Abstract
To develop an efficient gene-targeting system in Mortierella alpina 1S-4, we identified the ku80 gene encoding the Ku80 protein, which is involved in the nonhomologous end-joining pathway in genomic double-strand break (DSB) repair, and constructed ku80 gene-disrupted strains via single-crossover homologous recombination. The Δku80 strain from M. alpina 1S-4 showed no negative effects on vegetative growth, formation of spores, and fatty acid productivity, and exhibited high sensitivity to methyl methanesulfonate, which causes DSBs. Dihomo-γ-linolenic acid (DGLA)-producing strains were constructed by disruption of the Δ5-desaturase gene, encoding a key enzyme of bioconversion of DGLA to ARA, using the Δku80 strain as a host strain. The significant improvement of gene-targeting efficiency was not observed by disruption of the ku80 gene, but the construction of DGLA-producing strain by disruption of the Δ5-desaturase gene was succeeded using the Δku80 strain as a host strain. This report describes the first study on the identification and disruption of the ku80 gene in zygomycetes and construction of a DGLA-producing transformant using a gene-targeting system in M. alpina 1S-4.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Integration of exogenous DNA into the chromosome in all organisms follows two pathways of DNA double-strand break (DSB) repair: homologous recombination (HR) and nonhomologous end-joining (NHEJ) pathways (Kanaar et al. 1998). The repair of DSBs is induced by both exogenous and endogenous triggers and causes detrimental DNA lesions (Haber 2000). In the mechanism of the HR pathway, the homologous region is used as a template and the exogenous DNA is integrated into the chromosome. In contrast, in the mechanism of the NHEJ pathway, the strand ends of the exogenous DNA are directly ligated into DSBs without a requirement of sequence identity. These two mechanisms for DSB repair are independent of each other and are considered to function competitively (Van Dyck et al. 1999). The repair of DSBs requires many associated proteins, such as Rad protein group including Rad54, Rad51, Rad52, Mre11, and Xrs2 in the HR pathway (Kooistra et al. 2004; Krappmann 2007), and Ku70, Ku80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA ligase 4 (Lig4), and X-ray repair cross-complementing protein 4 (Xrcc4) in the NHEJ pathway (Critchlow and Jackson 1998; Daley et al. 2005). A pathway similar to HR has been confirmed ubiquitously in various organisms (bacteria, yeast, and human) (Krogh and Symington 2004; Shibata et al. 2001). In addition, important discoveries such as the Rad51-independent HR and Ku80-independent NHEJ pathways and the occurrence of all nonhomologous chromosomal integration under the control of Lig4 have been reported (Ishibashi et al. 2006).
The yeast Saccharomyces cerevisiae mainly utilizes the HR system for DSB repair. Accordingly, gene targeting through the HR pathway in S. cerevisiae exhibits quite high efficiency (Schiestl et al. 1994). In contrast, many other organisms, including mammals, plants, insects, and filamentous fungi, predominantly use the NHEJ pathway for DSB repair, and exogenous DNA, even if it consists of a long homologous sequence, can be integrated into nonspecific regions in chromosomes. Disruption of the ku70, ku80, or lig4 gene leads to an increase in the frequency of HR in filamentous fungi (Ishibashi et al. 2006; Ishidoh et al. 2014; Krappmann et al. 2006; Mizutani et al. 2008; Ninomiya et al. 2004; Takahashi et al. 2006; Tani et al. 2013). In particular, disruption of the ku80 and/or lig4 gene in Neurospora crassa and the lig4 gene in Aspergillus oryzae have led to 100 % targeting efficiency (Ishibashi et al. 2006; Mizutani et al. 2008; Ninomiya et al. 2004).
The oil-producing filamentous fungus Mortierella alpina 1S-4 is a producer of carbon 20 (C20) polyunsaturated fatty acids (PUFAs), such as arachidonic acid (20:4ω6, ARA) and eicosapentaenoic acid (20:5ω3, EPA), which are rich in triacylglycerols (Sakuradani et al. 2009). In addition, the lipid productivity of this fungus reaches 600 mg/g of dried mycelia. For these reasons, the fungus has been used as a model oleaginous microorganism for biosynthesis and accumulation of lipids, including PUFAs (Kawashima et al. 1995; Kikukawa et al. 2013; Sakuradani et al. 1999b, 2005, 2008, 2013). In previous studies, several techniques for gene manipulation in this fungus, such as a host–vector system (Ando et al. 2009a; Takeno et al. 2004a, 2005b), RNA interference (Takeno et al. 2005a), and transformation systems (Ando et al. 2009b; Takeno et al. 2004b), have been established. By use of such transformation systems, plasmid vectors are integrated randomly into the fungal genome.
