Abstract
Retrotransposons and retroviral insertions have molded the genomes of many eukaryotes. Since retroelements transpose via an RNA intermediate, the additive nature of the replication cycle can result in massive increases in copy number if left unchecked. Host organisms have countered with several defense systems, including domestication of retroelement genes that now act as restriction factors to minimize propagation. We discovered a novel truncated form of the Saccharomyces Ty1 retrotransposon capsid protein, dubbed p22 that inhibits virus-like particle (VLP) assembly and function. The p22 restriction factor expands the repertoire of defense proteins targeting the capsid and highlights a novel host–parasite strategy. Instead of inhibiting all transposition by domesticating the restriction gene as a distinct locus, Ty1 and budding yeast may have coevolved a relationship that allows high levels of transposition when Ty1 copy numbers are low and progressively less transposition as copy numbers rise. Here, we offer a perspective on p22 restriction, including its mode of expression, effect on VLP functions, interactions with its target, properties as a nucleic acid chaperone, similarities to other restriction factors, and future directions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retrotransposons and endogenous retroviruses dominate the chromosomal landscape in many eukaryotes and comprise over 40 % of the human genome (Goodier and Kazazian 2008; Jern and Coffin 2008). The Saccharomyces cerevisiae reference genome contains long terminal repeat (LTR) retrotransposons Ty1–Ty5, yet only 3 % of budding yeast’s compact genome comprises Ty sequences (Kim et al. 1998). Ty elements are effective models to understand retroviral and eukaryotic transposon biology because they inhabit the genome of a powerful model organism and are representative of a widely disseminated family of Copia/Ty1 LTR retrotransposons that are particularly abundant in plant genomes (Lee and Kim 2014). We study Ty1 elements because they are transcriptionally active and competent for retrotransposition, can cause mutations by insertional mutagenesis or genome rearrangement, and at 32 copies, are the most abundant retrotransposon in the reference strain [see review by (Curcio et al. 2015)]. Ty1 structure and replication cycle resembles that of retroviruses, except that Ty1 transposition is not infectious (Garfinkel et al. 1985) (Fig. 1). Ty1 is transcribed from LTR to LTR, forming a genomic RNA with the structural hallmarks of retroviral genomic RNA. Ty1 RNA serves as a template for protein synthesis and reverse transcription, which occurs after packaging of the RNA into VLPs. Translation of Ty1 POL requires a specific +1 frameshift event near the end of GAG (Farabaugh 1995), resulting in a large Gag-Pol precursor (p199). Cytoplasmic foci, termed T-bodies or retrosomes (Beliakova-Bethell et al. 2006; Checkley et al. 2010; Dutko et al. 2010; Malagon and Jensen 2008), contain Ty1 RNA, Gag, presumably Gag-Pol, and perhaps specific cellular proteins (Doh et al. 2014; Suzuki et al. 2011), and are sites where VLPs assemble. Cotranslational localization of Ty1 RNA to the endoplasmic reticulum (ER) followed by Gag retrograde translocation from the ER to the cytoplasm stabilizes Gag and facilitates VLP assembly (Doh et al. 2014). Our work suggests that Gag may encounter Ty1 RNA at an earlier step since Gag increases the stability and nuclear export of GAL1-promoted Ty1 RNA (Checkley et al. 2013). Reverse transcription takes place within mature VLPs following protein processing by an element-encoded protease (PR). Ty1 PR cleaves Gag-p49 near the C-terminus to generate Gag-p45 and Gag-Pol-p199 to form mature PR, integrase (IN), and reverse transcriptase (RT). Ty1 Gag is a multifunctional protein but unlike retroviral Gag is not cleaved into functionally distinct proteins such as matrix, capsid (CA), and nucleocapsid (NC), even though Ty1 Gag executes the same functions as retroviral CA and NC. A synthetic peptide containing sequences from the C-terminal region of Gag-p45 (Cristofari et al. 2000) and recombinant Gag derivatives (Nishida et al. 2015) display nucleic acid chaperone (NAC) activity in vitro, similar to NC. Like HIV-1 (Tekeste et al. 2015; Zhu et al. 2004), a complex formed between Ty1 RT and IN is required for reverse transcription (Wilhelm and Wilhelm 2006) and tRNA Meti prime minus-strand synthesis (Keeney et al. 1995). A pre-integration complex minimally containing IN and Ty1 cDNA returns to the nucleus via a classical import pathway that recognizes a nuclear localization signal near the C-terminus of IN (Kenna et al. 1998; McLane et al. 2008; Moore et al. 1998). Ty1 usually integrates upstream of genes actively transcribed by RNA Polymerase III (Bridier-Nahmias et al. 2015; Devine and Boeke 1996). Unlike other Ty elements, Ty1 integrates into genes transcribed by Pol II at a lower efficiency and displays a preference for promoter regions (Baller et al. 2012; Mularoni et al. 2012).
