Abstract
The goal of this work is to characterize membrane transporter genes in Cercospora fungi required for autoresistance to the photoactivated, active-oxygen-generating toxin cercosporin they produce for infection of host plants. Previous studies implicated a role for diverse membrane transporters in cercosporin resistance. In this study, transporters identified in a subtractive cDNA library between a Cercospora nicotianae wild type and a cercosporin-sensitive mutant were characterized, including two ABC transporters (CnATR2, CnATR3), an MFS transporter (CnMFS2), a uracil transporter, and a zinc transport protein. Phylogenetic analysis showed that only CnATR3 clustered with transporters previously characterized to be involved in cercosporin resistance. Quantitative RT-PCR analysis of gene expression under conditions of cercosporin toxicity, however, showed that only CnATR2 was upregulated, thus this gene was selected for further characterization. Transformation and expression of CnATR2 in the cercosporin-sensitive fungus Neurospora crassa significantly increased cercosporin resistance. Targeted gene disruption of CnATR2 in the wild type C. nicotianae, however, did not decrease resistance. Expression analysis of other transporters in the cnatr2 mutant under conditions of cercosporin toxicity showed significant upregulation of the cercosporin facilitator protein gene (CFP), encoding an MFS transporter previously characterized as playing an important role in cercosporin autoresistance in Cercospora species. We conclude that cercosporin autoresistance in Cercospora is mediated by multiple genes, and that the fungus compensates for mutations by up-regulation of other resistance genes. CnATR2 may be a useful gene, alone or in addition to other known resistance genes, for engineering Cercospora resistance in crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The purpose of this work is to characterize the role of membrane transporters in auto-resistance of plant pathogenic Cercospora fungi to the photoactivated toxin cercosporin they produce for infection of host plants. Cercospora species cause devastating leaf-blighting diseases on a wide range of important plant host species worldwide. Much of the reason for their success and broad host range is attributed to their production of the perylenequinone toxin cercosporin (Daub and Chung 2009; Daub et al. 2013). In the presence of light, cercosporin absorbs light energy and reacts with oxygen, generating reactive oxygen species (ROS) such as singlet oxygen (1O2) and superoxide (O2 −), which kill cells by damaging critical cellular components. Due to its generation of reactive oxygen species, particularly 1O2, cercosporin has broad-spectrum toxicity to almost all organisms including mice, bacteria, and many fungi in addition to host and non-host plants. The producing fungi are resistant, however. Thus, the mechanisms used by Cercospora fungi to tolerate cercosporin and the ROS it produces are of significant interest, as Cercospora auto-resistance genes may be of use in engineering crop plants for resistance to these pathogens. Similar photoactivated perylenequinones are also produced by other plant pathogens including species of Alternaria, Cladosporium, and Elsinoë. Thus, identification of resistance mechanisms against perylenequinones has the potential to provide control strategies against diverse fungal pathogens.
Multiple strategies have been used by our laboratory and others to identify the mechanisms used by Cercospora fungi for resistance to cercosporin. Studies on production of 1O2 quenchers, antioxidants, and antioxidant enzymes have documented an important role in resistance for these compounds, particularly for carotenoids and vitamin B6 (pyridoxal phosphate and its vitamers), which are potent 1O2 quenchers (Daub et al. 2013; Ehrenshaft et al. 1999). Transformation of tobacco to express Cercospora vitamin B6 biosynthetic genes, however, was unsuccessful in elevating levels of cercosporin resistance (Herrero and Daub 2007). Studies with redox-sensitive dyes, reducing agents, and with detection of reduced and oxidized forms of cercosporin (Daub et al. 1992, 2013) also documented the key role of redox potential and the maintenance of the non-photoactive reduced form of cercosporin within fungal hyphae as a core mechanism of cercosporin resistance. In support of these studies, Panagiotis and co-workers demonstrated that overexpression of a yeast FAD pyridine nucleotide reductase gene (Cpd1) increased cercosporin resistance in yeast and tobacco (Panagiotis et al. 2007; Ververidis et al. 2001).
Of all possible resistance mechanisms, the role of membrane transporters has been the most studied. Membrane transport proteins efflux a wide array of molecules across membranes (Dahl et al. 2004), and have been implicated in resistance to both endogenously produced and exogenously applied toxic agents. In filamentous fungi, there are two main families of transporters: the ATP-binding cassette (ABC) family and the major facilitator superfamily (MFS) transporters (Higgins 2007; Pao et al. 1998). Both these transporter families have common transmembrane (TM) domains arranged in two homologous halves (Jones and George 2002). ABC transporters also contain an ATP-binding cassette, which can transport both small molecules and macromolecules. Translocation is facilitated by energy from the ATP molecule which becomes bound to the ABC transporter during the efflux process (Higgins 1992). MFS transporters use chemiosmotic ion gradients for the translocation of small molecules (Pao et al. 1998). Both MFS and ABC transporters have been shown to provide resistance to cercosporin. These include the MFS transporters CFP (cercosporin facilitator protein) from C. kikuchii (Callahan et al. 1999), Bcmfs1 from Botrytis cinerea (Hayashi et al. 2002), and MgMfs1 from Mycosphaerella graminicola (Roohparvar et al. 2007). ABC transporters shown to play a role in cercosporin resistance include ATR1 from Cercospora nicotianae (Amnuaykanjanasin and Daub 2009) and Snq2p from yeast (Ververidis et al. 2001). The precise mechanism(s) by which these transporters impart resistance is not known, but is hypothesized to be due to transport of cercosporin out of the cell.
In our laboratory, studies to identify cercosporin resistance genes in Cercospora species began with the isolation and characterization of C. nicotianae mutants selected for sensitivity to cercosporin (Jenns and Daub 1995; Jenns et al. 1995). Complementation of the cercosporin-sensitive phenotype led to the identification of a gene encoding a zinc-cluster transcription factor named CRG1 (cercosporin resistance gene 1) (Chung et al. 2003). The cncrg1-null mutant (205C3) is highly sensitive to cercosporin and also reduced in cercosporin production. To identify putative cercosporin resistance genes regulated by CRG1, a subtractive cDNA library was generated between the C. nicotianae wild type (WT) and the cncrg1 null mutant (Herrero et al. 2007). The resulting library contained 185 expressed sequence tags (ESTs) representing genes altered in their regulation between WT and the cncrg1 mutant, as validated by quantitative RT-PCR (RT-qPCR) analysis. ESTs from the subtraction library were sequenced, classified into functional categories, and characterized as putative resistance genes based on the above-described known mechanisms of cercosporin resistance including reductive detoxification of cercosporin, antioxidants and quenchers of superoxide and singlet oxygen, and efflux out of the cell by membrane transporters. Interestingly, two genes in the Cercospora cercosporin biosynthetic cluster (CTB2, CTB5) were also found in the library. This cluster, called Cercosporin Toxin Biosynthesis (CTB), consists of eight genes encoding a polyketide synthase (CTB1), two methyltransferases (CTB2 and CTB3), three oxidoreductases (CTB5, CTB6 and CTB7), a membrane transporter (CTB4) and a transcription factor (CTB8) (Chen et al. 2007a). To date, however, there is no evidence to document a role of the biosynthetic cluster genes in resistance, as a ctb8 transcription factor null mutant, downregulated for all core cluster genes, is not altered in cercosporin resistance (Chen et al. 2007a).
The goal of this work is to further document the possible role of membrane transporters identified in the C. nicotianae subtractive library for their ability to impart cercosporin resistance. In total, eight genes encoding membrane transporters were identified in our subtractive library between C. nicotianae WT and the cercosporin-sensitive cncrg1-null mutant, three of which (the ABC transporter ATR1 and the MFS transporters CFP and CTB4) have previously been characterized (Amnuaykanjanasin and Daub 2009; Callahan et al. 1999; Choquer et al. 2007). Here, we report sequence and phylogenetic characterization of the five remaining predicted transporter genes recovered in the subtractive library: CnATR2 (ABC Transporter 2), CnATR3 (ABC Transporter 3), CnMFS2 (MFS transporter 2), a uracil transporter, and a zinc transport protein. Expression analysis under conditions of cercosporin toxicity indicated a probable role for CnATR2 in cercosporin resistance, so its role was characterized through expression in the cercosporin-sensitive fungus Neurospora crassa as well as by mutant analysis in C. nicotianae.