To construct a high-producing strain of beneficial PUFAs from this fungus by metabolic engineering, an efficient gene-targeting system using HR is necessary. However, gene targeting by HR in this fungus is rarely attempted, given that NHEJ is predominant and the efficiency of HR is low. In this study, to evaluate function of Ku80 to HR in M. alpina 1S-4, we identified the ku80 gene and constructed ku80 gene-disrupted strains via single-crossover HR. Moreover, to evaluate the gene-targeting efficiency in the ku80 gene disruptant, we constructed of a dihomo-γ-linolenic acid (DGLA)-producing strain by Δ5-desaturase (Δ5ds) gene disruption.
Materials and methods
Enzymes and chemicals
Restriction enzymes and other DNA-modifying enzymes were obtained from Takara Bio (Shiga, Japan). All other chemicals were of the highest purity commercially available.
Strains, media, and growth conditions
M. alpina 1S-4 is deposited in the Graduate School of Agriculture of Kyoto University, Japan (Sakuradani 2010) and the uracil auxotrophic strain (ura5 − strain) (Takeno et al. 2004b) was used as a host strain. Czapek–Dox agar medium containing 0.05 mg/ml uracil was used for sporulation of the ura5 − strain, as described previously (Takeno et al. 2004b). Synthetic complete (SC) medium was used as a uracil-free synthetic medium for cultivation of transformants derived from the M. alpina 1S-4 ura5 − strain at 28 °C (Takeno et al. 2004b). GY medium (20 mg/ml glucose and 10 mg/ml yeast extract) was used for fatty acid composition analysis and extraction of genomic DNA. GY agar medium containing 0.75 mg/ml 5-fluoroorotic acid (5-FOA) and 0.05 mg/ml uracil were used to confirm the growth of ku80-disrupted transformants (Boeke et al. 1984; Razanamparany and Bégueret 1986; Watrin et al. 1999). GY agar medium containing 100 μg/ml carboxin was used for selection of ku80-disrupted transformants. Escherichia coli strain DH5α was used for DNA manipulation and grown on LB agar plates containing 50 μg/ml kanamycin. All solid media contained 2 % agar.
Genomic DNA preparation
M. alpina 1S-4 host strain and transformants were cultivated in 100 ml of GY liquid medium at 28 °C for 5 days with shaking at 100 rpm. Fungal mycelia were harvested by suction filtration and washed with sterile water. Preparation of genomic DNA was performed by a previously described method (Okuda et al. 2014).
Cloning and identification of the ku80 gene from M. alpina 1S-4
Two highly degenerate primers, ku80 F and ku80 R (Table 1), were synthesized for cloning of the ku80 cDNA, based on the amino acid sequences of Ku80 homologs from two filamentous fungi, Rhizopus delemar (accession EIE88285) and Aspergillus clavatus (accession XP_001272945). The sequences of the primers correspond to regions that encode IAIQMIVT and PFAGDVNTY peptides. PCR amplification was performed in a total volume of 50 μl containing 1 μg of genomic DNA, 0.25 μl of Takara EX taq polymerase (Takara Bio), 5 μl of 10 × EX Taq buffer, 200 μM of each dNTP, and 5 pM of primers, and performed as 35 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min, followed by one cycle of extension at 72 °C for 5 min. The resulting 0.7-kb fragment was cloned into the pT7Blue T-Vector (Novagen, Merck KGaA, Darmstadt, Germany), and the nucleotide sequence was determined with a Beckman Coulter CEQ8000 system (Beckman Coulter, Fullerton, CA, USA). For cDNA synthesis and construction of a cDNA library, RNA extraction reagent Isogen (Nippon Gene, Tokyo, Japan) and a PrimeScript High Fidelity RT-PCR Kit (Takara Bio) were used, following the supplier’s instructions.
To isolate whole ku80 genomic DNA from M. alpina 1S-4, inverse PCR was performed with primers, ku80 IPCR F and ku80 IPCR R (Table 1). The SalI/XhoI-digested genomic fragment was self-ligated and then used as a template. PCR amplification was performed in a total volume of 50 μl containing 500 ng of the template, 0.25 μl of Takara EX taq polymerase (Takara Bio), 5 μl of 10 × EX Taq buffer, 200 μM of each dNTP, and 5 pM of each primer, and performed as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles of 94 °C for 40 s, 57 °C for 40 s, and 72 °C for 3 min and one cycle of extension period at 72 °C for 10 min. The amplified 3.3-kb fragment was cloned into the pT7Blue T-Vector and identified completely with the Beckman Coulter CEQ8000 system.