Ty1 retrotransposition. The functional organization of Ty1 is at the top and the replication cycle is depicted below. Boxed triangles denote long terminal repeats (LTRs). Ty1 is transcribed from LTR to LTR by RNA polymerase II to form a 5.7-kb transcript, which is packaged into VLPs or translated to produce Gag-p49 (shaded blue) or Gag-Pol-p199, which requires a programmed +1 frameshift event near the end of GAG. Gag accumulates in the cytoplasm to become the capsid of Ty1 VLPs and also mediates nucleic acid chaperone functions during VLP assembly and reverse transcription. The Gag-Pol precursor contains the enzymes required for Ty1 transposition: protease (PR; shaded orange), integrase (IN; shaded teal), and reverse transcriptase (RT; shaded gray). Prior to VLP formation, Gag and presumably Gag-Pol form mRNA/Gag foci termed T-bodies or retrosomes, which are sites where VLPs assemble. Ty1 mRNA is specifically packaged as a dimer into VLPs. Ty1 PR cleaves Gag-p49 and Gag-Pol-p199 precursors to form mature proteins, Gag-p45, PR-p23, IN-p71, and RT-p60. Like retroviruses, an IN/RT heteromer reverse transcribes Ty1 mRNA into a linear cDNA. A pre-integration complex (PIC) minimally containing Ty1 cDNA and IN is imported into the nucleus via a bipartite nuclear localization signal present in IN. Retrotransposition is completed by IN-mediated insertion into a new location in the host genome
Ty1 copy number control (CNC)
Ty1 transposition occurs at a low rate even though most elements are transcribed (Morillon et al. 2002) and contain intact open reading frames (Curcio and Garfinkel 1991b, 1994; Kim et al. 1998). Interestingly, S. cerevisiae and its close relatives such as S. paradoxus lack eukaryotic defense mechanisms such as RNAi or host restriction factors such as the APOBEC proteins (Drinnenberg et al. 2009, 2011; Harris et al. 2012; Malim and Bieniasz 2012; Zheng et al. 2012), suggesting that Ty1 retrotransposition is limited by a novel mechanism. Early on, one model proposed to explain “transpositional dormancy” is that a chromosomal or Ty1 encoded inhibitor is titrated when Ty1 is overexpressed from the GAL1 promoter on a multicopy pGTy1 plasmid (Boeke et al. 1985; Curcio and Garfinkel 1991a; Fink et al. 1986; Garfinkel et al. 1985). Expression of pGTy1 elements harboring several different coding sequence mutations, including a frameshift mutation adjacent to the GAG initiation codon, does not change the level of genomic Ty1 transposition, suggesting that the hypothetical inhibitor is not titrated by increasing the level of Ty1 RNA (Curcio and Garfinkel 1992; Garfinkel et al. 1985, 2003). Rather, the level of mature proteins IN and PR is best correlated with the massive increase in Ty1 transposition observed by pGTy1 induction, suggesting that Ty1 transposition is regulated posttranslationally (Curcio and Garfinkel 1992).
We discovered a novel form of CNC that minimizes Ty1 movement and began to explain transpositional dormancy (Garfinkel et al. 2003). CNC is defined as a decrease in transposition when additional elements are present in a genome, and variations on this theme are observed with P-elements in Drosophila (Craig 1990) and the Tn10 transposon in bacteria (Simons and Kleckner 1983). Ty1 CNC can be saturated by overexpression of Ty1 and, surprisingly, occurs in cells containing a multicopy pGTy1 plasmid that is repressed for transcription of Ty1 mRNA (Garfinkel et al. 2003; Matsuda and Garfinkel 2009). The CNC region encompasses part of GAG as determined by mutational analyses, and the major biochemical defects in CNC+ cells include a low level of mature IN and reverse transcription products (Garfinkel et al. 2003; Matsuda and Garfinkel 2009; Purzycka et al. 2013; Saha et al. 2015). These results suggest that a titratable trans-acting factor produced by Ty1 inhibits retrotransposition and mediates CNC.
Identifying the CNC factor
Ty1 mRNA does not encode the inhibitory factor, as shown by analyses with pGTy1 summarized above (Garfinkel et al. 2003; Matsuda and Garfinkel 2009). Therefore, subgenomic transcripts containing GAG sequence may encode a protein or noncoding RNA capable of inhibiting transposition in a dose-dependent manner. Ty1 antisense (AS) RNAs map within GAG and are implicated in transcriptional repression of Ty1 (Berretta et al. 2008; Servant et al. 2012). In contrast, Ty1 mRNA levels remain unchanged in the presence of the CNC factor produced from pGTy1, suggesting that CNC occurs posttranscriptionally (Garfinkel et al. 2003; Matsuda and Garfinkel 2009). Matsuda and Garfinkel (2009) provided support for the idea that Ty1AS transcripts are necessary for CNC and specifically associate with VLPs. However, additional findings suggest that the AS transcripts may not act alone, are not truly packaged into VLPs, do not anneal with Ty1 mRNA in VLPs, and may be present in lower molar amounts when compared with the mRNA in VLPs (Matsuda and Garfinkel 2009; Purzycka et al. 2013). Together, these results fail to support a model where Ty1 mRNA is the target for the AS transcripts and invite the possibility that a Ty1 protein mediates CNC.
