Abstract
Across many phyla, a common aspect of multicellularity is the organization of different cell types into spatial patterns. In the budding yeast Saccharomyces cerevisiae, after diploid colonies have completed growth, they differentiate to form alternating layers of sporulating cells and feeder cells. In the current study, we found that as yeast colonies developed, the feeder cell layer was initially separated from the sporulating cell layer. Furthermore, the spatial pattern of sporulation in colonies depended on the colony’s nutrient environment; in two environments in which overall colony sporulation efficiency was very similar, the pattern of feeder and sporulating cells within the colony was very different. As noted previously, under moderately suboptimal conditions for sporulation—low acetate concentration or high temperature—the number of feeder cells increases as does the dependence of sporulation on the feeder-cell transcription factor, Rlm1. Here we report that even under a condition that is completely blocked sporulation, the number of feeder cells still increased. These results suggest broader implications to our recently proposed “Differential Partitioning provides Environmental Buffering” or DPEB hypothesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
All species have evolved mechanisms by which individual organisms cooperate. This cooperation provides biological functions impossible to an individual organism. One aspect of cooperation is termed “task allocation” in which different members of the community perform different functions. Indeed, in social insects such as bees and ants, the allocation of workers performing a given task (e.g. foraging) changes based on the environment of the colony and on chemical signals between ants (Gordon 2013; Wilson and Holldobler 1988). Task allocation can be considered a community-wide type of phenotypic plasticity, the ability of an organism to change phenotype in response to environment.
Several lines of evidence suggest that cooperation between organisms extends to microbial species. In the first place, quorum-sensing signals in bacteria and fungi and protists allow these species to behave differently in dense cultures than in dilute cultures [reviewed in (Albuquerque and Casadevall 2012; Loomis 2014; Waters and Bassler 2005)]. In the second place, a variety of microorganisms form communities (e.g. colonies or biofilms) in which cells of different types are organized into spatial patterns [reviewed in (Honigberg 2011; Shapiro 1998; Vachova et al. 2012)]. For example in the budding yeast, S. cerevisiae, haploid colonies partition into two clearly differentiated cell layers, (U = upper, and L = lower) with each layer characterized by a distinct gene expression patterns, and metabolism (Cap et al. 2012; Vachova et al. 2013).
A second example of colony patterning occurs in diploid yeast. Once these colonies have ceased growth, they partition into layers of sporulating and non-sporulating cells (Piccirillo et al. 2010). In particular, as the colony begins to mature, a narrow horizontal layer of sporulating cells forms at the bottom of the colony (near the agar surface). Covering this narrow sporulation layer is a broader layer of unsporulated cells. In turn this unsporulated layer is covered by a second narrow layer of sporulating cells positioned approximately through the middle of the colony. Finally this unsporulated layer is blanketed by a second layer of unsporulated cells that extends to the surface of the colony. As the colony matures, this upper layer gradually but efficiently sporulates until the entire top half of the colony contains sporulated asci. In contrast, the bottom sporulation layer and the bottom unsporulated layer remain largely unchanged. The expansion of the sporulation region in the top half of the colony involves alkalization of the colony as sensed through the Rim101 pathway (Piccirillo et al. 2010). This same sporulation pattern is seen in all seven laboratory or wild yeast strain backgrounds on which it has been tested. In the wild strains in particular, this basic pattern is evident when colonies are grown on a range of carbon and nitrogen sources (Piccirillo and Honigberg 2010).
What is the function of partitioning colonies into sporulating and non-sporulating cell layers? A screen for mutants that are defective in sporulation in colonies but not in cultures implicated the cell-wall integrity (CWI) pathway in colony sporulation (Piccirillo et al. 2015). The CWI pathway is induced as colonies mature and one target of this pathway, the Rlm1 transcription factor, becomes active specifically in the underlying layer of non-sporulating cells, while stimulating sporulation in the overlying layer. As expected from this result, Rlm1 stimulates sporulation through a cell-nonautonomous mechanism (Piccirillo et al. 2015). Furthermore, Rlm1 activation in the unsporulated cell layer contributes to an increase in permeability in these cells, which nevertheless remain viable. Based on these observations, we termed cells in the unsporulated cell layer “feeder cells”, and we speculated that these feeder cells release nutrients or other signals that stimulate sporulation in the overlying cell layer.