Construction of the plasmid vector for ku80 gene targeting
A binary vector pKSUku80 was constructed for ku80 gene targeting on the backbone of pKSU, which is pBluescript II KS (+) (Stratagene, La Jolla, CA, USA) ligated with a ura5 gene marker cassette derived from the M. alpina 1S-4 transformation vector. A 4.3-kb fragment containing a partial ku80 gene, amplified with ku80 start and ku80 XhoI R primers (Table 1) using M. alpina 1S-4 genomic DNA as a template, was cloned into the pUC118 using a Mighty Cloning Kit (Blunt End) (Takara bio). The resulting plasmid (pUC118ku80) was digested with HindIII and the digested partial ku80 fragment was ligated into pKSU vector digested with the same restriction enzyme. The resulting plasmid was named pKSUku80 (Fig. 2a).
A pKSCD5 vector for the Δ5ds gene-targeting was constructed on the backbone of the M. alpina 1S-4 transformation vector pKSC. The CBXB gene expression cassette from pSBZNCBXB (Ando et al. 2009a), digested with EcoRI and XbaI, was ligated into pBluescript II KS (+) (Stratagene) digested with the same restriction enzymes, and the resulting plasmid was named pKSC. The partial Δ5ds gene (2024-bp) was amplified with two primers, Δ5 EcoRI F and Δ5 XhoI R primers (Table 1) and M. alpina 1S-4 genomic DNA as a template. The resulting fragment was digested with EcoRI and XhoI and ligated into the pKSC plasmid digested with the same restriction enzymes. The resulting plasmid was named pKSCD5 (Fig. 5a).
Transformation of M. alpina 1S-4
The gene-targeting vectors were introduced into spores of M. alpina 1S-4 by biolistic particle bombardment with PDS-1000/He Particle Delivery System (Bio-Rad, Hercules, CA, USA) (Takeno et al. 2004b). Given that linear plasmids are completely integrated into chromosomes by HR (Shiotani and Tsuge 1995), the vectors were digested with restriction enzymes and introduced into the spores: the pKSUku80 vector was digested with NcoI and the pKSCD5 vector was digested with NruI. Spores (1.5 × 108) were spread on an agar plate using SC uracil-free medium for transformation with pKSUku80 or GY containing 5-FOA for transformation with pKSCD5. After bombardment, the plates were incubated at 28 °C for 5 days. Transformants were transferred to a new GY plate containing 5-FOA.
Mutagen sensitivity
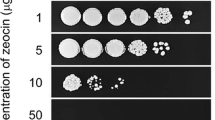
Sensitivity to chemical mutagen toxicity in gene-manipulated transformants was evaluated by spot test (Kato et al. 2004; Mizutani et al. 2008). Methyl methanesulfonate (MMS) was added to GY agar medium at final concentrations of 0, 0.01, 0.02, 0.025, 0.05, and 0.1 %.
Identification of gene disruption by PCR and southern blot analysis
The ku80 gene-disrupted candidates were evaluated by means of colony PCR using the extracted genomic DNA as a template and the primers ku80 start and ura5 stop R (Table 1). When integration into the genomic ku80 gene locus was successful, a 4.3-kb fragment was amplified using the primers and genomic DNA from the transformants.
Correct homologous integration in the genomic ku80 gene was confirmed by Southern blot analysis. The 1.0-kb partial ku80 gene amplified with primers ku80 F2 and ku80 R2 (Table 1) using M. alpina 1S-4 genomic DNA as a template was used as a probe for hybridization. Southern blot hybridization was performed as described previously (Sakuradani et al. 1999a). Genomic DNA (10 μg) digested with XhoI was size-fractioned by electrophoresis in 1 % agarose gel and transferred to Amersham Hybond-N+ membrane (GE Healthcare Ltd., Buckinghamshire, UK) using a VacuGene XL Vacuum Blotting System (GE Healthcare). Southern hybridization was performed using the Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare).
For Southern blotting analysis of Δ5ds gene disruption, 1.7 kb of the Δ5ds gene fragment amplified with primers Δ5 F and Δ5 R (Table 1) using M. alpina 1S-4 genomic DNA as a template was used as a probe for hybridization. Genomic DNA (10 μg) was digested with the pair NarI and XbaI or ClaI and XhoI. Southern blot hybridization was then performed as described above.
