plus Mg2+) and 0.5 μl of primers (forward and reverse) and was adjusted with Milli-Q H2O to a total volume of 25 μl. The PCR product underwent strict purification before bi-directional sequencing with ABI 3700 automated sequencer (Applied Biosystems, USA). The primer sequences are shown in Table 1.
RNA preparation and RT-PCR
Total RNA was extracted from PBMCs (3 × 106cells) using QIAamp RNA blood kits (QIAGEN, USA) according to the manufacturer’s instrctions. RT was performed using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas) in 20 μl final volume reactions containing 1 μg RNA. PCR amplificaton of cDNA aliquots was performed by adding 1.25 mM of each dNTP, 1.25 U Taq DNA polymerase (Takara) and 20 μM of sense and antisense primers. The reaction was performed in 25 μl of PCR buffer (15 mM MgCl2, 500 mM KCl, 100 mM Tris-HCl, pH 8.3). The sense and antisense primers used in this experiment were: IL23R: sense: 5′-CAC AAG CCT ACA GAC TAC AA-3′; antisense: 5′-CTG GAA TAG TTT CAC TGG GT-3′; β-actin: sense: 5′-GGA CTT CGA GCA AGA GAT GG-3′; antisense: 5′-TGT GTT GGC GTA CAG GTC TTT G-3′. Reactions were processed in a DNA thermal cycler through 30 cycles of 30 s of denaturation at 94°C, 45 s of annealing at 55°C for IL23R and at 58°C for β-actin, followed by 45 s of elongation at 72°C. PCR products were run on a 1.8% agarose gel and stained with ethidium bromide. The level of mRNA expression was presented as a ratio of IL23R PCR product over β-actin product.
Statistical analysis
Hardy–Weinberg equilibrium was performed by the χ2 analysis. Both genotypes and allele frequencies were calculated and compared by χ2 test using the SPSS 13.0 for Windows software. The magnitude of association was expressed as odds ratio with a 95% confidence interval (CI). Linkage disequilibrium and haplotype analysis were carried out by SHEsis. RT-PCR data are expressed as means ± SDs. Statistical analysis was performed using t test (SPSS 13.0).
Results
Genotypes and allele frequencies in controls and patients with AS
All the genotypes detected for SNPs rs11209026 and rs11465816 were homozygous, which was consistent with the HapMap data. Only one heterozygous genotype was detected for rs10889671. Consequently, SNPs rs11209032, rs6677188 and rs1343151 were included in the case–control study and all observed genotypes and allele frequencies met Hardy–Weinberg equilibrium. No significant differences were found in genotypes and allele frequencies of rs1343151 polymorphisms and no significant differences were found in allele frequencies of rs11209032 polymorphisms between cases and controls. The differences of the genotypes and allele frequencies of rs6677188 and the genotypes of rs11209032 between cases and controls were statistically significant (P < 0.001)(Table 2).
LD and haplotype frequency estimation
For the IL23R SNPs, pairwise linkage disequilibrium (LD) and haplotype were analyzed. The two SNPs, rs11209032 and rs6677188, were in strong LD (D′ = 0.925, r 2 = 0.561). A significantly higher proportion of GAC haplotype was noted in cases (odds ratio 2.048, 95%CI 1.404–2.986, P = 0.000179) and a significantly higher proportion of GTC haplotype was noted in controls (odds ratio 0.158, 95%CI 0.076–0.328, P = 3.58 × 10−8).
Expression of IL23R mRNA in PBMCs of AS patients and normal control subjects
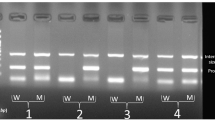
The expression of the IL23R mRNA was normalized by the respective β-actin mRNA. The average intensity ratio of IL23R PCR product/β-actin product (0.75 ± 0.25) in patients with AS was significantly higher than that in normal control subjects (0.44 ± 0.21; P < 0.001; Fig 1).