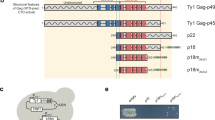
Genetic analyses of the CNC region demonstrated that several mutations abrogating CNC neither affect AS RNA levels nor map in the AS RNA transcription units (Saha et al. 2015) (Fig. 2). CNC− mutations T1108C, A1123G, A1296G, and ΔA1496 map downstream of two internal AUG codons (AUG1 and AUG2) about halfway into GAG, and all change GAG’s coding potential. A 5′ truncated Ty1 sense RNA, termed Ty1i RNA, is present in wild-type cells and Ty1i RNA is also produced by pGPOLΔ (a CNC-proficient pGTy1 lacking most of POL) used in the mutational analyses. Ty1i RNA contains 5′ ends 38 nts., upstream of AUG1. In addition, CNC− mutations T399C, Δ238–281, and Δ238–353 that map within the AS RNA transcription units (Matsuda and Garfinkel 2009) decrease the level of Ty1i RNA (Saha et al. 2015). The phenotypic screen for CNC− mutations also revealed a GAG separation-of-function allele A1123G that decreases CNC but not Ty1 transposition. Remarkably, Ty1 mobility increases rather than decreases in a Ty1-less strain repopulated with Ty1A1123G. Together, these results strongly support the idea that an altered form of Gag encoded by Ty1i RNA underlies CNC. Although these results predict that a 22-kDa protein (p22) encoded by the C-terminal half of GAG should be present, as well as a smaller protein (p18) that is produced by Ty1 PR cleavage, a commonly used VLP antiserum that reacts strongly with Gag-p49/p45 fails to reproducibly detect p22 (Adams et al. 1987; Saha et al. 2015). Therefore, we generated a new antiserum against recombinant p18 to show that both p22 and p18 are present in wild-type cells and encoded by the CNC region. Co-expression of p22/p18 and pGTy1his3-AI dramatically decreases Ty1 mobility 35,000-fold (Saha et al. 2015). Thus, the self-encoded p22 restriction factor is both necessary and sufficient for Ty1 CNC, and the postulated inhibitor responsible for transpositional dormancy.
The CNC region and Ty1 transcripts produced from pGPOLΔ. The CNC region covers GAG and the beginning of POL, as defined by deletion mapping. Locations of the GAL1 promoter (hatched rectangle), LTR (solid rectangle), Ty1, Ty1i, and Ty1 antisense (AS) transcripts, initiation codons present in Ty1i RNA, and CNC-defective mutations are noted. Included in these is the separation-of-function mutation A1123G, which affects CNC but not Ty1 transposition
Synthesis of p22/p18
Transcription and stability of Ty1i RNA is poorly understood but may respond differently to cellular genes used to produce Ty1 mRNA (Saha et al. 2015) (Fig. 2). For example, SPT3 encodes a subunit of the SAGA chromatin-remodeling complex that is implicated in TATA box recognition and is required for transcription of full-length Ty1 mRNA (Grant et al. 1997; Winston et al. 1984). Interestingly, deleting SPT3 does not alter the initiation site used for transcription of Ty1i RNA, and the level of Ty1i RNA increases in certain strains. These results suggest that Ty1 mRNA and Ty1i transcription responds differently to SPT3, and raise the possibility that there is differential recognition of the Ty1 and Ty1i promoters by SAGA. Ty1i RNA levels also increase, while Ty1 mRNA level slightly decreases in cells lacking XRN1, which encodes a 5′–3′ ribonuclease involved in RNA decay associated with P-bodies (Parker 2012), RNA transcription (Berretta et al. 2008; Haimovich et al. 2013), and assembly of functional Ty1 (Checkley et al. 2010; Dutko et al. 2010) and Ty3 (Beliakova-Bethell et al. 2006; Bilanchone et al. 2015) VLPs. Analyses of CNC− mutations within (Δ238–353, Δ238–281) or adjacent (T399C) to the 5′ LTR suggest that Ty1i and Ty1 mRNA utilize similar enhancer sequences for transcription (Curcio et al. 2015; Saha et al. 2015).
We used ribosome footprinting (Ingolia et al. 2009) and mutational analyses (Nishida et al. 2015; Saha et al. 2015) to determine whether initiation of p22 translation occurs from both AUG1 and AUG2. Ribo-seq read distribution from a published data set (Arribere and Gilbert 2013) provides evidence for initiation at AUG1 but not AUG2. However, mutational analyses using pGPOLΔ reveals that either AUG1 or AUG2 can be used to synthesize p22 proteins capable of inhibiting Ty1 mobility (Nishida et al. 2015). In vitro synthesis of p22 using a wheat germ extract confirms initiation at either AUG and also provides evidence for translation via a cap-dependent process. Together, our results suggest that translation from AUG1 involves canonical cap-dependent scanning, while leaky scanning is the most plausible mechanism for initiation at AUG2. Consistent with this interpretation is the observation that AUG2 is in a better sequence context for the yeast translational apparatus than AUG1 (Cavener and Ray 1991).