Fatty acid analysis
Fatty acid production and composition of transformants were analyzed as described previously (Kikukawa et al. 2013). In this study, mycelia of the transformants and the host strain were inoculated into 3 ml of GY liquid medium in a 20-ml Erlenmeyer flask and cultivated at 28 °C with reciprocal shaking at 120 rpm for 7 days. The fungal strains after cultivation were harvested by filtration and dried at 120 °C for 2 h. The dried cells were directly transmethylated with 10 % methanolic HCl at 55 °C for 2 h. The resulting fatty acid methyl esters were extracted with n-hexane, concentrated, and analyzed with a GC-17A gas chromatograph (GC; Shimadzu, Kyoto, Japan) equipped with an HR-SS-10 capillary column (Shinwa Chemical Industries, Kyoto, Japan). Fatty acids were quantified using tricosanoic acid as an internal standard. All experiments were performed in triplicate.
Nucleotide sequence accessions
The ku80 gene from M. alpina 1S-4 has been registered in the DNA Data Bank of Japan (DDBJ) database as accession LC009413. The Δ5ds genomic gene of M. alpina 1S-4 has been deposited in GenBank/EMBL/DDBJ as accession AB188307.
Results
Identification and phylogenetic analysis of the ku80 gene from M. alpina 1S-4
To isolate the ku80 partial gene fragment, a 0.7-kb gene fragment was amplified by PCR using highly degenerate primers and M. alpina 1S-4 genomic DNA as a template. The predicted amino acid sequence encoded by the partial gene fragment showed high similarity to those of Ku80 proteins from other organisms. To identify the whole ku80 gene from M. alpina 1S-4, inverse PCR was performed with M. alpina 1S-4 genomic DNA. The open reading frame of the ku80 gene from M. alpina 1S-4 was found to be 3366-bp length. Based on the whole ku80 gene information from genomic DNA, the full-length cDNA of the ku80 gene was obtained by PCR. The ku80 cDNA with 2511-bp length was predicted to encode a protein consisting of 836 amino acids. These results suggested that M. alpina 1S-4 ku80 genomic gene has nine exons (1–129, 257–311, 400–543, 653–821, 906–1467, 1580–1795, 1877–2043, 2141–3116, 3274–3366) and eight introns. The predicted amino acid sequence of M. alpina 1S-4 Ku80 shares low identities with those of metazoa (Mus musculus, 21 %; Rattus norvegicus, 21 %; Homo sapiens, 25 %; Tigriopus japonicas, 21 %), higher plants (Hordeum vulgare, 24 %; Triticum aestivum, 25 %; Oryza sativa, 25 %; Arabidopsis thaliana, 22 %), oleaginous yeast (Rhodosporidium toruloides, 24 %), fungi (Neurospora crassa, 29 %; N. tetrasperma, 29 %; Lecanicillium sp., 29 %; Aspergillus oryzae, 28 %; A. sojae, 28 %; A. fumigatus, 29 %; Penicillium digitatum, 28 %). Compared with various Ku80 proteins from these organisms, the Ku80 from M. alpina 1S-4 is located in the expected position in the phylogenetic tree (Fig. 1).
Phylogenetic tree of Ku80 proteins. The tree was created by the neighbor-joining (NJ) method with 10,000 bootstrap replicates using the sequence analysis software GENETYX 11.0 (Genetyx corp., Tokyo, Japan). MmKu80 (Mus musculus), AAH51660; RnKu80 (Rattus norvegicus), NP_803154; HsKu80 (Homo sapiens), P13010; TjKu80 (Tigriopus japonicus), AIL94178; HvKu80 (Hordeum vulgare subsp. vulgare), AEO86624; TaKu80 (Triticum aestivum), ADO00729; OsKu80 (Oryza sativa Japonica Group), Q75IP6; AtKu80 (Arabidopsis thaliana), AEE32242; MaKu80 (Mortierella alpina 1S-4), LC009413; RtKu80 (Rhodosporidium toruloides), AIA21644; NcKu80 (Neurospora crassa), AFM68948; NtKu80 (N. tetrasperma FGSC 2508), EGO57771; Lecanicillium sp. Ku80 (Lecanicillium sp. HF627), AHY22503; AoKu80 (Aspergillus oryzae), BAE78503; AsKu80 (A. sojae), BAE78504; AfKu80 (A. fumigatus Af293), Q4WI96; PdKu80 (Penicillium digitatum), AGT79985
Disruption of the ku80 gene of M. alpina 1S-4 with pKSUku80 vector
A vector for ku80 gene disruption, pSKUku80, digested with NcoI was delivered into spores of M. alpina 1S-4 ura5 − strain by biolistic particle bombardment with a PDS-1000/He Particle Delivery System. To confirm integration of a ura5 gene marker, all transformants grown on an SC uracil-free plate were inoculated onto GY medium containing 5-FOA. Finally, 77 transformants were obtained under these conditions. The transformants were selected by colony PCR with primers ku80 start and ura5 stop R (Table 1), and each genomic DNA as a template. Fragments of approximately 4.3 kb, which were formed presumably by integration via HR, were observed in only two transformants (3 and 6), but not in the host strain (Fig. S1 A and B).