Materials and methods
Fungal strains, culture conditions, and plasmids
All strains of C. nicotianae including the WT strain ATCC18366 were maintained on complete medium (CM) agar or potato dextrose agar (PDA; Difco, Sparks, MD) as previously described (Jenns and Daub 1995). Neurospora crassa ORS-6a (Fungal Genetics Stock Center) was maintained on Vogel’s medium (Vogel 1956). Cercospora nicotianae mutants used in this work include mutants for the CnATR1 ABC transporter (Amnuaykanjanasin and Daub 2009), the CnCRG1 zinc-cluster transcription factor (Chung et al. 2003), and the CnCTB1 polyketide synthase in the cercosporin biosynthesis cluster (Chen et al. 2007a).
Cloning, DNA plasmid isolation, restriction enzyme analysis, and ligation used standard molecular techniques (Deininger 1990). Standard PCR was performed using Taq polymerase (Denville Scientific Inc., NJ; Invitrogen, Carlsbad, CA, USA) or High Fidelity Taq polymerase (Invitrogen) and gene-specific primers (Table 1). The plasmids pGEM-T Easy (Promega, Madison, WI, USA) or pCB1636 (Sweigard et al. 1997), which harbors the hph gene encoding for hygromycin B phosphotransferase, were used as recipient vectors for construction of recombinant plasmids for cloning, sequencing and/or fungal transformation. Escherichia coli strain DH5-α was used to maintain all plasmids.
Cloning and sequencing of transporter genes
Different methods were performed to clone and sequence the full-length genomic sequences of the transporter genes based on partial EST sequences recovered in the subtractive library between C. nicotianae WT and the cncrg1 mutant (Herrero et al. 2007). Cloning of CnATR2 and CnMFS2 was performed using inverse PCR (Keim et al. 2004). For genes encoding the uracil transporter, the zinc transport protein and CnATR3, primers specific to homologs from the closely related maize pathogen Cercospora zeae-maydis (http://www.jgi.doe.gov/) were designed to amplify and sequence full-length copies of C. nicotianae transporter genes. Intronic regions in C. nicotianae were identified from genomic sequences in C. zeae-maydis, and Genescan (Burge 1998) was used to confirm the locations of introns.
DNA sequencing was performed by Eton Biosciences Inc. (San Diego, CA, USA) and Macrogen USA (Rockville, MD, USA). Sequence analyses were performed using the following programs: BLAST (the US National Center for Biotechnology Information); ExPASy Proteomics tools (Swiss Institute of Bioinformatics, Geneva, Switzerland); NEB cutter V2.0 (New England BioLabs, Beverly, MA, USA); and Search Launcher software (Baylor College of Medicine). Protein homologies were identified using protein BLAST. Nucleotide and amino acid sequences of CnATR2, CnATR3, CnMFS2, zinc transport protein gene and the uracil transporter gene are in the GenBank database with the accession numbers GU646036.1, KC959476, GU646037, KC959477, AF306523.1, respectively.
Phylogenetic analysis of transporter genes
CnATR2, CnATR3 and CnMFS2 protein sequences were searched for similarity to known proteins using tBLASTx (the US National Center for Biotechnology Information). Putative introns were predicted using similarity search results and the presence of the GT/AG 5′ and 3′ intron splicing sites (Jacobs and Stahl 1995) and were digitally removed. Deduced amino acid sequences of CnATR2, CnATR3 and CnMFS2 were aligned with known ABC and MFS transporters in the NCBI database using CLUSTALX package 1.81 (Thompson et al. 1997). Phylogenetic analysis was performed using the Neighbor Joining Method in the CLUSTAL X. The phylogenetic trees were viewed and edited with NJPLOT (Thompson et al. 1997).
Gene expression analysis in Cercospora
Differential expression of CnATR2 between WT and the cncrg1 mutant (Chung et al. 2003) was assayed as previously described (Amnuaykanjanasin and Daub 2009).
Assays for expression under conditions of cercosporin toxicity utilized disruption mutants for CnCTB1 (cnctb1), CnATR1 (cnatr1) (Amnuaykanjanasin and Daub 2009), and for CnATR2 (cnatr2). Homogenized mycelial plugs were used to inoculate 100 ml of PDB, and cultures were grown on a shaker (250 rpm) at room temperature for 3 days under dark conditions. Cultures were then treated with either 10 µM cercosporin or with 0.5 % acetone (used to solubilize cercosporin). After cercosporin or acetone treatment, cultures were incubated under high light intensities (55–65 µE m2/s) before harvesting mycelia for RNA extraction and RT-qPCR analysis.
Total RNA was extracted from lyophilized and homogenized mycelia with TRI-REAGENT (Sigma-Aldrich, St Louis, MO, USA) following the manufacturer’s recommendation with some modifications. Total nucleic acids were purified by phenol–chloroform extraction and ethanol precipitation following standard protocols. To obtain highly purified RNA, an LiCl and ethanol precipitation steps were performed, after which samples were treated with DNAse Turbo DNA-free™ Kit (Ambion, Austin, TX) according to the manufacturer’s recommendations.
Protocols for cDNA synthesis and qPCR were same as used in Beseli et al. (2015). Each sample was normalized against the following C. nicotianae-specific internal controls: 18S rRNA for cnctb1 and actin for cncrg1, cnatr1, cnatr2. Primers used to amplify each gene are shown in Table 1. Fold changes were calculated according to the 2−ΔΔCT method (Livak and Schmittgen 2001; Schmittgen et al. 2000; Winer 1999). Two independent experiments with three replications each were performed.
Transformation of N. crassa with CnATR2
The fungal transformation plasmid pTxA-1 (Amnuaykanjanasin and Daub 2009) containing the hph resistance gene under the control of a TrpC promoter (HygR cassette) was used to insert the CnATR2 gene under the control of the Pyrenophora tritici-repentis constitutive ToxA promoter (Lorang et al. 2001). A full-length copy of CnATR2 was amplified using primers 14c-start2 (containing XbaI) and 14c-3end (Table 1). This PCR fragment was digested with XbaI, and ligated to the pTxA-1 that was digested with XbaI and SmaI. The resulting plasmid (pTxA-14c) was sequenced to confirm the presence of an intact CnATR2 sequence. The pTxA-14c plasmid was transformed into N. crassa protoplasts. Protoplasts were generated as previously described (Beseli et al. 2015).
For transformation, a total of 2 × 107 protoplasts were incubated with 10 μg of plasmid DNA and regenerated in the presence of 100 µg/ml hygromycin. Single hygromycin (hyg)-resistant colonies that formed after 2–3 days were transferred to new plates and were continuously maintained under selection (100 µg/ml hygromycin) for subsequent manipulations. Transformation and presence of CnATR2 were confirmed in hyg-resistant colonies by PCR screening using CnATR2-specific primers ATR2-F1 and ATR2-R1 (Table 1). An NaOH DNA extraction method from (Wang et al. 1993) was used to extract gDNA for PCR analysis.
Screening of N. crassa CnATR2 transformants for cercosporin and photosensitizer resistance
The CnATR2 transformants confirmed by PCR analysis were tested for cercosporin resistance on split petri plates as previously described (Beseli et al. 2015). Radial growth of colonies was measured at 16 h after inoculation, and % growth on cercosporin was calculated relative to the acetone control on the same plate. Each transformant was replicated five times, and the experiment was repeated two times. The mean value of all 10 samples for each strain and the corresponding standard errors were calculated and significant differences between WT and each transformant were calculated using Dunnett’s t Tests (Dunnett 1955).