Another intriguing aspect of Ty1 CNC is the presence of p18, which is formed by cleavage of p22 by Ty1-PR at the same C-terminal site used to form Gag-p45 from Gag-p49 (Saha et al. 2015). The relative amount of p18 versus p22 is correlated with the level of PR produced from Ty1 (Tucker et al. 2015). When PR is expressed from the full ensemble of elements present in a genome or from pGTy1 induction, p18 is the predominant form, whereas in low copy strains or an spt3 mutant, only p22 is detected. Both p22 and p18 inhibit Ty1 transposition to similar extents (Nishida et al. 2015). How p22 gains access to PR within retrosomes or during VLP assembly is an interesting question, since PR is initially part of the Gag-Pol precursor in retrosomes and cleavage events to form IN and RT from Gag-Pol likely occur within VLPs.
Mechanism of restriction
Our results show that p22/p18 inhibits several steps in the process of retrotransposition prior to reverse transcription (Nishida et al. 2015; Saha et al. 2015; Tucker et al. 2015), which clarifies and greatly extends earlier work (Garfinkel et al. 2003; Matsuda and Garfinkel 2009) (Fig. 3). The challenge is to define the earliest and most inhibitory insults to the transposition process that account for the >30,000-fold decrease in mobility observed when Ty1 and p22/p18 are coexpressed in a Ty1-less strain (Saha et al. 2015). p22/p18 associates with VLPs through binding to Gag, alters other Ty1 proteins such as IN and RT, and affects VLP assembly. Ty1 Pol processing or stability and VLP yield decrease when pGTy1 and p22 are coexpressed. There is an accumulation of the POL-encoded precursors and less mature RT and IN. Aberrant forms of RT are detected and Gag appears to undergo more proteolysis as well. These unusual Ty1 proteins may result from altered processing by Ty1 PR, from cleavage by a cellular protease, or from differences in posttranslational modifications. Ty1 reverse transcriptase activity is lower and more broadly distributed following sucrose gradient sedimentation of cell extracts, reflecting alterations in VLP assembly as well as loss of mature IN. p22/p18 changes VLP morphology in a fraction of the particles as determined by negative staining. The restriction factor also disrupts pGTy1-induced (Saha et al. 2015) and endogenous retrosomes (unpublished data), and colocalizes with Gag.
Ty1 VLP functions inhibited by p22. An abbreviated Ty1 replication cycle is shown (also refer to Fig. 1) highlighting steps affected by p22 (shaded green). When p22 and Ty1 are co-expressed, p22 colocalizes with Gag and disrupts retrosome foci. p22 and p18, which is derived from p22 by Ty1 PR cleavage, associates with Ty1 VLPs and alters assembly and maturation
The functional organization of Ty1 Gag is not well understood (Fig. 4), therefore, mapping the regions responsible for Gag:p22/p18 interaction and CNC has revealed more information about GAG-encoded proteins (Nishida et al. 2015; Saha et al. 2015; Tucker et al. 2015). GST pulldown and co-immunoprecipitation analyses reveal interactions between p22/p18 and Gag. A segment of Gag encompassing p22 and additional N-terminal sequence, termed CTR, interacts with p18 as well as with a segment of p18 that lacks part of NAC region (sAUG1) (Nishida et al. 2015). Interestingly, only AUG1p22/p18, AUG1p18, and sAUG1 inhibit Ty1 mobility more than 5,000-fold, while expression of Gag-p45 or derivatives containing the N- and C-terminal regions of Gag produces comparable levels of protein but inhibits Ty1 mobility less than 10-fold.
Functional organization of Ty1 Gag. Gag-p49/p45 coding sequences are depicted at the top and show the TYA, CNCR, and UBN2 domains, a highly conserved W184 codon, the location of initiation codons responsible for p22 synthesis, and the nucleic acid chaperone region. Below are the restriction factor p22/p18, the Gag C-terminal region (CTR) that displays nucleic acid chaperone activity, and a short form of p18 (sAUG1) that inhibits Ty1 mobility and associates with the CTR but lacks nucleic acid chaperone activity
To define a target for p22/p18, we characterized missense mutations in Ty1 that confer partial resistance to CNC but do not markedly alter mobility in the absence of p22 (Tucker et al. 2015). The strongest CNCR mutations cluster in a region of GAG (termed the CNCR domain) (Fig. 4), which is present in the CTR and separate from p22 coding sequence, while other CNCR mutations map within the Gag UBN2 domain near the C-terminus. We did not recover resistance mutations in the yeast-specific TYA domain, which corresponds to an unstructured region encoded in the 5′ half of GAG. The CNCR domain contains a tryptophan residue (W184) conserved in domain A of Ty1/Copia elements from disparate organisms (Peterson-Burch and Voytas 2002). When W184 is changed to alanine, Ty1 mobility is abolished and VLP assembly is defective. The behavior of Ty1 Gag W184A and additional mutations in the adjacent UBN2 domain strongly suggest that p22 disturbs a central function of Gag during VLP assembly. Furthermore, CNCR mutations alter the cofractionation of p22/p18 and Gag, where the restriction factor remains mostly unprocessed and is excluded from fractions containing CNCR VLPs. Together, our results suggest that the CNCR alleles restore productive Gag–Gag interactions in the presence of p22 during VLP assembly. Furthermore, Gag:p22/p18 interactions during VLP assembly likely represent the earliest step in Ty1 transposition altered by p22.