The genome integration patterns of transformants 3 and 6 were confirmed by Southern blot analysis. Their genomic DNAs were digested with XhoI and a 1.0-kb fragment consisting of the partial ku80 gene was used as a probe (Fig. 2a). The 5.2-kb hybridization signal on the host strain was not detected in the two transformants (Fig. 2b). However, the expected 4.0- and 8.3-kb signals resulting from single-crossover HR were detected only in transformant 3. These results suggest that a single pKSUku80 vector was successfully integrated into the ku80 genomic DNA of transformant 3. In contrast, some of the introduced pKSUku80 vectors appear to have been integrated ectopically into the ku80 gene locus in transformant 6. Thus, transformant 3 was used as a host strain for Δ5ds gene disruption in the present research.
Construction scheme of ku80 gene disruptant and confirmation by Southern blot analysis. a The figure illustrates the homologous integration of pKSUku80 vector into the ku80 genomic gene locus in M. alpina 1S-4. Gray short bar indicates the position hybridized by the probe. Dotted lines indicate the position and base lengths of hybridization signals. b Southern hybridization analysis of ku80 gene-disrupted candidates. XhoI-digested genomic DNAs from transformants 3 and 6 and host strain were hybridized with the probe
Growth characteristics and mutagen sensitivity
Given that Ku70, Ku80, and Lig4 proteins are involved in DSB repair through NHEJ in diverse organisms (Hopfner et al. 2002; Lisby and Rothstein 2004) and telomere maintenance in some organisms (Hande 2004), the growth characteristics and mutagen sensitivity of M. alpina 1S-4 Δku80 strain were investigated. The growth rate of the Δku80 strain did not decrease, compared with that of the wild strain both on plate medium and in liquid medium (data not shown). In addition, the germination rate of its spores was similar to that of the wild strain (data not shown). Furthermore, given that the fatty acid productivity and composition of the Δku80 strain were similar to those of the wild strain, we infer that the ku80 gene disruption did not affect fatty acid productivity (Table 2). The sensitivity to chemical mutagens causing DSBs, MMS, of M. alpina 1S-4 Δku80 strain was evaluated as described previously (Ishibashi et al. 2006; Ninomiya et al. 2004). The Δku80 strain showed no sensitivity to low (≤0.02 %) concentrations of MMS, but showed high sensitivity to 0.05 % MMS (Fig. 3).
Construction and characterization of Δ5ds gene disruptant with pKSCD5 vector
To evaluate gene-targeting efficiency in the Δku80 strain and construct a strain producing valuable PUFAs, Δ5ds gene disruption causing an increase in DGLA production and a decrease in ARA production (Fig. 4) was performed in the Δku80 strain as a host strain (Fig. 5a). A vector for Δ5ds gene disruption, pKSCD5, which contains the CBXB marker, was digested with NruI to enhance gene-targeting efficiency and was introduced into spores of the Δku80 strain on GY medium containing 100 μg/ml carboxin by biolistic particle bombardment. After bombardment, the spores were cultivated at 28 °C for 5 days. Finally, 32 stable transformants were obtained.
Construction scheme of Δ5ds gene disruptant and confirmation by Southern blot analysis. a The figure illustrates the homologous integration of pKSCD5 vector into the Δ5ds genomic gene locus in M. alpina 1S-4. Gray short bar indicates the position hybridized by the probe. Dotted lines indicate the position and base lengths of hybridization signals. b Southern hybridization analysis of Δ5ds gene-disrupted candidates. NarI- and XbaI- or CraI- and XhoI-digested genomic DNAs were hybridized with the probe. Lane #15 and Δku80 indicate a Δ5ds gene-disrupted candidate and the host strain (ku80-disruptant), respectively
All stable transformants, the wild type, and the Δku80 strain were cultivated in 3 ml of GY medium at 28 °C for 7 days with reciprocal shaking, and their fatty acid productivities were determined by GC analysis. The ratio of DGLA to total fatty acids reached 36.8 % in transformant 15, whereas that of ARA was only 3.4 % (Fig. 6; Table 2). Transformant 15 exhibited the same fatty acid composition as that of the Δ5ds gene-defective mutant S14 isolated previously (Jareonkitmongkol et al. 1993) (Table 2). To confirm the disruption of the Δ5ds gene in transformant 15, Southern blot analyses were performed with genomic DNAs prepared from the Δku80 strain and transformant 15 (Fig. 5b). When the genomic DNAs were digested with ClaI and XhoI, the 4.3-kb hybridization signal corresponding to the original Δ5ds open reading frame was not detected in transformant 15, but the 3.3- and 7.5-kb signals were detected. When the genomic DNAs were digested with NarI and XbaI, the 3.7-kb signal was not detected in transformant 15, but the expected 4.2- and 6.0-kb signals were detected. These results showed that the Δ5ds gene in transformant 15 was successfully disrupted by integration of pKSCD5 vector. Unexpected signals in transformant 15, however, were observed on Southern blot. This finding may mean that several pKSCD5 vectors had been introduced into random sites in the genomic DNA of the Δku80 strain by biolistic particle bombardment.