In the current study, we investigated the effect of environment on partitioning of yeast colonies into sporulating and feeder cell layers. For this purpose, we examined the distribution of these cells at a relatively early stage of pattern formation, and also the effect of colony environment on sporulation patterns and on the relative allocation of feeder and sporulating cells in colonies. These results are discussed in the context of our recently proposed model for how differential partitioning of yeast colonies maximizes the efficiency of colony sporulation across a range of conditions (the DPEB hypothesis).
Results
Confocal images reveal separation of feeder and sporulation layers in colonies
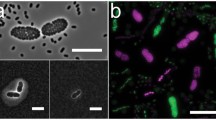
As a first step to investigate the effect of colony environment on sporulating and feeder cell populations, we examined cross-sections of colonies at an early stage of maturation (4 days) for the distribution of the two cell types. Feeder cells in these colonies were visualized by first staining the colonies with propidium iodide (PI), since feeder cells have increased permeability to this nucleic acid stain (Piccirillo et al. 2015). Sporulating cells were visualized by expression of the promoter of an early meiotic gene (IME2), fused to GFP (ime2Δ-GFP). Confocal images of these cross-sections revealed a layer of feeder cells fluorescing red underlying a layer of sporulating cells fluorescing green (Fig. 1). These images extend our previous conclusion based on conventional fluorescence microscopy on colony cross-section. These results are also consistent with experiments in which colonies were resuspended and the populations expressing ime2Δ-GFP and/or permeable to PI were determined (Piccirillo et al. 2015). All three approaches show that feeder and sporulating cell populations in colonies have little or no overlap.
Confocal image of PI-permeability and ime2Δ-GFP expression in colonies. Colonies of strain SH4793 were grown for 4 days, stained with PI, and visualized by confocal microscopy. Section shown is through the center of the colony, with the top of the colony at the top of the image. Red cells = PI, green cells = ime2Δ-GFP expression. PI and GFP images were collected separately and then merged (Image J). Scale bar 50 μ
The image shown was typical of those we captured for early stages of colony development in having a layer of PI-permeable cells that was clearly distinct and separate from the layer expressing ime2Δ-GFP. Indeed, the thick intervening cell layer that was neither permeable to PI nor expressing ime2Δ-GFP indicated that at early stages of colony development, colonies are partitioned into more than just two cell types, e.g. undifferentiated cells, feeder cells, and sporulating cells.
Colony sporulation pattern influenced by nutrient environment
As described in the Introduction, the basic pattern of partitioning sporulating colonies into feeder and sporulating cell layers is relatively robust (Piccirillo and Honigberg 2010). To expand on these observations, we examined this pattern in wild-type colonies grown on different media. In particular we compared our standard agar medium for investigating colonies (YA), to medium containing only half the yeast extract (LYA). We noted that colonies grown on LYA medium colonies were significantly shorter than those on YA medium (0.19 ± 0.02 mm from LYA vs. 0.30 ± 0.02 mm on YA, n = 10, P = 0.01). The overall level of sporulation on the two media was very similar (53 ± 1.4 % for YA and 47 ± 3 % for LYA), but when sections of colonies were examined, the sporulation pattern was quite different. For example on YA medium, a very sharply defined layer of feeder cells was apparent in 6 day colonies (Piccirillo et al. 2015) and 10 day colonies (Fig. 2a, b). In contrast, in LYA medium, even after 10 days, these shorter colonies contained much narrower and less well-defined feeder cell layers. Instead, sporulation in LYA colonies was distributed relatively evenly throughout the entire colony (Fig. 2c, d). Thus, the pattern of sporulating and feeder cells in the population varied in the two environments even though the overall sporulation levels were similar.