Strains of N. crassa CnATR2 transformants that demonstrated resistance to cercosporin were further tested for resistance to other photosensitizers. Assays for resistance were conducted as above using 10 µM of hypericin (Alfa Aesar, Ward Hill, MA, USA) and 10 µM of rose bengal (Sigma-Aldrich, St. Louis, MO, USA). Hypocrellin A (Abcam, Cambridge, MA, USA) was used at 5 µM due to its high toxicity. Hypocrellin A and hypericin were solubilized in acetone, and control medium contained the same amount of acetone (0.5 %). Rose bengal was dissolved in water. Each transformant and the WT N. crassa were replicated five times and the experiment was repeated twice.
For gene expression analysis, total RNA was extracted from lyophilized and homogenized mycelia of five randomly chosen transformants, and RT-qPCR analysis was conducted as previously described (Beseli et al. 2015). Each sample was normalized against the N. crassa-specific tubulin control.
Targeted gene disruption in C. nicotianae
For targeted disruption of the CnATR2 gene in WT C. nicotianae, a split-marker recombination technique was modified as previously described (You et al. 2009). The 5′ [from 200 base pairs (bp) upstream to 400 bp downstream of start codon] and 3′ (from 900 bp downstream to 500 bp upstream of stop codon) CnATR2 sequences were amplified from C. nicotianae gDNA by PCR with CnATR2-specific primers containing restriction enzyme linkers (Table 1). The two primer sets used to amplify the 5′ and 3′ ends of CnATR2 were: ATR2-5′-F1 w/ApaI, ATR2-5′-R1 w/ApaI and ATR2-3′-F1 w/EcoRI, ATR2-3′-R1 w/SacI. The PCR products were digested with the appropriate restriction enzymes (ATR2-5′ digested with ApaI; ATR2-3′ digested with EcoRI and SacI) for cloning into the receptor plasmid pCB1636 (Sweigard et al. 1997). Plasmid pCB1636 was first digested with ApaI and ligated with ApaI digested ATR2-5′. Recombinant plasmids isolated from this step were selected based on the insert’s correct orientation, and subsequently digested with EcoRI and SacI and ligated with ATR2-3′ to obtain a CnATR2 disruption construct (Supplemental Fig. 1). Using this construct as a template, two different overlapping PCR fragments were amplified using primers (Table 1) specific to the CnATR2 sequence and the HygR cassette (split marker 1: ATR2-5′-F1 and HYG-split 5A; split marker 2: ATR2-3′-R1 and HYG-split 3S). Each split marker PCR fragment was sequenced to confirm their identity.
Protoplasts of C. nicotianae were isolated and transformed as previously described (Amnuaykanjanasin and Daub 2009) except that cultures were grown in PDB. For transformation, 1 × 107 protoplasts were incubated with 10 µg of each of the two split marker PCR fragments in the presence of spermidine (3 mM) and heparin (0.5 mg/ml). Transformants were originally selected in medium containing 125 µg/ml of hygromycin. Hyg-resistant colonies were transferred to PDA amended with 125 µg/ml hygromycin, and were transferred a minimum of five transfers to fresh selection plates to ensure stability of transformation.
CnATR2 disruption was confirmed by PCR analysis using primer sequences shown in Table 1. Genomic DNA was extracted from lyophilized mycelia that were grown in PDB amended with 125 µg/ml hygromycin using the E.Z.N.A. Fungal DNA Mini kit (Omega Bio-tek, Norcross, GA, USA).
Screening of cnatr2 disruption mutants for cercosporin sensitivity, production, sporulation, and pathogenicity
Transformants confirmed to have a disrupted copy of CnATR2 were screened for cercosporin sensitivity as described above for N. crassa sensitivity tests, except that radial growth of colonies was measured 3 days after the inoculation. For assaying cercosporin production, disruptants were grown under cercosporin-producing conditions in PDA under constant light (14 µE m2/s) at room temperature. Cercosporin was extracted with 5 N KOH and concentrations were measured as previously described (Jenns et al. 1989). For sporulation, cultures were grown on V8 agar (30 % V8 juice, 0.045 M CaCO3, 2 % agar) at 18 °C. To induce conidiation, the cultures were brushed with a sterile paint brush after 4 days and incubated under the same conditions for four additional days. To recover spores, plates were flooded with water and brushed gently. Spores were quantified using a hemocytometer. For inoculations, spores were diluted to a final concentration of 3.3 × 104 cells/ml, and 50 ml of spore solution was atomized onto the upper leaves of 3-month-old ‘Burley 21′ tobacco plants (3 plants per strain). Plants were covered with plastic bags for 4 days to provide high humidity conditions. Disease ratings were taken 7 weeks after inoculation.
Results
Sequence analysis of transporters
Five genes encoding transporters recovered in a subtractive library between the C. nicotianae WT and the cercosporin-sensitive cncrg1 transcription factor mutant were analyzed. RT-qPCR analysis showed that EST 103cF (CnMFS2) was strongly downregulated in cncrg1, whereas 86cR (CnATR3) and 53cR (encoding the uracil transporter) were both upregulated in cncrg1 (Herrero et al. 2007). Preliminary macroarray analysis (unpublished data) of the expression of 184sR5 (encoding the zinc transport protein) and 14c (CnATR2) showed little difference in expression of either gene between the WT and the cncrg1 mutant.
Full-length sequences were obtained for the five transporter genes. CnATR2 has a 3926 bp genomic sequence, a 3864 bp open reading frame (ORF), and has a single intron. The 1288 amino acid CnATR2 protein consists of two similar halves, and each half is arranged as a transmembrane segment (TMS)6—nucleotide-binding domain (NBD), similar to most eukaryote transporters (Lamping et al. 2010). CnATR3 has a 4717 bp genomic sequence, 4404 bp ORF and six introns (GenBank accession# KC959476). As with CnATR2, the protein consists of two similar halves with transmembrane and nucleotide-binding domains. CnMFS2 (GenBank accession # GU646037) has a 1625 bp genomic sequence that consists of a 1464 bp ORF with three introns. The putative protein size is 488 amino acids with 12 TM domains arranged in two similar halves. The zinc transport protein gene has an 1865 bp genomic sequence, and a 1761 bp ORF with one intron (GenBank accession# KC959477). It includes the Zinc (Zn2+)–Iron (Fe2+) Permease (ZIP) Family (TC 2.A.5) domain with eight putative transmembrane spanners that have been shown to transport Zn2+ as well as Fe2+ in some cases (Marchler-Bauer et al. 2011). The uracil transporter gene has a 1176 bp genomic sequence and lacks introns (GenBank accession# AF306523.1). The gene includes solute binding domains found in Solute Carrier (SLC) families five and six and in nucleobase-cation-symport-1 (NCS1) transporters. Proteins with the SLC5 domain are involved in co-transport of Na+ with sugars, amino acids, inorganic ions or vitamins (Wright et al. 2004). The SLC6 domain is commonly found in Na+/Cl−2 dependent plasma membrane transporters (neurotransmitters) (Kristensen et al. 2011). NCS1 transporters are essential secondary active transporters for nucleobases, hydantoins, and vitamins and work using a sodium ion, or a proton as a mechanism of energization (Weyand et al. 2008). Various transporters with NCS1 transporter-like family domains are common in fungi such as Aspergillus nidulans, A. fumigatus, C. albicans and Saccharomyces cerevisiae (Pantazopoulou and Diallinas 2007). Homologs of each of the transporter proteins identified are shown in Table 2.
Phylogenetic analysis of CnATR2, CnATR3, and CnMFS2
As noted above, ABC and MFS transporters have been identified in previous studies to have roles in cercosporin production and resistance, thus we compared CnATR2, CnATR3, and CnMFS2 to these and other ABC and MFS transporters using phylogenetic analysis (Figs. 1, 2; Supplemental Table S1). For the ABC transporter analysis, the two ABC domains of each transporter were used due to their greater degree of conservation as compared to transmembrane domains. In the analysis, “ABC1” and “ABC2” refer to the first and second ATP-binding cassette domains from the N-terminus and C-terminus, respectively (Fig. 1). Overall, phylogenetic analysis showed a clear separation between the ABC-G (or PDR, pleiotropic drug resistance-like) subfamily and the ABC-C (or MRP, multidrug resistance-associated protein) subfamily (Fig. 1). In the PDR (ABC-G) group, all the ABC1 domains were clustered with 95 % bootstrap support, and all the ABC2 domains were grouped together with 100 % bootstrap support (Fig. 1a). With the exception of the slime mold Dictyostelium discoideum AbcG14 domains, which grouped with the fungi, the non-fungal ABC proteins, including Arabidopsis thaliana PDR4, D. discoideum AbcG16, and Oryza sativa PDR9, fell into a separate branch within each of the ABC1 or ABC2 clusters. ATR3 clustered with the fungal PDR proteins, along with two previously characterized transporters involved in cercosporin resistance, the C. nicotianae ATR1 (Amnuaykanjanasin and Daub 2009) and the yeast ScSnq2p (Servos et al. 1993) (Fig. 1a).