Discussion
To improve gene-targeting efficiency in M. alpina 1S-4, we cloned and identified the ku80 gene encoding the Ku80 protein, which forms a Ku-protein complex with Ku70 protein, and is involved in the NHEJ pathway. Ku80 homolog proteins of other organisms were classified by kingdom (Fig. 1). However, the predicted translation product of the ku80 gene of this fungus shares low (<30 %) identities with those of other organisms. We constructed a ku80 gene disruptant (transformant 3 in Fig. 2b) via HR using the pKSUku80 vector. In transformant 6, we speculated that some of the introduced pKSUku80 vectors were integrated ectopically into the ku80 gene locus (Fig. 2b). In general, one of the problems of biolistic particle bombardment is the delivery of many plasmids into cells. However, in view of the result for transformant 3, the biolistic particle bombardment method is applicable to the integration of a single vector into the genome via HR. The Δku80 strain showed no marked differences in vegetative growth, formation of spores, or fatty acid productivity compared with the host strain (Table 2). Thus, we expect the Δku80 strain to be a superior host strain for metabolic engineering for PUFA production. Furthermore, the Δku80 strain exhibited a sensitivity to 0.05 % MMS (Fig. 3) similar to those of N. crassa, A. fumigatus, and A. aculeatus (da Silva Ferreira et al. 2006; Ninomiya et al. 2004; Tani et al. 2013). Such sensitivity indicates that the NHEJ pathway in this strain is repressed. To evaluate gene-targeting efficiency and construct a beneficial PUFA-producing strain, the disruption of Δ5ds gene was performed using the Δku80 strain as a host strain. The Δ5ds gene disruptant, transformant 15, produced a large amount of DGLA. The DGLA productivity of transformant 15 was at the same level (Table 2) as that of a Δ5ds gene-defective mutant obtained by chemical mutagenesis (Jareonkitmongkol et al. 1993). However, chemical mutagens cause mutation in multiple locations in the genome and often suppress growth, spore germination, and PUFA production. The ARA production of the Δ5ds gene disruptant was drastically decreased. Though we isolated a single colony from spore of the Δ5ds gene disruptant to avoid contamination of intact spores, quite low level of ARA remained. Given that pKSCD5 vector was integrated via single-crossover HR, the incomplete Δ5-desaturase may act in catalyzing the conversion of DGLA to ARA. For this reason, future gene targeting should be performed via double-crossover HR. Southern blot analysis using the genomic DNA from the Δ5ds gene disruptant suggested that several vectors were integrated into ectopic sites on the chromosome. To obtain a complete disruptant with a single plasmid, more transformants should be isolated and checked, or gene targeting should be performed by an alternative transformation method such as an A. tumefaciens-mediated method, introducing a single vector into a spore of the host strain.
Only one of the 77 transformants was detected as a ku80 gene disruptant (Table 3). Using of the ku80 gene-disrupted strain as a host, one of the 32 transformants was detected as Δ5ds gene disruptant. In previous research, Δ6ds-gene disruption was attempted in Mortierellaceae, M. isabellina, by biolistic particle bombardment and more than 70 transformants were obtained. However, none was disrupted in its Δ6ds-gene (Zhang et al. 2007). In this study, we found that we obtained at least one of 32 transformant by using Δku80 strain. The efficiencies of gene targeting in Δku80 strains from A. sojae, A. oryzae, A. fumigatus, A. niger, and Lecanicillium sp. were significantly improved (da Silva Ferreira et al. 2006; Honda et al. 2011; Ishidoh et al. 2014; Takahashi et al. 2006). However, the efficiency in the Δku80 strain from M. alpina 1S-4 was hardly improved. In a previous study in N. crassa, chromosomal integration of exogenous DNA was achieved via two types of HR and two types of NHEJ, the Ku80-dependent major pathway and the Ku80-independent minor pathway (Ishibashi et al. 2006). In M. alpina 1S-4, it is suggested that the Ku80-independent pathway may play a major pathway of NHEJ and reduce gene-targeting efficiency. When the Δku80 strain was used as a host, the incomplete Ku80 protein formed a Ku-protein complex with the Ku70 protein, and pKSCD5 vector was integrated ectopically via the NHEJ pathway. The loss of Lig4 activity involved in both the major and minor NHEJ pathways raised the targeting efficiency to 100 % in A. oryzae and A. luchuensis (Mizutani et al. 2008; Takahashi et al. 2011). Further improvement in targeting efficiency in the Δku80 strain, such as by simultaneous disruption of the lig4 gene, might facilitate metabolic engineering and reverse genetic studies in M. alpina 1S-4.