Sections of yeast colonies grown on different concentrations of nitrogen source. a Sections of two colonies grown on YA medium (0.5 % yeast extract) for 10 days, b distribution of sporulated cells in colonies grown as in a determined by superimposing a grid containing nine equal-sized rectangles stacked on top of one another (long side horizontal) on each image scaled to just cover central region of colony from bottom to top. Distributions show percentage of the population recognized as asci in each of these rectangles from the top (T, left) to bottom (B, right) of the colony. c Sections of two colonies grown on LYA medium (0.25 % yeast extract) for 10 days. d Distribution of sporulated cells in colonies grown as in c determined as described in b. Scale bars in a and c are 50 μ. Error bars in b and d represent SEM, n = 3
Relative partitioning of colonies into feeder and sporulating cells is influenced by environment
Consistent with the results described above, conditions that decrease colony sporulation can lead to a higher fraction of feeder cells in the population and a greater dependence of sporulation on RLM1 (Piccirillo et al. 2015). For the current study we confirmed this effect and extended it to examine an environment where sporulation is almost completely absent. In particular, as seen in the earlier study, when the concentration of acetate in the medium was only one-quarter of standard levels, the colony sporulation frequency decreased significantly (~50 %, P = 0.0004, Fig. 3a), and the frequency of feeder cells in the colony (e.g. cells with active Rlm1) increased ~sevenfold (Fig. 3b). Similarly, when colonies were grown at 34 °C, rather than at the optimal temperature of 30 °C, the sporulation frequency decreased significantly (~40 %, P = 0.0006, Fig. 3a) and the frequency of feeder cells increased ~ninefold (Fig. 3b). Finally, we examined colonies that have both decreased acetate and increased temperature. We found that in this “dual-stress” environment the fraction of colonies allocated to feeder cells is approximately the same as in colonies grown at 34 °C at the standard acetate concentration; however, sporulation is almost completely eliminated in these dual-stress colonies (Fig. 3a, b). In summary, two stress conditions, as well as the combination of these two conditions, led to both a decreased frequency of sporulated cells and an increased frequency of feeder cells.
Effect of acetate concentrations and temperature on the fraction of sporulating and feeder cells in colonies and dependence of sporulation on RLM1. a Fraction of colony (SH3881) that formed spores at indicated potassium acetate concentrations (x-axis) and temperature (y-axis). b Fraction of colonies under same conditions as in a identifiable as feeder cells by expression of the UASRlm1-LacZ allele in strain SH5065. c Dependence of colony sporulation on RLM1 grown under the same conditions as in a. Dependence score = sporulation frequency in RLM1 colony (SH3881)/sporulation frequency in rlm1Δ colony (SH4708). Dependence score was not calculated for dual stress condition. Error bars are SEM, n = 3
Strikingly, for the stress conditions that display reduced but still measurable sporulation, this “residual” sporulation is much more dependent on RLM1 than in colonies grown under the standard condition (Piccirillo et al. 2015). An “Rlm1-dependency score” can be calculated for any given condition; this score is simply the sporulation frequency in RLM1 + colonies divided by the sporulation frequency in rlm1Δ colonies. Note that if sporulation is completely independent of RLM1 under a given condition, the dependency score will be 1.0, whereas greater dependency on RLM1 will lead to a higher score. Consistent with our earlier study, in the decreased acetate condition, the RLM1-dependence score increased significantly (~3.5-fold) relative to standard conditions (Fig. 3c). Similarly, colonies grown under high-temperature stress displayed RLM1-dependence scores ~3.3-fold higher than colonies grown under standard conditions (Fig. 3c). For the dual-stress condition, sporulation in both wild-type (1.1 ± 0.9 %, n = 3) and rlm1Δ colonies (0.4 ± 0.2 %, n = 3) was too low to confidently determine a dependency score. Thus, we confirmed that in environments that cause a higher proportion of feeder cells in colonies and a lower (but still accurately measurable) proportion of sporulated cells, sporulation becomes more dependent on the feeder-cell specific gene, RLM1.
Discussion
The principal conclusions presented above are as follows: (1) during an early stage of colony development, feeder and sporulating cell layers are separated by an intervening layer of cells that have not fully adopted either fate, (2) colony sporulation patterns depend strongly on environment even when comparing environments in which overall sporulation levels are similar, and (3) even in an environment in which sporulation was almost completely inhibited, a large proportion of the colony was still allocated to feeder cells.
The DPEB hypothesis
The correspondence between an increased frequency of feeder cells and an increased dependence of sporulation on RLM1 (hence feeder cell development) led us to propose the “Differential Partitioning provides Environmental Buffering” (DPEB) hypothesis (Piccirillo et al. 2015). According to this hypothesis, yeast evolved such that colonies are partitioned between two fates differently depending on their environment, and this “differential partitioning” allows sporulation to occur relatively efficiently across a wider range of environments. For example, an increased number of feeder cells could provide nutrients necessary for other cells in the colony to sporulate efficiently under suboptimal sporulation conditions. A corollary to the DPEB hypothesis is that in the absence of differential partitioning, the range of environments that could support sporulation would be much narrower.
Because the ability to sporulate across a wider range of environments would provide a selective advantage to S. cerevisiae communities that is not available to an individual yeast cell, DPEB would provide a selective advantage to multicellularity. Thus, DPEB could promote the evolution of species from unicellular to multicellular—a transition that arose multiple times in different phyla during evolution (Niklas and Newman 2013). To our knowledge, this particular advantage to multicellularity has not been proposed previously.