Phylogenetic analysis of ABC transporters. Fungal, plant, protist, and animal branches are noted in black circles as F, P, Pr, and A, respectively. Transporters from other taxonomic groups within those branches (e.g. Dictyostelium transporter clustering within the fungal group) are noted by hatched ovals. Transporters in this study as well as those previously shown to play a role in cercosporin resistance or production are shown in bold. CnATR2 and CnATR3 are marked by black squares, and the two previously reported cercosporin efflux pumps CnATR1 and ScSnq2p are marked by gray squares. CnATR1, CnATR3, and ScSnq2p grouped with the subfamily ABC-G (PDR) transporters (panel a). CnATR2 grouped with ABC-C (MRP) transporters (panel b). Bootstrap analysis was performed with 1000 replicates, and values of >60 % are given at the nodes. A bacterial ABC transporter, Bacillus subtilis BceA, was used as the outgroup. Protein names and accession numbers for each sequence are shown in supplemental Table S1
Phylogenetic analysis of MFS transporters. The phylogenetic tree includes known fungal MFS transporters, with target compounds, when known, indicated in parenthesis. Transporters in this study as well as those previously shown to be involved in cercosporin production or resistance are shown in bold. CnMFS2 clusters with the group of dityrosine transporters. There was a clear divide between dityrosine transporters in yeast and filamentous fungi. Two transporters previously shown to be involved in cercosporin resistance and export (BfMfs1, CFP) clustered together. CnCTB4, the transporter found in the cercosporin biosynthetic cluster, is also shown. Bootstrap analysis was performed with 1000 replicates, and values of >60 % are given at the nodes. A bacterial MFS transporter, Oxalobacter formigenes OxlT, was used as the outgroup in this tree. Protein names and accession numbers for each sequence are shown in supplemental Table S1
For the MRP-like (ABC-C) group, the ABC1 and ABC2 domains of fungal transporters were clustered with considerably lower bootstrap support (Fig. 1b). CnATR2 clustered with the MRP-like group for both the ABC1 and ABC2 domains. No previously characterized transporters with roles in cercosporin production or resistance were found in the MRP-like transporter group.
The MFS transporter MFS2 in C. nicotianae clustered with dityrosine transporters (Fig. 2, Supplemental Table S1) including the S. cerevisiae Dtr1p that has been experimentally verified for transport of bisformyldityrosine into the surface of ascospores (Felder et al. 2002). Filamentous fungal and yeast dityrosine transporters were clearly separated in the tree with strong bootstrap support (Fig. 2). Two MFS transporters previously shown to play a role in cercosporin resistance, CFP from Cercospora kikuchii and C. nicotianae (Amnuaykanjanasin and Daub 2009; Callahan et al. 1999) and Bcmfs1 from Botrytis cinerea (Hayashi et al. 2002), were clustered together in a distinct group with 100 % bootstrap support. CnCTB4, the MFS transporter found within the cercosporin biosynthetic cluster, is also shown.
Expression of transporters under conditions of cercosporin toxicity
To test for a possible role in cercosporin resistance, the expression of the five transporter genes along with the previously characterized CnCFP was assayed under conditions of cercosporin toxicity by treating the cercosporin-sensitive cnatr1 mutant (Amnuaykanjanasin and Daub 2009) with cercosporin under high light conditions. Both CnCFP and CnATR2 were strongly induced when cercosporin toxicity was induced (Fig. 3). CnCFP was induced more than 50-fold after both 1 and 3 h of treatment, and CnATR2 was upregulated 9–16-fold. By contrast, there was no induction of CnMFS2, CnATR3, or the uracil transporter or the zinc transport protein genes. Based on these gene expression results, CnATR2 was chosen for further characterization.
RT-qPCR analysis of gene expression of transporter genes in the C. nicotianae cnatr1 mutant treated with cercosporin in the light. Each sample was normalized against the actin control, and fold-change relative to no-cercosporin was calculated according to the 2−ΔΔCT method (Livak and Schmittgen 2001). Data represent the mean of two independent experiments each with three technical replications. Error bars represent 95 % confidence intervals
CnATR2 characterization
CnATR2 was one of six of the 185 ESTs recovered in the subtractive library that was found in both the forward (downregulated in cncrg1 relative to WT) and reverse (up-regulated in cncrg1 relative to WT) libraries (Herrero et al. 2007). As noted earlier, preliminary macroarray data (unpublished) demonstrated little difference in CnATR2 expression between WT and the cncrg1 mutant. We tested the expression of CnATR2 in WT and the cncrg1 mutant using qRT-PCR (Fig. 4a). As medium composition was previously shown to affect relative gene expression between C. nicotianae WT and the cncrg1 mutant in other transporters (CnCFP, CnCTB4, CnATR1) (Amnuaykanjanasin and Daub 2009), we tested cultures grown in both PDB and CM liquid medium. The C. nicotianae WT and the cncrg1 mutant both synthesize cercosporin when grown in CM medium, however, only WT synthesizes cercosporin in PDB medium. CnATR2 gene expression was downregulated in the cncrg1 mutant relative to WT when grown in PDB medium, but upregulated in CM medium. This pattern is the same as the expression pattern of the previously characterized CnATR1 gene that is involved in cercosporin resistance (Amnuaykanjanasin and Daub 2009). To test directly if CnATR2 expression is regulated by the presence of cercosporin, we assayed CnATR2 expression in a C. nicotianae strain mutant for the CnCTB1 polyketide synthase in the cercosporin biosynthetic pathway and thus defective in cercosporin production (Choquer et al. 2005). CnATR2 expression was strongly upregulated within 15 min of the addition of cercosporin, and then decreased (Fig. 4b).
Expression analysis of CnATR2. a Differential expression of CnATR2 in the cercosporin-sensitive C. nicotianae cncrg1 mutant, deficient for the CRG1 transcription factor. Cultures were grown for 6 days in two different culture media: potato dextrose broth (PDB) and CM medium. Gene expression was quantified using RT-qPCR, with each sample normalized against the actin control. b Expression of CnATR2 in response to cercosporin in a non-cercosporin-producing C. nicotianae mutant deficient for the CTB1 polyketide synthase in the cercosporin biosynthetic pathway. Gene expression was quantified using RT-qPCR normalized against the 18S rRNA control. Cultures were treated with cercosporin or 0.5 % acetone, and RNA was extracted at 15 min, and 1, 2 and 6 h after addition of cercosporin. Expression in response to cercosporin is shown relative to the acetone control. Data for both analyses were calculated according to the 2−ΔΔCT method (Livak and Schmittgen 2001). Error bars represent 95 % confidence intervals
Ability of CnATR2 to impart resistance in N. crassa
The ability of CnATR2 to impart cercosporin resistance was tested by transforming the cercosporin-sensitive fungus N. crassa to express CnATR2, an assay previously used to identify putative cercosporin resistance genes (Beseli et al. 2015). Colonies were selected on medium containing hygromycin, and putative transformants were screened for the presence of CnATR2 by PCR. PCR-positive transformants were assayed for resistance to cercosporin by measuring radial growth on cercosporin-containing medium relative to growth on control medium. Cercosporin at 10 µM inhibits radial growth of N. crassa, resulting in approximately 30 % of the radial growth on medium lacking cercosporin (Fig. 5a). Of the 23 PCR-positive transformants tested, six were found to be significantly more resistant to cercosporin than WT (P < 0.05). Mean resistance of the remaining transformants varied from 40 to 60 %.