In addition to its function as the CA for VLPs, Ty1 Gag probably contains retroviral-like NAC functions that participate in forming dimeric Ty1 RNA, packaging RNA into VLPs, annealing the tRNA Meti primer to the RNA template, and enhancing DNA strand transfers during reverse transcription. Both full-length Gag and p22/p18 contain the NAC region originally defined by the activity of a synthetic peptide (Cristofari et al. 2000) (Fig. 4). We analyzed the NAC activity of Gag and p18 derivatives and determined whether p18 impacts Ty1 RNA transactions necessary for retrotransposition using a variety of approaches (Nishida et al. 2015). The CTR displays robust NAC activity comparable to that observed with mature Gag-p45 (data not shown). Interestingly, AUG1p18 and AUG2p18 proteins display different properties even though they both contain the NAC region but differ by only 10 residues. AUG1p18 shows highly reduced NAC activity but specific binding to RNA, whereas AUG2p18 shows the converse behavior. However, the two forms of the restriction protein may possess additional properties when expressed together in vivo. AUG2p18 lacking NAC activity (sAUG2) inhibits CTR-mediated annealing of tRNA Meti to Ty1 RNA, and p22/p18 antagonizes Ty1 RNA dimerization and packaging. Our results support the view that p22/p18 also inhibits essential Ty1 RNA structural transitions through p22/p18 interactions with Gag and possibly with Ty1 RNA.
A new paradigm for inhibiting gag function
p22/p18-mediated restriction may have evolved quickly after an ancestral Saccharomyces lineage lost the evolutionarily conserved RNAi pathway (Drinnenberg et al. 2009, 2011), since expression of p22 is likely all that is required to establish CNC. In contrast, other retroelement genes have been domesticated by their host to provide new cellular functions (Kaneko-Ishino and Ishino 2012). The prototypic Gag-like restriction factors Fv1 and enJS56A1 block replication of murine leukemia virus (MLV) and Jaagsiekte sheep retrovirus (JSRV), respectively, by interacting with viral proteins during infection (Arnaud et al. 2007; Best et al. 1996; Hilditch et al. 2011), and share features in common with CNC of Ty1 by p22/p18. Fv1 is derived from the GAG gene of a member of the HERV-L family of human and murine endogenous retroviruses (Benit et al. 1997; Best et al. 1996; Qi et al. 1998). Fv1 inhibits progression of the MLV life cycle following infection and reverse transcription, but prior to integration. Although the infecting viral Gag protein as well as Fv1 determines the level of restriction, an ordered assembly of Gag is required for efficient Fv1 binding (Goldstone et al. 2014; Hilditch et al. 2011). In contrast, p22/p18 affects VLP assembly and function, whereas Fv1 inhibits a different step in the replication cycle that occurs early post-infection prior to integration. Conceptually similar to MLV-Fv1 restriction, the sheep genome harbors about 20 copies of endogenous (en) JSRVs and these sequences are homologous with exogenous JSRV virus that causes lung cancer. Certain endogenous copies have evolved a trans-dominant Gag protein enJS56A that like Ty1 p22 blocks replication posttranslationally. The JSRV:enJS56A interaction prevents Gag from entering into an endosome trafficking pathway, and results in aggregation and turnover by the proteasome (Arnaud et al. 2007; Murcia et al. 2007). Indeed, Dolly the cloned sheep contracted lung cancer caused by JSRV because she lacked enJS56A (Leroux et al. 2007).
Retroviral studies involving sensitivity and escape from host restriction factors show similarities to Ty1 CNCR mutants. Although Gag CNCR mutations rescue Ty1 retrotransposition in the presence of p22, they do not interfere with Gag:p22 binding (Tucker et al. 2015). Similarly, Mx2 restriction of HIV-1 requires Mx2:CA binding, yet known Mx2 escape mutations in the CA gene do not significantly alter binding between Mx2 and CA (Fricke et al. 2014; Schulte et al. 2015). It is clear in both cases that escape mutations can promote retroelement replication in ways distinct from the disruption of restriction factor-target binding. Separately, we found that artificial adjustments of Gag:p22 ratios by altering the level of expression influence the robustness of CNC. Resistance to the sheep restriction factor enJS56A1 is achieved by the provirus enJSRV26 simply by increasing proviral expression due to a mutation in the envelope glycoprotein (Armezzani et al. 2011). Interestingly, GAG CNCR mutations do not affect steady-state Gag levels, although it seems logical that this evasion strategy would prove fruitful. However, it is still possible that the ratio of Gag:p22 is specifically higher within retrosomes comprising CNCR Gag as compared to wild-type Gag.