Discussion
IL-23 is a heterodimeric protein that is a member of the IL-12 family of cytokines. It is composed of a p19 subunit in addition to a p40 subunit, which is also a component of IL-12. IL-23 also has one receptor subunit in common with IL-12, IL12Rβ1. Although IL-23 has its own unique subunit IL23R, together with IL12Rβ1, forms the signaling complex [13]. Recent studies have identified IL-23 as a key player in both innate and adaptive immune systems. Most IL-23 is secreted by activated dendritic cells, monocytes and macrophages following their exposure to pathogen-derived molecules that bind to toll-like receptors. IL-23 stimulates a unique CD4+ helper T-cell population characterized by the production of IL-17, tumor necrosis factor and IL-6, known as Th17 cells. These cells play a central role in driving autoimmune inflammation in a number of animal models. IL-17 stimulates monocytes and endothelial cells to produce proinflammatory mediators, which in turn promote rapid neutrophil recruitment [14]. IL-23p19 mRNA expression was found to be elevated in psoriatic lesions and biopsy specimens from inflamed areas in both Crohn’s disease (CD) and ulcerative colitis (UC) [15, 16]. TNF antagonists have been approved for the treatment of psoriasis and CD [17]. All these findings suggest that the IL23R pathway may have important implications in the pathogenesis of chronic inflammatory disease.
IL23R gene polymorphisms have been recently reported to be associated with an autoimmune inflammatory disease, IBD, [18] and this association has been replicated in different populations [9, 19–21]. Although it seems that IL23R plays an important role in the susceptibility to IBD and other autoimmune diseases, recent studies did not find an association with rheumatoid arthritis (RA) or with systemic lupus erythematosus (SLE) in a Spanish population [22, 23].
Given the clinical and immunological overlap between IBD and AS, studies sought to examine the association between IL23R polymorphisms and AS and indicated that IL-23R was a major genetic determinant of the susceptibility to AS [12]. Our study attempted to determine the contribution of IL23R polymorphisms in AS in Chinese population. The genotypes of SNPs rs11209026 and rs11465816 that we detected were all homozygous, which is consistent with the HapMap data (http://www.hapmap.org). We did not find significant difference between cases and controls in the rs1343151, which was associated with AS in the North American data set [12]. The difference between these two studies may be a result of different ethnic backgrounds or disease heterogeneity. The most significant SNP in our study was rs6677188; we noted a significant difference between AS patients and controls for the genotypes and allele frequencies. We also noted a significant difference for genotypes of rs11209032 between AS patients and controls. These two SNPs, which locate in the intergenic region and encode no amino acids, in particular, confer susceptibility to AS. Besides, they may correlate to the shearing of mRNA or function in the interaction of IL23R and its adjacent gene IL12RB2 by an unknown mechanism. TagSNPs in linkage disequilibrium with these SNPs might reveal more functionally putative variants in the future. Furthermore, we reported for the first time the distribution of rs6677188 since it was discovered. We subsequently analyzed pairwise LD and haplotype of rs11209032, rs6677188 and rs1343151 and found that rs11209032 and rs6677188 are in strong LD. Haplotype GTC was protective in AS, while haplotype GAC may be a risk factor for AS. All these data added to our knowledge of the involvement of IL23R polymorphisms as a genetic component in AS.
Until now, no research has been done on the expression of IL23R related to autoimmune diseases. To explore whether IL23R expression was different between AS patients and controls, we examined the IL23R mRNA expression in PBMCs using RT-PCR. Fortunately, we found significantly higher levels of IL23R in AS patients than in controls. This suggests that further analysis for IL23R in AS and other autoimmune diseases should be valuable.
In conclusion, we suggest that the IL23R gene polymorphisms play a relevant role in the susceptibility to AS in the Chinese population, which is consistent with the results previously obtained in other populations. These results may propose a new understanding of the etiology and pathogenesis of AS and suggest that the Th17 T-cell subset is at the center of the process, which leads to the development of the disease. These data suggest that IL-23/IL23R system may be a novel potential therapeutic target for AS.