Transformation of the cercosporin-sensitive fungus Neurospora crassa with CnATR2. a Cercosporin resistance of N. crassa wild type (gray bar) and CnATR2-transformed strains (white bars). Data are radial growth on 10 µM cercosporin relative to the acetone control. Data are the results of two independent experiments with five replications each. Strains marked with three asterisks have significantly greater resistance than wild type (P < 0.05). Error bars represent standard error. b RT-qPCR analysis of gene expression of CnATR2 transformants #8, 11, 13, 21 and 31. Each sample was normalized against the tubulin control, and fold-change relative to the non-transformed wild type Neurospora crassa control was calculated according to the 2−ΔΔCT method (Livak and Schmittgen 2001). Error bars represent 95 % confidence intervals
Five randomly chosen CnATR2 transformants screened for resistance were selected to analyze the expression of CnATR2 (Fig. 5b). RT-qPCR analysis of CnATR2 expression in all five of the CnATR2 transformants showed high levels of expression (tubulin-normalized) relative to the WT control. Transformants # 8 and # 21 that were not statistically more resistant than WT had the lowest expression (99-fold and 54-fold, respectively). The cercosporin-resistant transformants (# 11, 13, 31) had the highest expression (669-fold, 235-fold, and 505-fold, respectively). Thus, CnATR2 expression correlated with resistance in the transformants tested.
To test if expression of CnATR2 could impart resistance to other 1O2-generating photosensitizers, we tested response of the cercosporin-resistant N. crassa transformants to other photosensitizers: hypocrellin A (a perylenequinone analog from the fungus Hypocrella bambusae); hypericin [an extended quinone from the plant Hypericum perforatum (St. John'sWort)], and the structurally unrelated photosensitizer rose bengal. Assay of toxicity to the N. crassa WT in the dark showed a lack of dark toxicity by hypericin and hypocrellin A, similar to cercosporin; however, rose bengal suppressed N. crassa growth in the dark by approximately 50 % (data not shown). Results of the toxicity assays in the light with the WT N. crassa and the cercosporin-resistant CnATR2 transformants are shown in Fig. 6. Hypocrellin A was the most toxic, and was used at a lower concentration (5 µM) due to its high toxicity. Hypericin showed similar levels of toxicity as does cercosporin, and rose bengal was the least toxic. For all compounds, the CnATR2 transformants were not significantly more resistant than WT N. crassa.
Growth of Neurospora crassa CnATR2 WT (gray bar) and cercosporin-resistant transformants (white bars) on medium containing 1O2-generating photosensitizers: a 5 µM hypocrellin A, b 10 µM hypericin and c 10 µM rose bengal. Data shown are relative to growth on the acetone control (hypocrellin A, hypericin) or water control (rose bengal). Data are the results of two independent experiments with five replications each. Each data point is reported as mean (±SE)
atr2 disruption mutants of C. nicotianae
A split marker strategy was used to generate C. nicotianae disruption mutants by homologous recombination. A total of 191 hyg-resistant colonies were screened to confirm CnATR2 disruption using PCR. Primer sets from the HygR marker sequences, genomic regions outside the split marker sequences, and genomic sequences within the deleted region (Table 1) were used to confirm disruption as shown in Fig. 7. A total of nine transformants were confirmed as being disrupted for CnATR2.
Targeted gene disruption of CnATR2 in C. nicotianae wild type (WT) using a split-marker recombination method (You et al. 2009). a Diagrams of C. nicotianae ATR2 genomic DNA, split marker PCR fragments, and resulting disruption sequence. Hatched bars show regions of homology between genomic DNA and split marker PCR fragments. Homologous recombination results in an intact HygR cassette replacing a region (dotted bar) of the CnATR2 sequence. Primers used to confirm disruption are shown. b Example of PCR amplification to confirm disruption. Primers and expected band sizes are shown in the table. Top gel image: primer set A; middle gel image: primer set B; Bottom gel image: primer set C. Lane 1: 1 kb marker; lanes 2–7: hyg-resistant transformants; lane 8: WT. Transformants # 17, # 65 were confirmed as disruptants due to the presence of the 780 bp (top gel) and 1700 bp (middle gel) bands and lack of the wild type 2500 bp (bottom gel). One transformant (# 29) contained both the disrupted sequences and an intact wild type sequence. Seven additional disruptants (not shown) were identified from a total of 191 hyg-resistant transformants screened
The nine cnatr2-disruption mutants were tested for cercosporin sensitivity by growing them on cercosporin-containing medium in the light. WT and two transformed, but not disrupted, strains were used as controls. Results are shown in Fig. 8. There was no statistically significant difference in cercosporin resistance between the WT, the non-disrupted transformants, or the cnatr2 disruption mutants in radial growth on cercosporin.
Cercosporin resistance of C. nicotianae wild type (gray bar), CnATR2 non-disruption transformants (black bars) and disruption mutants (white bars). Data are the results of 10 replications each and the mean of two separate experiments. The mean values are included at the top of each bar. Error bars represent upper and lower confidence intervals (P < 0.05). There were no significant differences in cercosporin sensitivity between wild type, non-disruption hyg-resistant transformants and cnatr2 disruptants
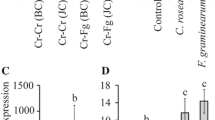
As other transporters (ATR1, CFP) have shown a dual role in both cercosporin resistance and production, eight of the cnatr2 disruption mutants (# 17, # 62, # 65, # 75, # 78, # 151, # 173 and # 181), two non-disrupted transformants (# 58 and # 59) and WT C. nicotianae were selected for assay of cercosporin production (Fig. 9a). There was no significant difference between cercosporin production between WT and the cnatr2 transformants suggesting that CnATR2 is not involved in cercosporin production. All eight cnatr2 disruption mutants, two non-disrupted transformants and WT C. nicotianae were also tested for changes in sporulation (Fig. 9b). Average production of spores ranged from 0.8 to 2.6 × 105 spores/plate, and there was no significant difference in the number of spores produced by the strains. All eight disruption mutants, two non-disrupted transformants and WT C. nicotianae were then used for pathogenicity assays on tobacco. Disease severity was assayed by counting the number of coalesced lesions on inoculated leaves; there were no significant differences between the disruption mutants and WT C. nicotianae (Fig. 9c).
Cercosporin production, spore production, and pathogenicity of Cercospora nicotianae atr2 disruption mutants (white bars), wild type (gray bars), and non-disruption transformants (black bars). a Cercosporin production by cnatr2 disruption mutants. Cercosporin was extracted in 5 N KOH from 7-day-old colonies grown on potato dextrose agar, and quantified by measuring absorbance of the KOH extract at 480 nm. Data are results of 2 experiments with four replicates per strain per experiment. Error bars represent upper and lower confidence intervals (P < 0.05). b Sporulation assay. Bars represent mean spore number per petri dish. Data are the average of 7 plates. Each data point is reported as mean (±SE). c Pathogenicity assay. Bars represent mean number of coalesced lesions on four infected leaves per plant from three plants. Each data point is reported as mean (±SE). Although variation was seen between strains in all three assays, there were no statistically significant differences between wild type and the cnatr2 disruption mutants for cercosporin production, spore production, or pathogenicity
Expression of other transporters and cercosporin biosynthetic genes in cnatr2 disruption mutants
Expression of CnATR2 in N. crassa demonstrated that CnATR2 can provide cercosporin resistance; however, atr2 mutants of C. nicotianae were not more sensitive to cercosporin than WT. We thus assayed for expression of other transporters previously shown to impart cercosporin resistance (CnATR1, CnCFP) as well as expression of the other library transporters to determine if they are upregulated in the cnatr2 mutant background (Fig. 10a). Expression of CnATR1, CnMFS2, CnATR3, the uracil transporter, and the zinc transport protein was not increased. By contrast, CnCFP expression was strongly increased, over 700-fold at 1 h and over 170-fold at 3 h. These results suggest that the lack of sensitivity of the cnatr2 mutant may be due to compensation by overexpression of CnCFP.