In summary, this report is the first to date to describe the identification and disruption of the ku80 gene in Mucoromycotina fungi. We succeeded in gene targeting in M. alpina 1S-4. On the other hand, though the ku80 gene disruptant showed normal growth, germination, and lipid production, gene-targeting efficiency was hardly improved. On the other hand, we achieved to construct a Δ5ds gene disruptant using the Δku80 strain as a host strain. This gene-targeting system may contribute to the construction of various PUFA-producing strains via metabolic engineering.
References
Ando A, Sakuradani E, Horinaka K, Ogawa J, Shimizu S (2009a) Transformation of an oleaginous zygomycete Mortierella alpina 1S-4 with the carboxin resistance gene conferred by mutation of the iron-sulfur subunit of succinate dehydrogenase. Curr Genet 55:349–356
Ando A, Sumida Y, Negoro H, Suroto DA, Ogawa J, Sakuradani E, Shimizu S (2009b) Establishment of Agrobacterium tumefaciens-mediated transformation of an oleaginous fungus, Mortierella alpina 1S-4, and its application for eicosapentaenoic acid producer breeding. Appl Environ Microbiol 75:5529–5535
Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346
Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23:394–398
da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Härtl A, Heinekamp T, Brakhage AA, Goldman GH (2006) The akuB KU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211
Daley JM, Palmbos PL, Wu DL, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451
Haber JE (2000) Partners and pathways: repairing a double-strand break. Trends Genet 16:259–264
Hande MP (2004) DNA repair factors and telomere-chromosome integrity in mammalian cells. Cytogenet Genome Res 104:116–122
Honda Y, Kobayashi K, Kirimura K (2011) Increases in gene-targeting frequencies due to disruption of kueA as a ku80 homolog in citric acid-producing Aspergillus niger. Biosci Biotechnol Biochem 75:1594–1596
Hopfner KP, Putnam CD, Tainer JA (2002) DNA double-strand break repair from head to tail. Curr Opin Struct Biol 12:115–122
Ishibashi K, Suzuki K, Ando Y, Takakura C, Inoue H (2006) Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc Natl Acad Sci USA 103:14871–14876
Ishidoh K, Kinoshita H, Ihara F, Nihira T (2014) Efficient and versatile transformation systems in entomopathogenic fungus Lecanicillium species. Curr Genet 60:99–108
Jareonkitmongkol S, Sakuradani E, Shimizu S (1993) A novel Δ5-desaturase-defective mutant of Mortierella alpina 1S-4 and its dihomo-γ-linolenic acid productivity. Appl Environ Microbiol 59:4300–4304
Kanaar R, Hoeijmakers JH, van Gent DC (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8:483–489
Kato A, Akamatsu Y, Sakuraba Y, Inoue H (2004) The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr Genet 45:37–44
Kawashima H, Akimoto K, Fujita T, Naoki H, Konishi K, Shimizu S (1995) Preparation of 13C-labeled polyunsaturated fatty acids by an arachidonic acid-producing fungus Mortierella alpina 1S-4. Anal Biochem 229:317–322
Kikukawa H, Sakuradani E, Kishino S, Park S-B, Ando A, Shima J, Ochiai M, Shimizu S, Ogawa J (2013) Characterization of a trifunctional fatty acid desaturase from oleaginous filamentous fungus Mortierella alpina 1S-4 using a yeast expression system. J Biosci Bioeng 116:672–676
Kooistra R, Hooykaas PJ, Steensma HY (2004) Efficient gene targeting in Kluyveromyces lactis. Yeast 21:781–792
Krappmann S (2007) Gene targeting in filamentous fungi: the benefits of impaired repair. Fungal Biol Rev 21:25–29
Krappmann S, Sasse C, Braus GH (2006) Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot Cell 5:212–215
Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38:233–271
Lisby M, Rothstein R (2004) DNA repair: keeping it together. Curr Biol 14:R994–R996
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol 45:878–889
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101:12248–12253
Okuda T, Ando A, Sakuradani E, Kikukawa H, Kamada N, Ochiai M, Shima J, Ogawa J (2014) Selection and characterization of promoters based on genomic approach for the molecular breeding of oleaginous fungus Mortierella alpina 1S-4. Curr Genet 60:183–191
Razanamparany V, Bégueret J (1986) Positive screening and transformation of ura5 mutants in the fungus Podospora anserina: characterization of the transformants. Curr Genet 10:811–817