Our results also illustrate the delicate balance between resistance to p22 and fitness of Ty1 (Tucker et al. 2015). Since all p22 sequences are present in Gag-p49 (Fig. 4), surfaces or protein domains that interact with p22 may be the same or overlap with domains important for Gag function, and would likely constrain an “arms race” between p22 and its target protein Gag. In fact, several CNCR GAG mutations result in decreased Ty1 fitness. This is similar to studies involving HIV-1 CA resistance to the independently encoded restriction factors TRIM5α and Mx2, which demonstrate that resistance mutations in CA can have detrimental effects on HIV fitness (Fricke et al. 2014; Rihn et al. 2013; Soll et al. 2013). HIV-1 CA’s sensitivity to mutation or genetic fragility is comparable to what we see with Ty1 Gag, as our CNCR analysis supports the idea that Ty1 Gag is under strict structural constraint (Tucker et al. 2015). Lastly, since Ty1 GAG or p22 sequences have not yet been detected as a domesticated gene, the graduated rate provided by CNC may benefit Saccharomyces and Ty1, such as extending chronological lifespan (VanHoute and Maxwell 2014).
Future directions
There is much to be learned about the mechanism of Ty1/p22 restriction and whether this strategy is employed by retroelements elsewhere. Investigating VLP assembly and p22, Gag-p45, and VLP structure will help us understand how p22 alters VLP functions. In particular, cryo-electron microscopy approaches have advanced tremendously (Bai et al. 2015) since Ty1 VLPs were last examined (AL-Khayat et al. 1999; Burns et al. 1992; Palmer et al. 1997). Further understanding of VLP assembly and CNC will be gained by identifying cellular proteins that interact with Gag or p22 and may provide clues as to whether CNC is regulated. Ty1-Gag:p22 restriction is perfectly suited for investigating how RNA structure dictates function, and reinforces the idea that both strong and weaker defects mediated by p22/p18 contribute to the severe inhibition of Ty1 movement. We are also determining how Ty1 CNC is distributed in wild S. paradoxus and S. cerevisiae isolates using functional tests and high-throughput sequencing, and discovering a Ty1-like restriction factor in unrelated organisms is a reasonable expectation given the track record of the Ty/yeast paradigm.
References
Adams SE, Mellor J, Gull K, Sim RB, Tuite MF, Kingsman SM, Kingsman AJ (1987) The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49:111–119
AL-Khayat HA, Bhella D, Kenney JM, Roth JF, Kingsman AJ, Martin-Rendon E, Saibil HR (1999) Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J Mol Biol 292:65–73
Armezzani A, Arnaud F, Caporale M, di Meo G, Iannuzzi L, Murgia C, Palmarini M (2011) The signal peptide of a recently integrated endogenous sheep betaretrovirus envelope plays a major role in eluding Gag-mediated late restriction. J Virol 85:7118–7128. doi:10.1128/JVI.00407-11
Arnaud F, Murcia PR, Palmarini M (2007) Mechanisms of late restriction induced by an endogenous retrovirus. J Virol 81:11441–11451. doi:10.1128/JVI.01214-07
Arribere JA, Gilbert WV (2013) Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23:977–987. doi:10.1101/gr.150342.112
Bai X-C, McMullan G, Scheres SHW (2015) How cryo-EM is revolutionizing structural biology. Trend Biochem Sci 40:49–57. doi:10.1016/j.tibs.2014.10.005
Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF (2012) A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res 22:704–713. doi:10.1101/gr.129585.111
Beliakova-Bethell N, Beckham C, Giddings TH, Winey M, Parker R, Sandmeyer S (2006) Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94–101. doi:10.1261/rna.2264806
Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T (1997) Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a Gag coding sequence closely related to the Fv1 restriction gene. J Virol 71:5652–5657
Berretta J, Pinskaya M, Morillon A (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Gene Dev 22:615–626. doi:10.1101/gad.458008
Best S, Le Tissier P, Towers G, Stoye JP (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi:10.1038/382826a0
Bilanchone V, Clemens K, Kaake R, Dawson AR, Matheos D, Nagashima K, Sitlani P, Patterson K, Chang I, Huang L, Sandmeyer S (2015) Ty3 retrotransposon hijacks mating yeast RNA processing bodies to infect new genomes. PLoS Genet 11:e1005528
Boeke JD, Garfinkel DJ, Styles CA, Fink GR (1985) Ty elements transpose through an RNA intermediate. Cell 40:491–500
Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Saïb A, Werner M, Voytas DF, Lesage P (2015) An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348:585–588. doi:10.1126/science.1259114
Burns N, Saibil H, White N, Pardon J, Timmins P, Richardson S, Richards B, Adams S, Kingsman S, Kingsman A (1992) Symmetry, flexibility and permeability in the structure of yeast retrotransposon virus-like particles. EMBO J 11:1155–1164