RT-qPCR analysis of gene expression in cnatr2 mutant (disruptant# 17) treated with cercosporin in the light. Each sample was normalized against the actin control, and fold-change relative to no-cercosporin treatment was calculated according to the 2−ΔΔCT method (Livak and Schmittgen 2001). a Expression of transporter genes. Data are the mean of two independent experiments. The mean values are included at the top of each bar. Error bars represent 95 % confidence intervals. b Expression of genes in the cercosporin biosynthetic cluster (CTB genes). Data are the mean of two independent experiments. Error bars represent 95 % confidence intervals
The subtractive library of putative cercosporin resistance genes also includes two genes from the cercosporin biosynthetic pathway: CnCTB2, encoding an O-methyltransferase, and CnCTB5, encoding an O2, FAD/FMN-dependent oxidoreductase (Daub and Chung 2009; Herrero et al. 2007). Analysis of expression of cercosporin biosynthetic genes in the cnatr2 mutant showed significant upregulation of CnCTB5 as well as CnCTB7, encoding a second FAD/FMN-dependent oxidoreductase (Fig. 10b); expression of CnCTB2 and the other biosynthetic genes including the CnCTB4 transporter was not altered.
Discussion
Cercospora fungi are highly successful plant pathogens due to their production of the photosensitizing toxin cercosporin. Cercosporin shows almost universal toxicity to cells because it produces reactive oxygen species including the highly toxic 1O2. For many years, our work has been focused on understanding the genes and mechanisms involved in cercosporin autoresistance (Daub et al. 2013), as these genes may have utility in engineering crop plants for resistance. In this work, we focused on the study of membrane transporters in Cercospora nicotianae. Transporters have previously been shown to provide cercosporin resistance in several fungal species. Also, genes encoding membrane transporters were recovered in a subtractive library between the C. nicotianae WT strain and a cercosporin-sensitive mutant deficient for the CRG1 transcription factor required for resistance (Herrero et al. 2007).
Membrane transporters shown to play a role in resistance to cercosporin are diverse. In Cercospora, both the ABC transporter ATR1 (Amnuaykanjanasin and Daub 2009) and the MFS transporter CFP (Callahan et al. 1999) were found to play a role in resistance; both of these were also recovered in the subtractive library. In addition, studies in other fungal systems confirmed the importance of transporters in resistance to cercosporin and to structurally related compounds. The yeast ABC transporter Snq2p increased cercosporin resistance in yeast when overexpressed (Ververidis et al. 2001). MFS transporters from B. cinerea (Bcmfs1) and Mycosphaerella graminicola (MgMfs1) have also been shown to provide resistance to cercosporin and to camptothecin, a quinone alkaloid produced by Camptotheca acuminata (Hayashi et al. 2002, Roohparvar et al. 2007). The role of transporters has also been shown in medical studies using the structurally similar photosensitizer hypericin, where increased expression of ABC transporters in cancer cells reduced utility of hypericin as an anti-cancer agent (Jendzelovsky et al. 2009).
For this study, we cloned and conducted bioinformatics analysis to identify the remaining transporter genes in our subtractive library. We identified two additional ABC transporters (CnATR2, CnATR3), an MFS transporter (CnMFS2), a uracil transporter, and a zinc transport protein. As both ABC and MFS transporters have previously been implicated in cercosporin resistance and production, we conducted a phylogenetic analysis of the ABC and MFS transporters along with other transporters previously implicated in cercosporin resistance or production. ABC transporters previously characterized to play a role in cercosporin resistance (the C. nicotianae ATR1 and S. cerevisiae Snq2p) were found in the PDR (pleiotropic drug resistance) subfamily (Fig. 1A). Our phylogenetic analysis showed that CnATR3 clusters in this group. The two ABC domains of CnATR3 cluster most closely with the domains of AtrE from the human fungal pathogen Trichophyton rubrum; AtrE is involved in drug resistance to terbinafine, 4-nitroquinoline 1-oxide, and ethidium bromide (Fachin, et al. 1996). The ABC2 domain of CnATR3 clusters with Snq2p as well as with the Schizosaccharomyces pombe BFR1 and the S. cerevisiae PDR10. BFR1 confers brefeldin A and cycloheximide resistance (Nagao et al. 1995). PDR10 has not been shown to be involved in resistance, but rather in maintenance of plasma membrane asymmetry of budding yeast (Rockwell et al. 2009). The two ABC domains of the previously characterized CnATR1 clustered within their own group in the PDR subfamily, along with transporters in the Leotiomycetes Monilinia fructicola and B. cinerea. Transporters from these fungi confer resistance to many major classes of fungicides such as myclobutanil and propiconazole (Schnabel et al. 2003) as well as some natural toxic compounds such as camalexin (Stefanato et al. 2009). CnATR1’s role in cercosporin resistance makes this group of ABC transporters strong candidates for toxin resistance. In our study, however, CnATR3 expression was unchanged under conditions of cercosporin toxicity; thus, it is not known at this time what role CnATR3 plays in C. nicotianae.
CnATR2 clustered with the MRP (multidrug resistance-associated protein-like) transporters (Fig. 1b). The ABC domains of CnATR2 cluster with those of Mycosphaerella populorum MRP3, Mycosphaerella fijiensis MDR, and Zymoseptoria tritici ABC. These fungi are in the Dothidiomycetes, the main taxonomic group in which perylenequinone producers are found (Daub and Chung 2009; Daub et al. 2013), and M. fijiensis is resistant to cercosporin (unpublished results). Our results demonstrate that CnATR2 can impart cercosporin resistance to N. crassa, suggesting that the homologous Mycosphaerella and Zymoseptoria transporters would be candidates for investigating possible roles in resistance to perylenequinones.
Two of the transporters previously shown to have a role in cercosporin resistance are MFS transporters, Bcmfs1 from B. cinerea (Hayashi et al. 2002) and CFP from C. nicotianae and C. kikuchii (Callahan et al. 1999). CnMFS2, investigated in this study, did not cluster with these transporters, nor did any of these cluster with CnCTB4, the MFS transporter in the cercosporin biosynthetic cluster. CnCTB4 has been shown to be regulated by CRG1 (Daub et al. 2013). However, in this study we found that CnCTB4 was not induced under conditions of cercosporin toxicity, a result consistent with previous studies documenting no role for CnCTB4 in cercosporin resistance (Choquer et al. 2007; Daub et al. 2013). CnMFS2 clusters with a transporter protein from Dothistroma septosporum (teleomorph = Mycosphaerella pini), a fungus also in the Dothidiomycetes which produces the photoactivated toxin dothistromin (De Wit et al. 2012; Stoessl et al. 1990). The Dothistroma transporters cluster with dityrosine transporters involved in diverse roles in cells including yeast spore formation (Felder et al. 2002) and in protein localization and zinc transport (Salazar et al. 2009) and with no known role in protection against toxic compounds. We found that CnMFS2 was not upregulated under conditions of cercosporin toxicity.
To test for a role of these transporters in cercosporin resistance, we assayed for changes in expression when the cercosporin-sensitive cnatr1 transporter mutant was exposed to cercosporin toxicity. Only two of the transporter genes were upregulated. The previously characterized CnCFP (Callahan et al. 1999) was induced almost 100-fold at both time points tested (1 and 3 h). CnATR2 was also induced approximately 10-fold at both time points. As indicated above, there was no induction of CnATR3, CnMFS2, or genes encoding the uracil transporter or the zinc transport protein.