Feeder cells stimulate sporulation but are neither necessary nor sufficient under all conditions
It is interesting that colonies exposed to the dual-stress condition—decreased acetate and increased temperature—had a much more pronounced sporulation defect than either individual perturbation; indeed, the decrease in the dual-stress condition was much more than the sum of the defects from the individual perturbations. This comparison suggests that the inhibitory effects of high temperature and low acetate may converge on a common target required for sporulation. For example, both increased temperature and decreased acetate have been implicated in limiting respiration (Blank and Sauer 2004; Cortassa et al. 2000), which is essential for sporulation (Jambhekar and Amon 2008). Following this reasoning, the dual-stress condition may decrease respiratory rates below the threshold required for sporulation, i.e. a respiratory level too low to allow sporulation regardless of the increased numbers of feeder cells.
In contrast to stressed colonies, colonies grown on LYA medium sporulate efficiently without forming a distinct feeder cell layer. Colonies grown on this medium are significantly shorter relative to our standard medium. Hence, more of the cells in the LYA colony are close to the surface of the nutrient agar. We speculate that it is for this reason that sporulation occurs efficiently in LYA colonies without the need for a distinct feeder cell layer. In summary, feeder cells are essential for efficient sporulation under some conditions, unnecessary for sporulation under other conditions, and insufficient for sporulation under still other conditions. These results, together with the confocal images, suggest that the partitioning of colonies into feeder and non-feeder cells may happen even before cells begin to sporulate.
Broader relevance of DPEB hypothesis
In principle, the DPEB hypothesis can be extended to the partitioning of other cell fates within yeast communities. For example, pseudohyphal differentiation occurs in a subset of the population of a yeast colony, typically at the edge of these communities, and this type of differentiation is influenced by both temperature (McCusker et al. 1994a, b) and nutrient environment [reviewed in (Cullen 2015; Gagiano et al. 2002)]. In addition, by analogy to the current study, it is possible that the relative size of the layers of metabolically distinct U and L cells in haploid colonies (Cap et al. 2012; Vachova et al. 2013) may change in response to environmental cues. Thus, DPEB in microorganisms is analogous to task allocation in social organisms (see “Introduction”); that is, the community benefits by reallocating tasks to individual organisms as environmental conditions change.
DPEB is an example of phenotypic plasticity, i.e. a beneficial change to an organism’s phenotype in response to its environment, in this respect DPEB may be relevant to the phenotypic plasticity observed across many metazoan species—where environment affects both differentiation and development. For example, external temperature during embryonic development determines sex in many reptile, fish, and amphibian species (Baroiller et al. 2009; Eggert 2004; Merchant-Larios and Diaz-Hernandez 2013). Furthermore, the maternal diet during early development is hypothesized to influence the timing of sexual maturation, and the long-term health of the ensuing adults (Brakefield et al. 2005; Danielsen et al. 2013). Finally, diet can regulate the proliferation and differentiation of adult stem cell populations (Ables et al. 2012). Thus, it is within the realm of possibility that differential partitioning of cell types in developing organisms help to promote successful development across a range of environments.
Methods
Strains
The wild-type (IME2 + RLM1 +) strain used throughout the study is SH3881 (MAT a /MATα ade2/ade2 can1:ADE2:CAN1/can1:ADE2:CAN1 his3-11,15/his3-11,15 lys2(3′Δ):HIS3:lys2(5′ Δ)/LYS2 URA3/ura3-1). The ime2-GFP strain used for confocal microscopy is SH4793 (isogenic to SH3881 except leu2-3,112/LEU+ trp1-1/trp1-3′del ura3-1/ura3-1 ime2::GFP-TRP1/ime2::GFP-TRP1 rlm1D::URA/RLM1+). The rlm1Δ strains used in Fig. 3 are SH4708 and SH4767 (isogenic to SH3881 except ura3-1/ura3-1 rlm1Δ::URA3/rlm1Δ::URA3). The UASRlm1-LacZ strain used in Fig. 3 is SH5065 [isogenic to SH3881 except leu2-3, 112/leu2-3, 112 trp1-1/trp1-3′Δ ura3-1/ura3-1 ime2D::LEU2/ime2::GFP-TRP1 pS778(URA3)]. The plasmid pS778 contains two copies of the Rlm1 binding sites attached to the CYC1 promoter and fused to the LacZ ORF (UASRlm1-LacZ) (Jung et al. 2002). All strains are available on request.