Cavener DR, Ray SC (1991) Eukaryotic start and stop translation sites. Nucleic Acid Res 19:3185–3192
Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ (2010) P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol 30:382–398. doi:10.1128/MCB.00251-09
Checkley MA, Mitchell JA, Eizenstat LD, Lockett SJ, Garfinkel DJ (2013) Ty1 Gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 14:57–69. doi:10.1111/tra.12013
Craig NL (1990) P element transposition. Cell 62:399–402
Cristofari G, Ficheux D, Darlix J-L (2000) The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 275:19210–19217. doi:10.1074/jbc.M001371200
Curcio MJ, Garfinkel DJ (1991a) Regulation of retrotransposition in Saccharomyces cerevisiae. Mol Microbiol 5:1823–1829
Curcio MJ, Garfinkel DJ (1991b) Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA 88:936–940
Curcio MJ, Garfinkel DJ (1992) Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol 12:2813–2825
Curcio MJ, Garfinkel DJ (1994) Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136:1245–1259
Curcio MJ, Lutz S, Lesage P (2015) The Ty1 LTR-retrotransposon of budding yeast. Microbiol spectr 3:1–35. doi:10.1128/microbiolspec.MDNA3-0053-2014
Devine SE, Boeke JD (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Gene Dev 10:620–633
Doh JH, Lutz S, Curcio MJ (2014) Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet 10:e1004219. doi:10.1371/journal.pgen.1004219
Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel D (2009) RNAi in budding yeast. Science 326:544–550. doi:10.1126/science.1176945
Drinnenberg IA, Fink GR, Bartel DP (2011) Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333:1592. doi:10.1126/science.1209575
Dutko JA, Kenny AE, Gamache ER, Curcio MJ (2010) 5′ and 3′ mRNA decay factors colocalize with Ty1 Gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol 84:5052–5066. doi:10.1128/JVI.02477-09
Farabaugh PJ (1995) Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem 270:10361–10364
Fink GR, Boeke JD, Garfinkel DJ (1986) The mechanism and consequences of retrotransposition. Trends in Genetics. 2:118–123
Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F (2014) MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi:10.1186/PREACCEPT-6453674081373986
Garfinkel DJ, Boeke JD, Fink GR (1985) Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:502–517
Garfinkel DJ, Nyswaner K, Wang J, Cho J-Y (2003) Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165:83–99
Goldstone DC, Walker PA, Calder LJ, Coombs PJ, Kirkpatrick J, Ball NJ, Hilditch L, Yap MW, Rosenthal PB, Stoye JP, Taylor IA (2014) Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci USA 111:9609–9614. doi:10.1073/pnas.1402448111
Goodier J, Kazazian H (2008) Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23–35
Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Gene Dev 11:1640–1650
Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M (2013) Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153:1000–1011. doi:10.1016/j.cell.2013.05.012
Harris RS, Hultquist JF, Evans DT (2012) The restriction factors of human immunodeficiency virus. J Biol Chem 287:40875–40883. doi:10.1074/jbc.R112.416925
Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA, Stoye JP (2011) Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci USA 108:5771–5776. doi:10.1073/pnas.1100118108
Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. doi:10.1126/science.1168978
Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732
Kaneko-Ishino T, Ishino F (2012) The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front Microbiol 3:262. doi:10.3389/fmicb.2012.00262
Keeney JB, Chapman KB, Lauermann V, Voytas DF, Aström SU, von Pawel-Rammingen U, Bystrom A, Boeke JD (1995) Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol 15:217–226
Kenna MA, Brachmann CB, Devine SE, Boeke JD (1998) Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol 18:1115–1124
Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8:464–478
Lee S-I, Kim N-S (2014) Transposable elements and genome size variations in plants. Genom Inf 12:87–97
Leroux C, Girard N, Cottin V, Greenland T, Mornex J-F, Archer F (2007) Jaagsiekte sheep retrovirus (JSRV): from virus to lung cancer in sheep. Vet Res 38:211–228. doi:10.1051/vetres:2006060
Malagon F, Jensen TH (2008) The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 28:6022–6032. doi:10.1128/MCB.00684-08
Malim MH, Bieniasz PD (2012) HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. doi:10.1101/cshperspect.a006940
Matsuda E, Garfinkel DJ (2009) Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci USA 106:15657–15662. doi:10.1073/pnas.0908305106
McLane LM, Pulliam KF, Devine SE, Corbett AH (2008) The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acid Res 36:4317–4326. doi:10.1093/nar/gkn383
Moore SP, Rinckel LA, Garfinkel DJ (1998) A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol 18:1105–1114