Upregulation of CnATR2 in response to cercosporin toxicity led us to further characterize its role in resistance. We showed that CnATR2 is transiently upregulated in response to cercosporin, and that its regulation in the cncrg1 transcription factor mutant was similar to that of CnATR1, previously shown to play a role in both cercosporin resistance and production. We then tested the ability of CnATR2 to increase cercosporin resistance in the cercosporin-sensitive fungus N. crassa. We used this assay successfully in a previous study to characterize library genes that are homologs of 1O2-resistance genes in the photosynthetic bacterium Rhodobacter sphaeroides (Beseli et al. 2015). Under the conditions of the assay, growth of WT N. crassa is inhibited by about 70 % whereas growth of the resistant C. nicotianae is inhibited by about 40 %. Assay of N. crassa CnATR2 transformants identified six that had significantly more resistance to cercosporin than WT, confirming the ability of CnATR2 to impart cercosporin resistance. Not all of the transformants had significantly increased resistance. Random insertion of transgenes into the fungal genome is known to affect gene expression (Smith et al. 2008); thus, lack of resistance may reflect differences in transgene expression. To test this hypothesis, expression of CnATR2 in selected N. crassa transformants was analyzed, and a correlation was found between expression of the transgene and resistance of the transformant. We thus hypothesize that ATR2 imparts resistance, but that the effect is small and high levels of expression are required. Assays with the other photosensitizers also suggest that resistance is specific to cercosporin.
It is important to note that N. crassa has 17 ABC transporters that function in efflux (Toone 2011). The primers used for amplification of CnATR2 in the N. crassa transformants were selected based on lack of similarity to sequences in WT N. crassa. Specificity of the primers used to screen for CnATR2 were tested by PCR, and lack of amplification in WT N. crassa was confirmed. The N. crassa ABC transporter with the closest homology to CnATR2 has only 49 % identity; thus, no close homologs are present. N. crassa has some resistance to cercosporin, but we have no evidence that this is due to any of the ABC transporters.
The role of CnATR2 in cercosporin resistance was further characterized by creating disruption mutants. Previous studies with cnatr1 and ckcfp transporter mutants in C. nicotianae and C. kikuchii confirmed that mutants were more sensitive to cercosporin and also showed decreases in cercosporin production, sporulation and pathogenicity. When CnATR1 was disrupted in C. nicotianae, cercosporin resistance was decreased by 20–30 % (Amnuaykanjanasin and Daub 2009). In Cercospora kikuchii ckcfp disruption mutants, resistance was decreased 40–45 % (Callahan et al. 1999). Production of cercosporin was also significantly decreased in both ckcfp and cnatr1 disruption mutants, and ckcfp disruption mutants were shown to be less pathogenic. These results suggest that these two transporter genes have an important role in the regulation of cercosporin biosynthesis as well as resistance against cercosporin. In contrast, there was no difference between the cnatr2 disruptants and WT C. nicotianae or the non-disrupted transformants in either cercosporin resistance or production of cercosporin. Sporulation and pathogenicity were also unaffected.
The discrepancy between the ability of CnATR2 to impart cercosporin resistance to N. crassa and the lack of increased sensitivity of C. nicotianae cnatr2 mutants led us to test if induction of other transporters compensates for the cnatr2 mutation, as is seen in the cnatr1 mutant where both CnCFP and CnATR2 are induced under conditions of cercosporin toxicity. We tested the expression of CnATR1, CnATR3, CnCFP, CnMFS2, and the uracil transporter and zinc transfer protein genes in the cnatr2 mutant under conditions of cercosporin toxicity. Of these genes, only CnCFP was induced, but it was strongly induced (200–700-fold). Given the documented role of CFP in cercosporin resistance, we hypothesize that upregulation of CnCFP may compensate for mutations in both CnATR1 and CnATR2. Further research with double mutants of CnCFP with CnATR1 or CnATR2 will be needed to investigate this hypothesis.
In addition to induction of transporters in the cnatr2 mutant, we also tested induction of the CTB genes in the cercosporin biosynthetic pathway. We have hypothesized that resistance genes may be found within the biosynthetic cluster, but gene disruption experiments in C. nicotianae have not shown decreases in resistance in cnctb mutants (Chen et al. 2007b). Based on our hypothesis in this study that upregulation of other resistance genes compensates for mutations, we tested whether the cnctb genes were upregulated in the cnatr1 and cnatr2 mutants under conditions of cercosporin toxicity. In the cnatr1 mutant, all genes in the biosynthetic cluster (CTB1-8) were upregulated (data not shown), a result that was unexpected due to the cercosporin deficiency caused by the loss of CnATR1. Cercosporin production is not affected in the cnatr2 mutant, but we found that two of the genes, CnCTB5 and CnCTB7, were upregulated. These two genes encode putative FAD/FMN- or NADPH-dependent oxidoreductases (Chen et al. 2007b), and reduction of cercosporin is a major mechanism of cercosporin resistance (Daub et al. 1992, 2013). It is possible that CnCTB5 and CnCTB7 may contribute to cercosporin resistance but, as with CnATR2, mutations are compensated for by upregulation of other genes. Further research will be needed to better define these enzymes and their function.
In summary, we investigated transporters identified in a subtractive library of genes putatively involved in C. nicotianae resistance to the photoactivated toxin cercosporin. Phylogenetic analysis did not show close homologies with other transporters previously shown to play a role in cercosporin resistance; however, analysis of gene expression in response to cercosporin and under conditions of cercosporin toxicity identified CnATR2 as a putative resistance gene, a result confirmed by the ability of this gene to impart resistance to N. crassa. Disruption mutants were not altered in resistance, but other known and putative resistance genes were upregulated in the mutants and may compensate for the cnatr2 mutation. We hypothesize that cercosporin resistance in Cercospora is mediated by multiple mechanisms that are carefully regulated and redundant to ensure resistance. This work adds to previous studies in Cercospora and in other fungi showing the importance of membrane transporters in resistance to cercosporin. Current work in the laboratory is focused on transforming tobacco with the CnATR1 and CnCFP genes to impart Cercospora disease resistance. Based on our results, CnATR2 is a good candidate to be included in our plant transformation studies.
References
Amnuaykanjanasin A, Daub ME (2009) The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae. Fung Genet Biol 46:146–158
Beseli A, Goulart da Silva M, Daub ME (2015) The role of Cercospora zeae-maydis homologs of Rhodobacter sphaeroides 1O2-resistance genes in resistance to the photoactivated toxin cercosporin. FEMS Microbiol Lett 362:1–7
Burge CB (1998) Modeling dependencies in pre-mRNA splicing signals. In: Salzberg S, Searls D, Kasif S (eds) Computational methods in molecular biology, vol 8. Elsevier Science, Amsterdam, pp 127–163
Callahan T, Rose M, Meade M, Ehrenshaft M, Upchurch R (1999) CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol Plant Microbe Interact 12:901–910
Chen HQ, Lee MH, Daub ME, Chung KR (2007a) Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae. Mol Microbiol 64:755–770
Chen HQ, Lee MH, Chung KR (2007b) Functional characterization of three genes encoding putative oxidoreductases required for cercosporin toxin biosynthesis in the fungus Cercospora nicotianae. Microbiology 153:2781–2790
Choquer M, Lahey KA, Chen HL, Cao L, Ueng PP, Daub ME, Chung KR (2005) The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin toxin biosynthesis and fungal virulence in Cercospora nicotianae. Molec Plant Microbe Interact 18:468–476
Choquer M, Lee MH, Bau HJ, Chung KR (2007) Deletion of a MFS transporter-like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Lett 581:489–494
Chung KR, Daub ME, Kuchler K, Schaller C (2003) The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem Biophys Res Commun 302:302–310