Media and growth conditions
Colonies for confocal were grown by plating 300 cells on YA medium (2 % potassium acetate, 0.5 % yeast extract pH 6.0) and incubated for 4 days. Colonies for embedding/sectioning were grown for 10 days on either YA or LYA medium [2 % potassium acetate, 0.25 % yeast extract, a combination of amino acids (SS-21), pH 7.0]. For comparison of colonies in different acetate concentrations and different temperatures, colonies were inoculated with 0.5 μl of a cell suspension (spot colonies) as described (Piccirillo et al. 2015) on either SP2 % (2 % potassium acetate, 0.5 % yeast extract pH 7.0) or SP0.5 % (0.5 % potassium acetate, 0.5 % yeast extract pH 7.0).
Colony sections
For confocal microscopy of colonies, colonies were embedded in agar containing propidium iodide, incubated for 24 h at 30 °C as described previously for conventional fluorescent microscopy (Piccirillo et al. 2015), and then transferred to a glass coverslip (Fisher Scientific catalog#12-544-D). The block was then cleaved using a razor blade attached to a micromanipulator, the two halves separated using a razor blade, and the resulting cross-sections tipped gently so that the exposed side of the colony contacted the coverslip. Samples were imaged immediately using a Leica SP5 confocal microscope and LAS AF LITE software. For light microscopy of colony sections, colonies were embedded in Spurr’s and sectioned as described previously (Piccirillo and Honigberg 2011).
Measuring colony height
Colonies were visualized with a Leica CME light microscope and a 10× objective. We recorded the rotation of the fine-focus knob necessary to shift the focal plane from the base to the top of the colony. As noted by the manufacturer, each division of the fine focus knob corresponds to movement of the objective by 3 μ. To increase contrast and enable focusing, the agar at the base of the colony and the top of the colony were stained with ink using a Sharpie. The method was verified by measuring the heights of stacks of coverslips of known thickness.
Assays for frequency of feeder cells and sporulated cells in colonies
Spot colonies were incubated for 6 days, and then resuspended in 2 M sorbitol and examined by light microscopy to determine the frequency of asci (sporulated cells). Alternatively 6 days colonies were embedded in agar containing X-gal and incubated as described (Piccirillo et al. 2015) and then these colonies were cleaved with a razor blade, cells recovered in 2 M sorbitol, and examined by light microscopy for the fraction of cells that were blue.
Statistics and reproducibility
All quantitative data in the study is expressed as the mean of three biological replicas with error bars representing the SEM. P values are from unpaired Student’s t test (raw). Experiments with rlm1Δ mutants were performed on two independently derived mutants (SH4708 and SH4767). All experiments comparing two or more strains based on scoring cells (e.g. spore formation, UASRlm1-LacZ expression) were performed as double-blind experiments. For Fig. 2, colonies grown on the two different media were grown, embedded and sectioned in two separate experiments. All other experiments were repeated at least once on a different day. For experiments involving calculating the frequency of a given cell population after re-suspending colonies, at least 250 cells were counted for each sample.
References
Ables ET, Laws KM, Drummond-Barbosa D (2012) Control of adult stem cells in vivo by a dynamic physiological environment: diet-dependent systemic factors in Drosophila and beyond. Wiley Interdiscip Rev Dev Biol 1(5):657–674. doi:10.1002/wdev.48
Albuquerque P, Casadevall A (2012) Quorum sensing in fungi—a review. Med Mycol 50(4):337–345. doi:10.3109/13693786.2011.652201
Baroiller JF, D’Cotta H, Saillant E (2009) Environmental effects on fish sex determination and differentiation. Sex Dev 3(2–3):118–135. doi:10.1159/000223077
Blank LM, Sauer U (2004) TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 150(Pt 4):1085–1093. doi:10.1099/mic.0.26845-0
Brakefield PM, Gems D, Cowen T, Christensen K, Grubeck-Loebenstein B, Keller L, Westendorp RG (2005) What are the effects of maternal and pre-adult environments on ageing in humans, and are there lessons from animal models? Mech Ageing Dev 126(3):431–438. doi:10.1016/j.mad.2004.07.013
Cap M, Stepanek L, Harant K, Vachova L, Palkova Z (2012) Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 46(4):436–448. doi:10.1016/j.molcel.2012.04.001