Sakuradani E (2010) Advances in the production of various polyunsaturated fatty acids through oleaginous fungus Mortierella alpina breeding. Biosci Biotechnol Biochem 74:908–917
Sakuradani E, Kobayashi M, Shimizu S (1999a) Δ6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Gene cloning and its heterologous expression in a fungus, Aspergillus. Gene 238:445–453
Sakuradani E, Kobayashi M, Shimizu S (1999b) Δ9-fatty acid desaturase from arachidonic acid-producing fungus. Unique gene sequence and its heterologous expression in a fungus. Aspergillus. Eur J Biochem 260:208–216
Sakuradani E, Abe T, Iguchi K, Shimizu S (2005) A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl Microbiol Biotechnol 66:648–654
Sakuradani E, Murata S, Kanamaru H, Shimizu S (2008) Functional analysis of a fatty acid elongase from arachidonic acid-producing Mortierella alpina 1S-4. Appl Microbiol Biotechnol 81:497–503
Sakuradani E, Ando A, Ogawa J, Shimizu S (2009) Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl Microbiol Biotechnol 84:1–10
Sakuradani E, Ando A, Shimizu S, Ogawa J (2013) Metabolic engineering for the production of polyunsaturated fatty acids by oleaginous fungus Mortierella alpina 1S-4. J Biosci Bioeng 116:417–422
Schiestl RH, Zhu J, Petes TD (1994) Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol 14:4493–4500
Shibata T, Nishinaka T, Mikawa T, Aihara H, Kurumizaka H, Yokoyama S, Ito Y (2001) Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material. Proc Natl Acad Sci USA 98:8425–8432
Shiotani H, Tsuge T (1995) Efficient gene targeting in the filamentous fungus Alternaria alternata. Mol Gen Genet 248:142–150
Takahashi T, Masuda T, Koyama Y (2006) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics 275:460–470
Takahashi T, Mizutani O, Shiraishi Y, Yamada O (2011) Development of an efficient gene-targeting system in Aspergillus luchuensis by deletion of the non-homologous end joining system. J Biosci Bioeng 112:529–534
Takeno S, Sakuradani E, Murata S, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S (2004a) Cloning and sequencing of the ura3 and ura5 genes, and isolation and characterization of uracil auxotrophs of the fungus Mortierella alpina 1S-4. Biosci Biotechnol Biochem 68:277–285
Takeno S, Sakuradani E, Murata S, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S (2004b) Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl Microbiol Biotechnol 65:419–425
Takeno S, Sakuradani E, Tomi A, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S (2005a) Improvement of the fatty acid composition of an oil-producing filamentous fungus, Mortierella alpina 1S-4, through RNA interference with Δ12-desaturase gene expression. Appl Environ Microbiol 71:5124–5128
Takeno S, Sakuradani E, Tomi A, Inohara-Ochiai M, Kawashima H, Shimizu S (2005b) Transformation of oil-producing fungus, Mortierella alpina 1S-4, using Zeocin, and application to arachidonic acid production. J Biosci Bioeng 100:617–622
Tani S, Tsuji A, Kunitake E, Sumitani J, Kawaguchi T (2013) Reversible impairment of the ku80 gene by a recyclable marker in Aspergillus aculeatus. AMB Express 3:4
Van Dyck E, Stasiak AZ, Stasiak A, West SC (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398:728–731
Watrin L, Lucas S, Purcarea C, Legrain C, Prieur D (1999) Isolation and characterization of pyrimidine auxotrophs, and molecular cloning of the pyrE gene from the hyperthermophilic archaeon Pyrococcus abyssi. Mol Gen Genet 262:378–381
Zhang X, Li M, Wei D, Wang X, Chen X, Xing L (2007) Disruption of the fatty acid Δ6-desaturase gene in the oil-producing fungus Mortierella isabellina by homologous recombination. Curr Microbiol 55:128–134
Acknowledgments
This work was partially supported by Grants-in Aid for Scientific Research of Japan (Numbers 22380051 to E. Sakuradani and 23248014 to J. Ogawa), the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) of Japan, and the Advanced Low Carbon Technology Research and Development Program (ALCA) of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Goldman.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kikukawa, H., Sakuradani, E., Nakatani, M. et al. Gene targeting in the oil-producing fungus Mortierella alpina 1S-4 and construction of a strain producing a valuable polyunsaturated fatty acid. Curr Genet 61, 579–589 (2015). https://doi.org/10.1007/s00294-015-0481-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0481-2