References
Brown MA, Kennedy LG, MacGregor AJ, Darke C, Duncan E, Shatford JL et al (1997) Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum 40:1823–1828. doi:10.1002/art.1780401015
Timms AE, Crane AM, Sims AM, Cordell HJ, Bradbury LA, Abbott A et al (2004) The interleukin 1 gene cluster contains a major susceptibility locus for ankylosing spondylitis. Am J Hum Genet 75:587–595. doi:10.1086/424695
Brown MA, Edwards S, Hoyle E, Campbell S, Laval S, Daly AK et al (2000) Polymorphisms of the CYP2D6 gene increase susceptibility to ankylosing spondylitis. Hum Mol Genet 9:1563–1566. doi:10.1093/hmg/9.11.1563
Tsui FW, Tsui HW, Cheng EY, Stone M, Payne U, Reveille JD et al (2003) Novel genetic markers in the 5′-flanking region of ANKH are associated with ankylosing spondylitis. Arthritis Rheum 48:791–797. doi:10.1002/art.10844
Trinchieri G, Pflanz S, Kastelein RA (2003) The IL-12 family of heterodimeric cytokines: new players in the regulation of T-cell responses. Immunity 19:641–644. doi:10.1016/S1074-7613(03)00296-6
Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J et al (2002) A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rβ1 and a novel cytokine receptor subunit, IL-23R. J Immunol 168:5699–5708
Belladonna ML, Renauld JC, Bianchi R, Vacca C, Fallarino F, Orabona C et al (2002) IL-23 and IL-12 have overlapping, but distinct effects on murine dendritic cells. J Immunol 168:5448–5454
Zhang XY, Zhang HJ, Zhang Y, Fu YJ, He J, Zhu LP et al (2006) Identification and expression analysis of alternatively spliced isoforms of human interleukin-23 receptor gene in normal lymphoid cells and selected tumor cells. Immunogenetics 57:934–943. doi:10.1007/s00251-005-0067-0
Tremelling M, Cummings F, Fisher SA, Mansfield J, Gwilliam R, Keniry A et al (2007) IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology 132:1657–1664. doi:10.1053/j.gastro.2007.02.051
Capon F, Di Meglio P, Szaub J, Prescott NJ, Dunster C, Baumber L et al (2007) Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum Genet 122:201–206. doi:10.1007/s00439-007-0397-0
Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP et al (2007) A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet 80:273–290. doi:10.1086/511051
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A et al (2007) Association scan of 14500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 39:1329–1337. doi:10.1038/ng.2007.17
Hunter CA (2005) New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol 5:521–531. doi:10.1038/nri1648
McKenzie BS, Kastelein RA, Cua DJ (2006) Understanding the IL-23-IL-17 immune pathway. Trends Immunol 27:17–23. doi:10.1016/j.it.2005.10.003
Lee E, Trepicchio WL, Oestreicher JL, Pittman D, Wang F, Chamian F et al (2004) Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med 199:125–130. doi:10.1084/jem.20030451
Schmidt C, Giese T, Ludwig B, Mueller-Molaian I, Marth T, Zeuzem S et al (2005) Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-p27p28 in Crohn’s disease but not in ulcerative colitis. Inflamm Bowel Dis 11:16–23. doi:10.1097/00054725-200501000-00003
Ware CF (2005) Network communications: lymphotoxins, LIGHT, and TNF. Annu Rev Immunol 23:787–819. doi:10.1146/annurev.immunol.23.021704.115719
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ et al (2006) A genome-wide association study identifies IL-23R as an inflammatory disease gene. Science 314:1461–1463. doi:10.1126/science.1135245
Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Davies G et al (2007) IL23R Arg381Gln is associated with childhood onset inflammatory bowel disease in Scotland. Gut 56:1173–1174. doi:10.1136/gut.2007.122069
Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D et al (2007) Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet 20:e58. doi:10.1371/journal.pgen.0030058
Oliver J, Rueda B, López-Nevot MA, Gomez-Garcia M, Martin J (2007) Replication of an association between IL23R gene polymorphism with inflammatory bowel disease. Clin Gastroenterol Hepatol 5:977–981. doi:10.1016/j.cgh.2007.05.002
Orozco G, Rueda B, Robledo G, García A, Martín J (2007) Investigation of the IL23R gene in a Spanish rheumatoid arthritis cohort. Hum Immunol 68:681–684. doi:10.1016/j.humimm.2007.05.008
Sanchez E, Rueda B, Callejas JL, Sabio JM, Ortego-Centeno N, Jimenez-Alonso J et al (2007) Analysis of interleukin-23 receptor (IL23R) gene polymorphisms in systemic lupus erythematosus. Tissue Antigens 70:233–237. doi:10.1111/j.1399-0039.2007.00881.x
Acknowledgments
We wish to thank all the participants who contributed their blood samples to our study. Dr. Gu’s work was funded by Research Grant 30325019 of National Science in China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, X., Huang, J., Lin, Z. et al. Single-nucleotide polymorphisms and expression of IL23R in Chinese ankylosing spondylitis patients. Rheumatol Int 30, 955–959 (2010). https://doi.org/10.1007/s00296-009-1085-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-009-1085-2