Dahl SG, Sylte I, Ravna AW (2004) Structures and models of transporter proteins. J Pharmacol Exp Ther 309:302–310
Daub ME, Chung KR (2009) Photoactivated perylenequinone toxins in plant pathogenesis. In: Deising H (ed) The mycota V plant relationships, 2nd edn. Springer-Verlag, Berlin
Daub ME, Leisman GB, Clark RA, Bowden EF (1992) Reductive detoxification as a mechanism of fungal resistance to singlet-oxygen-generating photosensitizers. Proc Natl Acad Sci USA 89:9588–9592
Daub ME, Herrero S, Chung KR (2013) Reactive oxygen species in plant pathogenesis: the role of perylenequinone photosensitizers. Antiox Redox Signal 19:970–989
De Wit PJ, Van der Burgt A, Ökmen B, Stergiopoulos I, Abd-Elsalam KA, Aerts AL, Bahkali AH, Beenen HG, Chettri P, Cox MP, Datema E, De Vries RP, Dhillon B, Ganley AR, Griffiths SA, Guo Y, Hamelin RC, Henrissat B, Kabir MS, Jashni MK, Kema G, Klaubauf S, Lapidus A, Levasseur A, Lindquist E, Mehrabi R, Ohm RA, Owen TJ, Salamov A, Schwelm A, Schijlen E, Sun H, Van den Burg HA, Van Ham RC, Zhang S, Goodwin SB, Grigoriev IV, Collemare J, Bradshaw RE (2012) The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. doi:10.1371/journal.pgen.1003088
Deininger P (1990) Molecular cloning: a laboratory manual. Anal Biochem 186:182–183
Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121
Ehrenshaft M, Bilski P, Li M, Chignell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96:9374–9378
Fachin AL, Maffei CM, Martinez-Rossi NM (1996) In vitro susceptibility of Trichophyton rubrum isolates to griseofulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs. Mycopathologia 135:141–143
Felder T, Bogengruber E, Tenreiro S, Ellinger A, Sá-Correia I, Briza P (2002) Dtr1p, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot Cell 1(5):799–810
Hayashi K, Schoonbeek HJ, De Waard MA (2002) Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl Environ Microbiol 68:4996–5004
Herrero S, Daub ME (2007) Genetic manipulation of vitamin B-6 biosynthesis in tobacco and fungi uncovers limitations to up-regulation of the pathway. Plant Sci 172:609–620
Herrero S, Amnuaykanjanasin A, Daub ME (2007) Identification of genes differentially expressed in the phytopathogenic fungus Cercospora nicotianae between cercosporin toxin-resistant and -susceptible strains. FEMS Microbiol Lett 275:326–337
Higgins CF (1992) ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8:67–113
Higgins CF (2007) Multiple molecular mechanisms for multidrug resistance transporters. Nature 446:749–757
Jacobs M, Stahl U (1995) Gene regulation in mycelial fungi. In: Kueck U (ed) The mycota, II; genetics and biotechnology. Springer-Verlag, New York, pp 155–167
Jendzelovsky R, Mikes J, Koval J, Soucek K, Prochazkova J, Kello M, Sackova V, Hofmanova J, Kozubik A, Fedorocko P (2009) Drug efflux transporters, MRP1 and BCRP, affect the outcome of hypericin-mediated photodynamic therapy in HT-29 adenocarcinoma cells. Photochem Photobiol Sci 8:1716–1723
Jenns AE, Daub ME (1995) Characterization of mutants of Cercospora-nicotianae sensitive to the toxin cercosporin. Phytopathology 85:906–912
Jenns AE, Daub ME, Upchurch RG (1989) Regulation of cercosporin accumulation in culture by medium and temperature manipulation. Phytopathology 79:213–219
Jenns AE, Scott DL, Bowden EF, Daub ME (1995) Isolation of mutants of the fungus Cercospora nicotianae altered in their response to singlet-oxygen-generating photosensitizers. Photochem Photobiol Sci 61:488–493
Jones PM, George AM (2002) Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc Natl Acad Sci 99:12639–12644
Keim M, Williams RS, Harwood AJ (2004) An inverse PCR technique to rapidly isolate the flanking DNA of dictyostelium insertion mutants. Mol Biotechnol 26:221–224
Kristensen AS, Andersen J, Jörgensen TN, Sörensen L, Eriksen J, Loland CJ, Strömgaard K, Gether U (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63:585–640
Lamping E, Baret PV, Holmes AR, Monk BC, Goffeau A, Cannon RD (2010) Fungal PDR transporters:phylogeny, topology, motifs and function. Fung Genet Biol 47:127–142
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–403
Lorang JM, Tuori RP, Martinez JP, Sawyer TL, Redman RS, Rollins JA, Wolpert JT, Johnson KB, Rodriguez RJ, Dickman MB, Ciuffetti LM (2001) Green fluorescent protein is lighting up fungal biology. Appl Environ Microb 67:1987–1994
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH (2011) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D384–D389
Nagao K, Taguchi Y, Arioka M, Kadokura H, Takatsuki A, Yoda K, Yamasaki M (1995) BFR1+, a novel gene of Schizosaccharomyces pombe which confers brefeldin A resistance, is structurally related to the ATP-binding cassette superfamily. J Bacteriol 177:1536–1543
Panagiotis M, Kritonas K, Irini NO, Kiriaki C, Nicolaos P, Athanasios T (2007) Expression of the yeast cpd1 gene in tobacco confers resistance to the fungal toxin cercosporin. Biomolec Eng 24:245–251
Pantazopoulou A, Diallinas G (2007) Fungal nucleobase transporters. FEMS Microbiol Rev 31:657–675
Pao SS, Paulsen IT, Saier MH Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34
Rockwell NC, Wolfger H, Kuchler K, Thorner J (2009) ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J Membr Biol 229:27–52
Roohparvar R, De Waard MA, Kema GHJ, Zwiers LH (2007) MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fung Genet Biol 44:378–388
Salazar G, Falcon-Perez JM, Harrison R, Faundez V (2009) SLC30A3 (ZnT3) oligomerization by dityrosine bonds regulates its subcellular localization and metal transport capacity. PLoS One 4(6):e5896. doi:10.1371/journalpone0005896
Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW (2000) Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 285:194–204
Schnabel G, Dai Q, Paradkar MR (2003) Cloning and expression analysis of the ATP binding cassette transporter MFABC1 gene and the alternative oxidase gene MfAOX1 from Monilinia fructicola. Pest Manag Sci 59:1143–1151
Servos J, Haase E, Brendel M (1993) Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet 236:214–218
Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, Freitag M, Motamedi MR, Selker EU (2008) The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenet Chromatin 1:5
Stefanato F, Abou-Mansour E, Buchala A, Kretschmer M, Mosbach A, Hahn M, Bochet C, Metraux JP, Schoonbeek HJ (2009) The ABC-transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J 58:499–510
Stoessl A, Abramowski Z, Lester HH, Rock GL, Towers GHN (1990) Further toxic properties of the fungal metabolite dothistromin. Mycopathologia 112:179–186
Sweigard J, Chumley F, Carroll A, Farrall L, Valent B (1997) A series of vectors for fungal transformation. Fung Genet Newslett 44:52–53
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Toone EJ (2011) Advances in Enzymology and Related Areas of Molecular Biology. In: Barabote D, Thekkiniath J, Strauss RE, Vediyappan G (eds) Xenobiotic efflux in bacteria and fungi: a genomics update. John Wiley and Sons Inc., Hoboken, p 271
Ververidis P, Davrazou F, Diallinas G, Georgakopoulos D, Kanellis AK, Panopoulos N (2001) A novel putative reductase (Cpd1p) and the multidrug exporter Snq2p are involved in resistance to cercosporin and other singlet oxygen-generating photosensitizers in Saccharomyces cerevisiae. Curr Genet 39:127–136
Vogel HJ (1956) A convenient growth medium for Neurospora (medium N). Microbial Genet Bull 13:42–43
Wang H, Qi M, Cutler A (1993) A simple method of preparing plant samples for PCR. Nucleic Acids Res 21:4153–4154
Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O’Reilly J, Ma P, Saidijam M, Patching SG, Hope RJ, Norbertczak HT, Roach PC, Iwata S, Henderson PJ, Cameron AD (2008) Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322:709–713
Winer J (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270:41–49
Wright E, Loo DD, Hirayama BA, Turk E (2004) Surprising versatility of Na+/glucose cotransporters (SLC5). Physiology 19:370–376
You B, Lee MH, Chung KR (2009) Gene-specific disruption in the filamentous fungus Cercospora nicotianae using a split-marker approach. Arch Microbiol 191:615–622
Acknowledgments
We thank Drs. Necip Baris Kacar and Consuelo Arellano for assistance with statistical analyses and Dr. Marilia Goulart da Silva for technical help. This research was supported in part from the USDA cooperative agreement# 58-3148-1-161.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Beseli, A., Amnuaykanjanasin, A., Herrero, S. et al. Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr Genet 61, 601–620 (2015). https://doi.org/10.1007/s00294-015-0486-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0486-x