Cortassa S, Aon JC, Aon MA, Spencer JF (2000) Dynamics of metabolism and its interactions with gene expression during sporulation in Saccharomyces cerevisiae. Adv Microb Physiol 43:75–115. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10907555
Cullen PJ (2015) Evaluating yeast filamentous growth at the single-cell level. Cold Spring Harb Protoc 2015(3):272–275. doi:10.1101/pdb.prot085084
Danielsen ET, Moeller ME, Rewitz KF (2013) Nutrient signaling and developmental timing of maturation. Curr Top Dev Biol 105:37–67. doi:10.1016/B978-0-12-396968-2.00002-6
Eggert C (2004) Sex determination: the amphibian models. Reprod Nutr Dev 44(6): 539–549. http://www.ncbi.nlm.nih.gov/pubmed/15762298
Gagiano M, Bauer FF, Pretorius IS (2002) The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res 2(4):433–470
Gordon DM (2013) The rewards of restraint in the collective regulation of foraging by harvester ant colonies. Nature 498(7452):91–93. doi:10.1038/nature12137
Honigberg SM (2011) Cell signals, cell contacts, and the organization of yeast communities. Eukaryot Cell 10(4):466–473. doi:10.1128/EC.00313-10
Jambhekar A, Amon A (2008) Control of meiosis by respiration. Curr Biol 18(13):969–975. doi:10.1016/j.cub.2008.05.047
Jung US, Sobering AK, Romeo MJ, Levin DE (2002) Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol 46(3): 781–789. http://www.ncbi.nlm.nih.gov/pubmed/12410835
Loomis WF (2014) Cell signaling during development of Dictyostelium. Dev Biol 391(1):1–16. doi:10.1016/j.ydbio.2014.04.001
McCusker JH, Clemons KV, Stevens DA, Davis RW (1994a) Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136(4):1261–1269. http://www.ncbi.nlm.nih.gov/pubmed/8013903
McCusker JH, Clemons KV, Stevens DA, Davis RW (1994b) Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 degrees C and form pseudohyphae. Infect Immun 62(12):5447–5455. http://www.ncbi.nlm.nih.gov/pubmed/7960125
Merchant-Larios H, Diaz-Hernandez V (2013) Environmental sex determination mechanisms in reptiles. Sex Dev 7(1–3):95–103. doi:10.1159/000341936
Niklas KJ, Newman SA (2013) The origins of multicellular organisms. Evol Dev 15(1):41–52. doi:10.1111/ede.12013
Piccirillo S, Honigberg SM (2010) Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res Microbiol 161(5):390–398. doi:10.1016/j.resmic.2010.04.001
Piccirillo S, Honigberg SM (2011) Yeast colony embedding method. J Vis Exp. doi:10.3791/2510
Piccirillo S, White MG, Murphy JC, Law DJ, Honigberg SM (2010) The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184(3):707–716. doi:10.1534/genetics.109.113480
Piccirillo S, Morales R, White MG, Smith K, Kapros T, Honigberg SM (2015) Cell differentiation and spatial organization in yeast colonies: role of cell-wall integrity pathway. Genetics 201(4):1427–1438. doi:10.1534/genetics.115.180919
Shapiro JA (1998) Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi:10.1146/annurev.micro.52.1.81
Vachova L, Cap M, Palkova Z (2012) Yeast colonies: a model for studies of aging, environmental adaptation, and longevity. Oxid Med Cell Longev 2012:601836. doi:10.1155/2012/601836
Vachova L, Hatakova L, Cap M, Pokorna M, Palkova Z (2013) Rapidly developing yeast microcolonies differentiate in a similar way to aging giant colonies. Oxid Med Cell Longev 2013:102485. doi:10.1155/2013/102485
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi:10.1146/annurev.cellbio.21.012704.131001
Wilson EO, Holldobler B (1988) Dense heterarchies and mass communication as the basis of organization in ant colonies. Trends Ecol Evol 3(3):65–68. doi:10.1016/0169-5347(88)90018-3
Acknowledgments
We are grateful for the use of the confocal microscope in the University Missouri, Kansas City School of Dentistry Confocal Microscopy Core. This facility is supported by the UMKC Office of Research Services, the UMKC Center of Excellence in Dental and Musculoskeletal Tissues, and NIH grant S10RR027668. We are grateful to Dr. David Levin (Boston University) for the UASRlm1-LacZ plasmid. We thank Mr. Benjamin Iwai for sectioning embedded colonies. Research in this publication was funded by the National Institutes of General Medical Sciences of the NIH under award number R15GM094770.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Piccirillo, S., Kapros, T. & Honigberg, S.M. Phenotypic plasticity within yeast colonies: differential partitioning of cell fates. Curr Genet 62, 467–473 (2016). https://doi.org/10.1007/s00294-015-0558-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0558-y