Morillon A, Bénard L, Springer M, Lesage P (2002) Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol Cell Biol 22:2078–2088
Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD (2012) Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res 22:693–703. doi:10.1101/gr.129460.111
Murcia PR, Arnaud F, Palmarini M (2007) The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J Virol 81:1762–1772. doi:10.1128/JVI.01859-06
Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, Saha A, Gumna J, Garfinkel DJ, Purzycka KJ (2015) Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acid Res 43:7414–7431. doi:10.1093/nar/gkv695
Palmer KJ, Tichelaar W, Myers N, Burns NR, Butcher SJ, Kingsman AJ, Fuller SD, Saibil HR (1997) Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J Virol 71:6863–6868
Parker R (2012) RNA degradation in Saccharomyces cerevisiae. Genetics 191:671–702. doi:10.1534/genetics.111.137265
Peterson-Burch BD, Voytas DF (2002) Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol Biol Evol 19:1832–1845
Purzycka KJ, Legiewicz M, Matsuda E, Eizentstat LD, Lusvarghi S, Saha A, Le Grice SF, Garfinkel DJ (2013) Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acid Res 41:463–473. doi:10.1093/nar/gks983
Qi CF, Bonhomme F, Buckler-White A, Buckler C, Orth A, Lander MR, Chattopadhyay SK, Morse HC (1998) Molecular phylogeny of Fv1. Mamm Genome 9:1049–1055
Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD (2013) Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog 9:e1003461. doi:10.1371/journal.ppat.1003461
Saha A, Mitchell JA, Nishida Y, Hildreth JE, Ariberre JA, Gilbert WV, Garfinkel DJ (2015) A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 89:3922–3938. doi:10.1128/JVI.03060-14
Schulte B, Buffone C, Opp S, Di Nunzio F, De Souza Augusto, Aranha Vieira D, Brandariz-Nunez A, Diaz-Griffero F (2015) Restriction of HIV-1 requires the N-terminal region of MxB/Mx2 as a capsid-binding motif but not as a nuclear localization signal. J Virol. doi:10.1128/JVI.00753-15
Servant G, Pinson B, Tchalikian-Cosson A, Coulpier F, Lemoine S, Pennetier C, Bridier-Nahmias A, Todeschini A-L, Fayol H, Daignan-Fornier B, Lesage P (2012) Tye7 regulates yeast Ty1 retrotransposon sense and antisense transcription in response to adenylic nucleotides stress. Nucleic Acid Res 40:5271–5282. doi:10.1093/nar/gks166
Simons RW, Kleckner N (1983) Translational control of IS10 transposition. Cell 34:683–691
Soll SJ, Wilson SJ, Kutluay SB, Hatziioannou T, Bieniasz PD (2013) Assisted evolution enables HIV-1 to overcome a high TRIM5alpha-imposed genetic barrier to rhesus macaque tropism. PLoS Pathog 9:e1003667. doi:10.1371/journal.ppat.1003667
Suzuki K, Morimoto M, Kondo C, Ohsumi Y (2011) Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell 21:358–365. doi:10.1016/j.devcel.2011.06.023
Tekeste SS, Wilkinson TA, Weiner EM, Xu X, Miller JT, Le Grice SFJ, Clubb RT, Chow SA (2015) Interaction between reverse transcriptase and integrase is required for reverse transcription during HIV-1 replication. J Virol 89:12058–12069
Tucker JM, Larango ME, Wachsmuth LP, Kannan N, Garfinkel DJ (2015) The Ty1 retrotransposon restriction factor p22 targets Gag. PLoS Genet 11:e1005571. doi:10.1371/journal.pgen.1005571
VanHoute D, Maxwell PH (2014) Extension of Saccharomyces paradoxus chronological lifespan by retrotransposons in certain media conditions is associated with changes in reactive oxygen species. Genetics 198:531–545. doi:10.1534/genetics.114.168799
Wilhelm M, Wilhelm F-X (2006) Cooperation between reverse transcriptase and integrase during reverse transcription and formation of the preintegrative complex of Ty1. Eukaryot Cell 5:1760–1769. doi:10.1128/EC.00159-06
Winston F, Durbin KJ, Fink GR (1984) The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675–682
Zheng YH, Jeang KT, Tokunaga K (2012) Host restriction factors in retroviral infection: promises in virus-host interaction. Retrovirology 9:112. doi:10.1186/1742-4690-9-112
Zhu K, Dobard C, Chow SA (2004) Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol 78:5045–5055
Acknowledgments
This work was supported by the National Science Center, Poland [2011/01/D/NZ1/03478, 2012/06/A/ST6/00384]; Foundation for Polish Science [HOMING PLUS/2012-6/12 to K.J.P.]; Ministry of Science and Higher Education, Poland [0492/IP1/2013/72 to K.J.P.]; National Institutes of Health [GM095622 to D.J.G.]; National Science Foundation Graduate Fellowship [1011RH25213 to J.M.T.]; and the University of Georgia Research Foundation [to D.J.G.]. We also thank Wioletta Czaja for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Garfinkel, D.J., Tucker, J.M., Saha, A. et al. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces . Curr Genet 62, 321–329 (2016). https://doi.org/10.1007/s00294-015-0550-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0550-6